Seasonal Patterns of Dominant Microbes Involved in Central Nutrient Cycles in the Subsurface
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. Bacterial Cell Lysis and Protein Extraction
2.3. SDS-PAGE, Proteolytic Digestion, and Peptide Extraction
2.4. Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
2.5. Data Analysis
2.6. Dissolved Organic Matter (DOM) Composition Measurement
3. Results
3.1. A Global View on the Microbial Spatial-Temporal Distribution
3.2. Taxonomic Characterization of the Subsurface Microbial Community
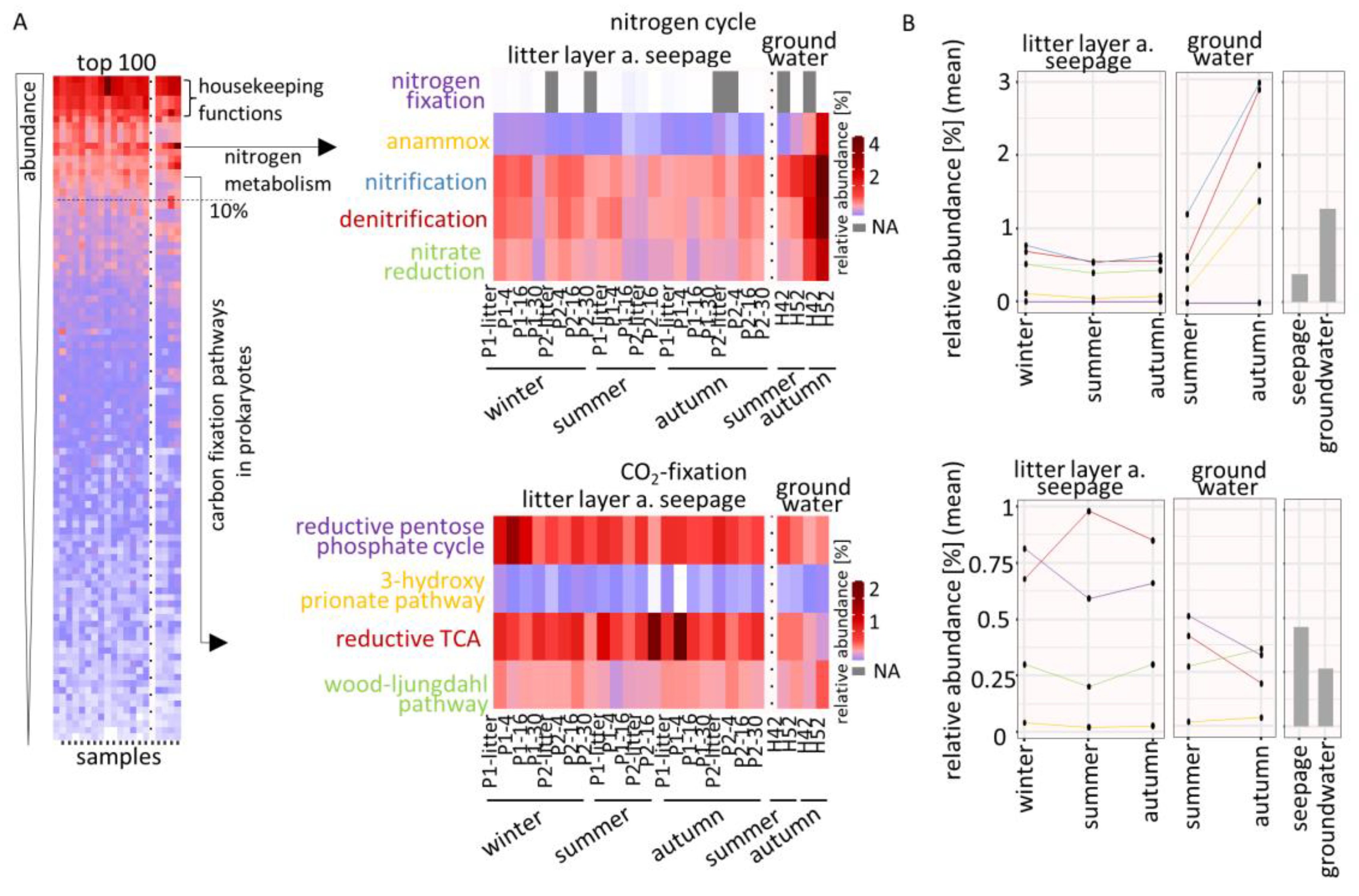
3.3. Functional Analysis of Pathways Relevant for Nutrient Cycles
3.4. Seasonal Effects on Dominant Families Involved in Nitrogen Cycle and CO2-Fixation
4. Discussion
4.1. Spatial-Temporal Distribution of the Community
4.2. Microbial Community Composition is Changed between Seepage Water and Groundwater
4.3. Seasonal Transition Promotes the Adaption of Microbial Dominances Responsible for Nutrient Cycles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Akob, D.M.; Kusel, K. Where microorganisms meet rocks in the Earth’s Critical Zone. Biogeosciences 2011, 8, 3531–3543. [Google Scholar] [CrossRef]
- Küsel, K.; Totsche, K.U.; Trumbore, S.E.; Lehmann, R.; Steinhäuser, C.; Herrmann, M. How Deep Can Surface Signals Be Traced in the Critical Zone? Merging Biodiversity with Biogeochemistry Research in a Central German Muschelkalk Landscape. Front. Earth Sci. 2016, 4. [Google Scholar] [CrossRef]
- Lin, H. Earth’s Critical Zone and hydropedology: Concepts, characteristics, and advances. Hydrol. Earth Syst. Sci. 2020, 14, 25–45, Erratum in. 2010, 14, 157. [Google Scholar] [CrossRef]
- Geesink, P.; Wegner, C.; Probst, A.J.; Herrmann, M.; Dam, H.T.; Kaster, A.; Küsel, K. Genome-inferred spatio-temporal resolution of an uncultivated Roizmanbacterium reveals its ecological preferences in groundwater. Environ. Microbiol. 2019, 22, 726–737. [Google Scholar] [CrossRef]
- Wilpiszeski, R.L.; Aufrecht, J.A.; Retterer, S.T.; Sullivan, M.B.; Graham, D.E.; Pierce, E.M.; Zablocki, O.D.; Palumbo, A.V.; Elias, D.A. Soil Aggregate Microbial Communities: Towards Understanding Microbiome Interactions at Biologically Relevant Scales. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Shank, E.A. Considering the Lives of Microbes in Microbial Communities. MSystems 2018, 3. [Google Scholar] [CrossRef]
