Transcriptomic Analysis of Shiga Toxin-Producing Escherichia coli during Initial Contact with Cattle Colonic Explants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Cattle Colonic Explants
2.3. Bacterial Adherence to Explants
2.4. RNA Extraction, Library Generation, and Sequencing
2.5. RNA-seq Analysis and Detection of Differential Gene Expression
2.6. Statistical Analysis
3. Results and Discussion
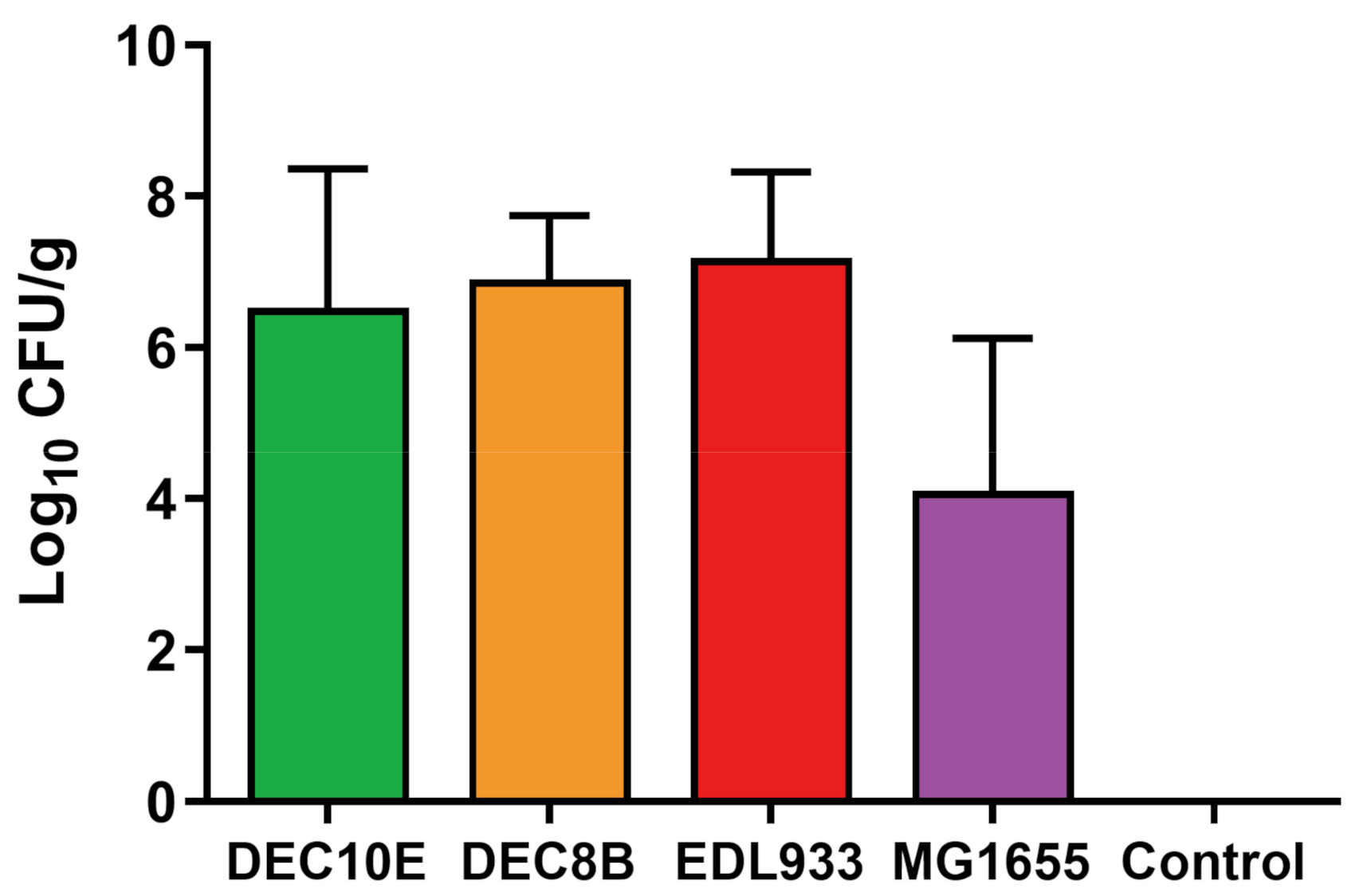
3.1. STEC Strains and MG1655 Adhered to Colonic Explants
3.2. H7 Flagellin (fliC) and Lon Protease (lon) Upregulated during Incubation with Colonic Explants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, A.; Cointe, A.; Mariani Kurkdjian, P.; Rafat, C.; Hertig, A. Shiga toxin-associated hemolytic uremic syndrome: A narrative review. Toxins 2020, 12, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spinale, J.M.; Ruebner, R.L.; Copelovitch, L.; Kaplan, B.S. Long-term outcomes of Shiga toxin hemolytic uremic syndrome. Pediatr. Nephrol. 2013, 28, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Tarr, P.I.; Gordon, C.A.; Chandler, W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005, 365, 1073–1086. [Google Scholar] [CrossRef]
- Gould, L.H.; Mody, R.K.; Ong, K.L.; Clogher, P.; Cronquist, A.B.; Garman, K.N.; Lathrop, S.; Medus, C.; Spina, N.L.; Webb, T.H. Increased recognition of non-O157 Shiga toxin–producing Escherichia coli infections in the United States during 2000–2010: Epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog. Dis. 2013, 10, 453–460. [Google Scholar] [CrossRef]
- Crim, S.M.; Griffin, P.M.; Tauxe, R.; Marder, E.P.; Gilliss, D.; Cronquist, A.B.; Cartter, M.; Tobin-D’Angelo, M.; Blythe, D.; Smith, K. Preliminary incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 US sites, 2006–2014. MMWR Morb. Mortal. Wkly Rep. 2015, 64, 495. [Google Scholar]
- Pires, S.M.; Majowicz, S.; Gill, A.; Devleesschauwer, B. Global and regional source attribution of Shiga toxin-producing Escherichia coli infections using analysis of outbreak surveillance data. Epidemiol. Infect. 2019, 147, e236. [Google Scholar] [CrossRef] [Green Version]
- Scallan, E.; Hoekstra, R.; Mahon, B.; Jones, T.; Griffin, P. An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol. Infect. 2015, 143, 2795–2804. [Google Scholar] [CrossRef] [Green Version]
- Moxley, R.A.; Stromberg, Z.R.; Lewis, G.L.; Loy, J.D.; Brodersen, B.W.; Patel, I.R.; Gangiredla, J. Haemorrhagic colitis associated with enterohaemorrhagic Escherichia coli O165: H25 infection in a yearling feedlot heifer. JMM Case Rep. 2015, 2, e005004. [Google Scholar] [CrossRef]
- Moxley, R.A.; Smith, D.R. Attaching-effacing Escherichia coli infections in cattle. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 29–56. [Google Scholar] [CrossRef]
- Sapountzis, P.; Segura, A.; Desvaux, M.; Forano, E. An overview of the elusive passenger in the gastrointestinal tract of cattle: The Shiga toxin producing Escherichia coli. Microorganisms 2020, 8, 877. [Google Scholar] [CrossRef] [PubMed]
- Gaytán, M.O.; Martínez-Santos, V.I.; Soto, E.; González-Pedrajo, B. Type three secretion system in attaching and effacing pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 129. [Google Scholar] [CrossRef] [Green Version]
- Baehler, A.A.; Moxley, R.A. Escherichia coli O157:H7 induces attaching-effacing lesions in large intestinal mucosal explants from adult cattle. FEMS Microbiol. Lett. 2000, 185, 239–242. [Google Scholar] [CrossRef]
- Phillips, A.; Navabpour, S.; Hicks, S.; Dougan, G.; Wallis, T.; Frankel, G. Enterohaemorrhagic Escherichia coli O157: H7 target Peyer’s patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 2000, 47, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, A.; Currie, C.G.; Mackie, S.; Tree, J.; McAteer, S.; McKendrick, I.; McNeilly, T.N.; Roe, A.; La Ragione, R.M.; Woodward, M.J. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157: H7 with bovine intestinal epithelium. Cell. Microbiol. 2009, 11, 121–137. [Google Scholar] [CrossRef] [Green Version]
- Gonyar, L.A.; Kendall, M.M. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157: H7. Infect. Immun. 2014, 82, 193–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venegas-Vargas, C.; Henderson, S.; Khare, A.; Mosci, R.E.; Lehnert, J.D.; Singh, P.; Ouellette, L.M.; Norby, B.; Funk, J.A.; Rust, S. Factors associated with Shiga toxin-producing Escherichia coli shedding by dairy and beef cattle. Appl. Environ. Microbiol. 2016, 82, 5049–5056. [Google Scholar] [CrossRef] [Green Version]
- Chase-Topping, M.; Gally, D.; Low, C.; Matthews, L.; Woolhouse, M. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 2008, 6, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Arthur, T.M.; Brichta-Harhay, D.M.; Bosilevac, J.M.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Super shedding of Escherichia coli O157:H7 by cattle and the impact on beef carcass contamination. Meat Sci. 2010, 86, 32–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stromberg, Z.R.; Lewis, G.L.; Schneider, L.G.; Erickson, G.E.; Patel, I.R.; Smith, D.R.; Moxley, R.A. Culture-based quantification with molecular characterization of non-O157 and O157 enterohemorrhagic Escherichia coli isolates from rectoanal mucosal swabs of feedlot cattle. Foodborne Pathog. Dis. 2018, 15, 26–32. [Google Scholar] [CrossRef]