- Anantharaman, K.; Brown, C.T.; Hug, L.A.; Sharon, I.; Castelle, C.J.; Probst, A.J.; Thomas, B.C.; Singh, A.; Wilkins, M.J.; Karaoz, U.; et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 2016, 7, 13219. [Google Scholar] [CrossRef] [PubMed]
- Dai, A. Increasing drought under global warming in observations and models. Nat. Clim. Chang. 2013, 3, 52–58. [Google Scholar] [CrossRef]
- Malik, A.A.; Swenson, T.; Weihe, C.; Morrison, E.W.; Martiny, J.B.H.; Brodie, E.L.; Northen, T.R.; Allison, S.D. Drought and plant litter chemistry alter microbial gene expression and metabolite production. ISME J. 2020, 14, 2236–2247. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, S.; Fu, Q. A Drier Future? Science 2014, 343, 737–739. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Ma, L.; Williams, M.A. Drought-induced soil microbial amino acid and polysaccharide change and their implications for C-N cycles in a climate change world. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Preece, C.; Verbruggen, E.; Liu, L.; Weedon, J.T.; Peñuelas, J. Effects of past and current drought on the composition and diversity of soil microbial communities. Soil Biol. Biochem. 2019, 131, 28–39. [Google Scholar] [CrossRef]
- Hueso, S.; García, C.; Hernández, T. Severe drought conditions modify the microbial community structure, size and activity in amended and unamended soils. Soil Biol. Biochem. 2012, 50, 167–173. [Google Scholar] [CrossRef]
- Evans, S.E.; Wallenstein, M.D. Climate change alters ecological strategies of soil bacteria. Ecol. Lett. 2013, 17, 155–164. [Google Scholar] [CrossRef]
- Huntington, T.G. Evidence for intensification of the global water cycle: Review and synthesis. J. Hydrol. 2006, 319, 83–95. [Google Scholar] [CrossRef]
- Schaeffer, S.M.; Homyak, P.M.; Boot, C.M.; Roux-Michollet, D.; Schimel, J.P. Soil carbon and nitrogen dynamics throughout the summer drought in a California annual grassland. Soil Biol. Biochem. 2017, 115, 54–62. [Google Scholar] [CrossRef]
- Benk, S.A.; Yan, L.; Lehmann, R.; Roth, V.-N.; Schwab, V.F.; Totsche, K.U.; Küsel, K.; Gleixner, G. Fueling Diversity in the Subsurface: Composition and Age of Dissolved Organic Matter in the Critical Zone. Front. Earth Sci. 2019, 7. [Google Scholar] [CrossRef]
- Yan, S.-F.; Yu, S.-E.; Wu, Y.-B.; Pan, D.-F.; She, D.-L.; Ji, J. Seasonal Variations in Groundwater Level and Salinity in Coastal Plain of Eastern China Influenced by Climate. J. Chem. 2015, 2015, 1–8. [Google Scholar] [CrossRef]