- Shridhar, P.B.; Noll, L.W.; Cull, C.A.; Shi, X.; Cernicchiaro, N.; Renter, D.G.; Bai, J.; Nagaraja, T. Spiral plating method to quantify the six major non-O157 Escherichia coli serogroups in cattle feces. J. Food Prot. 2017, 80, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Vlisidou, I.; Dziva, F.; La Ragione, R.M.; Best, A.; Garmendia, J.; Hawes, P.; Monaghan, P.; Cawthraw, S.A.; Frankel, G.; Woodward, M.J. Role of intimin-tir interactions and the tir-cytoskeleton coupling protein in the colonization of calves and lambs by Escherichia coli O157: H7. Infect. Immun. 2006, 74, 758–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, S.D.; Betting, D.J.; Whittam, T.S. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J. Clin. Microbiol. 1999, 37, 2719–2722. [Google Scholar] [CrossRef] [Green Version]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Hazen, T.H.; Sahl, J.W.; Redman, J.C.; Morris, C.R.; Daugherty, S.C.; Chibucos, M.C.; Sengamalay, N.A.; Fraser-Liggett, C.M.; Steinsland, H.; Whittam, T.S.; et al. Draft genome sequences of the diarrheagenic Escherichia coli collection. J. Bacteriol. 2012, 194, 3026–3027. [Google Scholar] [CrossRef] [Green Version]
- Latif, H.; Li, H.J.; Charusanti, P.; Palsson, B.Ø.; Aziz, R.K. A gapless, unambiguous genome sequence of the enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Genome Announc. 2014, 2, e00821-14. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Farfan, M.J.; Torres, A.G. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 2012, 80, 903–913. [Google Scholar] [CrossRef] [Green Version]
- McWilliams, B.D.; Torres, A.G. Enterohemorrhagic Escherichia coli adhesins. Microbiol. Spectr. 2014, 2, EHEC00032013. [Google Scholar] [CrossRef] [Green Version]
- Ross, B.N.; Rojas-Lopez, M.; Cieza, R.J.; McWilliams, B.D.; Torres, A.G. The role of long polar fimbriae in Escherichia coli O104: H4 adhesion and colonization. PLoS ONE 2015, 10, e0141845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letourneau, J.; Levesque, C.; Berthiaume, F.; Jacques, M.; Mourez, M. In vitro assay of bacterial adhesion onto mammalian epithelial cells. J. Vis. Exp. 2011, e2783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Zhu, M.-J. Defects in polynucleotide phosphorylase impairs virulence in Escherichia coli O157: H7. Front. Microbiol. 2015, 6, 806. [Google Scholar] [CrossRef] [PubMed]
- Girard, F.; Dziva, F.; van Diemen, P.; Phillips, A.D.; Stevens, M.P.; Frankel, G. Adherence of enterohemorrhagic Escherichia coli O157, O26, and O111 strains to bovine intestinal explants ex vivo. Appl. Environ. Microbiol. 2007, 73, 3084–3090. [Google Scholar] [CrossRef] [Green Version]
- Cookson, A.L.; Woodward, M.J. The role of intimin in the adherence of enterohaemorrhagic Escherichia coli (EHEC) O157:H7 to HEp-2 tissue culture cells and to bovine gut explant tissues. Int. J. Med. Microbiol. 2003, 292, 547–553. [Google Scholar] [CrossRef]
- Katwal, P.; Thomas, M.; Uprety, T.; Hildreth, M.B.; Kaushik, R.S. Development and biochemical and immunological characterization of early passage and immortalized bovine intestinal epithelial cell lines from the ileum of a young calf. Cytotechnology 2019, 71, 127–148. [Google Scholar] [CrossRef] [Green Version]