- Zhao, M.; Xue, K.; Wang, F.; Liu, S.; Bai, S.; Sun, B.; Zhou, J.; Yang, Y. Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J. 2014, 8, 2045–2055. [Google Scholar] [CrossRef]
- Flynn, T.M.; Sanford, R.A.; Ryu, H.; Bethke, C.M.; Levine, A.D.; Ashbolt, N.J.; Domingo, J.W.S. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol. 2013, 13, 1–15. [Google Scholar] [CrossRef]
- Tölli, J.; King, G.M. Diversity and Structure of Bacterial Chemolithotrophic Communities in Pine Forest and Agroecosystem Soils. Appl. Environ. Microbiol. 2005, 71, 8411–8418. [Google Scholar] [CrossRef] [PubMed]
- Wallenstein, M.D.; Hall, E.K. A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochemistry 2011, 109, 35–47. [Google Scholar] [CrossRef]
- Isobe, K.; Ohte, N. Ecological Perspectives on Microbes Involved in N-Cycling. Microbes Environ. 2014, 29, 4–16. [Google Scholar] [CrossRef]
- Haase, D.; Larondelle, N.; Andersson, E.; Artmann, M.; Borgström, S.; Breuste, J.; Gomez-Baggethun, E.; Gren, Å.; Hamstead, Z.A.; Hansen, R.; et al. A Quantitative Review of Urban Ecosystem Service Assessments: Concepts, Models, and Implementation. AMBIO 2014, 43, 413–433. [Google Scholar] [CrossRef] [PubMed]
- Yeager, C.M.; Northup, D.E.; Grow, C.C.; Barns, S.M.; Kuske, C.R. Changes in Nitrogen-Fixing and Ammonia-Oxidizing Bacterial Communities in Soil of a Mixed Conifer Forest after Wildfire. Appl. Environ. Microbiol. 2005, 71, 2713–2722. [Google Scholar] [CrossRef]
- Lamba, S.; Bera, S.; Rashid, M.; Medvinsky, A.B.; Sun, G.-Q.; Acquisti, C.; Chakraborty, A.; Li, B.-L. Organization of biogeochemical nitrogen pathways with switch-like adjustment in fluctuating soil redox conditions. R. Soc. Open Sci. 2017, 4, 160768. [Google Scholar] [CrossRef]
- Florio, A.; Felici, B.; Migliore, M.; Dell’Abate, M.T.; Benedetti, A. Nitrogen losses, uptake and abundance of ammonia oxidizers in soil under mineral and organo-mineral fertilization regimes. J. Sci. Food Agric. 2015, 96, 2440–2450. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.A.; Cape, J.N.; Reis, S.; Sheppard, L.J.; Jenkins, A.; Grizzetti, B.; Galloway, J.N.; et al. The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20130164. [Google Scholar] [CrossRef]