- Keen, J.E.; Laegreid, W.W.; Chitko-McKown, C.G.; Durso, L.M.; Bono, J.L. Distribution of Shiga-toxigenic Escherichia coli O157 in the gastrointestinal tract of naturally O157-shedding cattle at necropsy. Appl. Environ. Microbiol. 2010, 76, 5278–5281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Zhang, H.; Wang, H.; Hu, J.; Du, M.; Zhu, M.-J. Host inflammatory response inhibits Escherichia coli O157:H7 adhesion to gut epithelium through augmentation of mucin expression. Infect. Immun. 2014, 82, 1921–1930. [Google Scholar] [CrossRef] [Green Version]
- Rossez, Y.; Wolfson, E.B.; Holmes, A.; Gally, D.L.; Holden, N.J. Bacterial flagella: Twist and stick, or dodge across the kingdoms. PLoS Pathog. 2015, 11, e1004483. [Google Scholar] [CrossRef] [PubMed]
- Erdem, A.L.; Avelino, F.; Xicohtencatl-Cortes, J.; Girón, J.A. Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J. Bacteriol. 2007, 189, 7426–7435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bretschneider, G.; Berberov, E.M.; Moxley, R.A. Reduced intestinal colonization of adult beef cattle by Escherichia coli O157:H7 tir deletion and nalidixic-acid-resistant mutants lacking flagellar expression. Vet. Microbiol. 2007, 125, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeilly, T.N.; Naylor, S.W.; Mahajan, A.; Mitchell, M.C.; McAteer, S.; Deane, D.; Smith, D.G.; Low, J.C.; Gally, D.L.; Huntley, J.F. Escherichia coli O157:H7 colonization in cattle following systemic and mucosal immunization with purified H7 flagellin. Infect. Immun. 2008, 76, 2594–2602. [Google Scholar] [CrossRef] [Green Version]
- Nagy, A.; Mowery, J.; Bauchan, G.R.; Wang, L.; Nichols-Russell, L.; Nou, X. Role of extracellular structures of Escherichia coli O157:H7 in initial attachment to biotic and abiotic surfaces. Appl. Environ. Microbiol. 2015, 81, 4720–4727. [Google Scholar] [CrossRef] [Green Version]
- Goulter, R.M.; Taran, E.; Gentle, I.R.; Gobius, K.S.; Dykes, G.A. Escherichia coli strains expressing H12 antigens demonstrate an increased ability to attach to abiotic surfaces as compared with E. coli strains expressing H7 antigens. Colloids Surf. B Biointerfaces 2014, 119, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.K.; Ishida, H.; Liu, Z.; Vogel, H.J. Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J. Bacteriol. 2013, 195, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Bernhardt, T.G.; De Boer, P.A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 2005, 18, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Rosen, R.; Biran, D.; Gur, E.; Becher, D.; Hecker, M.; Ron, E.Z. Protein aggregation in Escherichia coli: Role of proteases. FEMS Microbiol. Lett. 2002, 207, 9–12. [Google Scholar] [CrossRef]
- Christensen, S.K.; Mikkelsen, M.; Pedersen, K.; Gerdes, K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 2001, 98, 14328–14333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffith, K.L.; Shah, I.M.; Richard, E.W., Jr. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 2004, 51, 1801–1816. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, S.A.; Rivera-Rivera, I.; Sauer, R.T.; Baker, T.A. The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease. Mol. Microbiol. 2010, 75, 1539–1549. [Google Scholar] [CrossRef] [Green Version]
- Babin, B.M.; Kasperkiewicz, P.; Janiszewski, T.; Yoo, E.; Dra̧g, M.; Bogyo, M. Leveraging peptide substrate libraries to design inhibitors of bacterial Lon protease. ACS Chem. Biol. 2019, 14, 2453–2462. [Google Scholar] [CrossRef] [PubMed]
- Mellies, J.L.; Elliott, S.J.; Sperandio, V.; Donnenberg, M.S.; Kaper, J.B. The Per regulon of enteropathogenic Escherichia coli: Identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 1999, 33, 296–306. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, M.A.; Morgan, J.K.; Ares, M.A.; Yáñez-Santos, J.A.; Riordan, J.T.; Girón, J.A. The two-component system CpxRA negatively regulates the locus of enterocyte effacement of enterohemorrhagic Escherichia coli involving σ32 and Lon protease. Front. Cell. Infect. Microbiol. 2016, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bando, S.Y.; Iamashita, P.; Silva, F.N.; Costa, L.d.F.; Abe, C.M.; Bertonha, F.B.; Guth, B.E.; Fujita, A.; Moreira-Filho, C.A. Dynamic gene network analysis of Caco-2 cell response to Shiga toxin-producing Escherichia coli-associated hemolytic–uremic syndrome. Microorganisms 2019, 7, 195. [Google Scholar] [CrossRef] [Green Version]
- Wang, O.; McAllister, T.A.; Plastow, G.; Stanford, K.; Selinger, B. Host mechanisms involved in cattle Escherichia coli O157 shedding: A fundamental understanding for reducing foodborne pathogen in food animal production. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stromberg, Z.R.; Masonbrink, R.E.; Mellata, M. Transcriptomic Analysis of Shiga Toxin-Producing Escherichia coli during Initial Contact with Cattle Colonic Explants. Microorganisms 2020, 8, 1662. https://doi.org/10.3390/microorganisms8111662
Stromberg ZR, Masonbrink RE, Mellata M. Transcriptomic Analysis of Shiga Toxin-Producing Escherichia coli during Initial Contact with Cattle Colonic Explants. Microorganisms. 2020; 8(11):1662. https://doi.org/10.3390/microorganisms8111662
Chicago/Turabian StyleStromberg, Zachary R., Rick E. Masonbrink, and Melha Mellata. 2020. "Transcriptomic Analysis of Shiga Toxin-Producing Escherichia coli during Initial Contact with Cattle Colonic Explants" Microorganisms 8, no. 11: 1662. https://doi.org/10.3390/microorganisms8111662
APA StyleStromberg, Z. R., Masonbrink, R. E., & Mellata, M. (2020). Transcriptomic Analysis of Shiga Toxin-Producing Escherichia coli during Initial Contact with Cattle Colonic Explants. Microorganisms, 8(11), 1662. https://doi.org/10.3390/microorganisms8111662