- Kumar, S.; Herrmann, M.; Thamdrup, B.; Schwab, V.F.; Geesink, P.; Trumbore, S.E.; Totsche, K.-U.; Küsel, K. Nitrogen Loss from Pristine Carbonate-Rock Aquifers of the Hainich Critical Zone Exploratory (Germany) Is Primarily Driven by Chemolithoautotrophic Anammox Processes. Front. Microbiol. 2017, 8, 1951. [Google Scholar] [CrossRef]
- Berg, I.A. Ecological Aspects of the Distribution of Different Autotrophic CO2Fixation Pathways. Appl. Environ. Microbiol. 2011, 77, 1925–1936. [Google Scholar] [CrossRef]
- Long, P.E.; Williams, K.H.; Hubbard, S.S.; Banfield, J.F. Microbial Metagenomics Reveals Climate-Relevant Subsurface Biogeochemical Processes. Trends Microbiol. 2016, 24, 600–610. [Google Scholar] [CrossRef]
- Herrmann, M.; Rusznyák, A.; Akob, D.M.; Schulze, I.; Opitz, S.; Totsche, K.U.; Küsel, K. Large Fractions of CO2-Fixing Microorganisms in Pristine Limestone Aquifers Appear To Be Involved in the Oxidation of Reduced Sulfur and Nitrogen Compounds. Appl. Environ. Microbiol. 2015, 81, 2384–2394. [Google Scholar] [CrossRef]
- Wilmes, P.; Heintz-Buschart, A.; Bond, P.L. A decade of metaproteomics: Where we stand and what the future holds. Proteomics 2015, 15, 3409–3417. [Google Scholar] [CrossRef]
- Lohmann, P.; Schäpe, S.S.; Haange, S.-B.; Oliphant, K.; Allen-Vercoe, E.; Jehmlich, N.; Von Bergen, M. Function is what counts: How microbial community complexity affects species, proteome and pathway coverage in metaproteomics. Expert Rev. Proteom. 2020, 17, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Kohlhepp, B.; Lehmann, R.; Seeber, P.; Kusel, K.; Trumbore, S.E.; Totsche, K.U. Aquifer configuration and geostructural links control the groundwater quality in thin-bedded carbonate–siliciclastic alternations of the Hainich CZE, central Germany. Hydrol. Earth Syst. Sci. 2017, 21, 6091–6116. [Google Scholar] [CrossRef]
- Potthast, K.; Meyer, S.; Crecelius, A.C.; Schubert, U.S.; Tischer, A.; Michalzik, B. Land-use and fire drive temporal patterns of soil solution chemistry and nutrient fluxes. Sci. Total. Environ. 2017, 605, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.S.; Lehmann, R.; Stoll, W.; Rosenberger, J.; Totsche, K.U.; Küsel, K. The endolithic bacterial diversity of shallow bedrock ecosystems. Sci. Total. Environ. 2019, 679, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Starke, R.; Müller-Nurasyid, M.; Gaspar, M.; Marz, M.; Küsel, K.; Totsche, K.U.; Von Bergen, M.; Jehmlich, N. Candidate Brocadiales dominates C, N and S cycling in anoxic groundwater of a pristine limestone-fracture aquifer. J. Proteom. 2017, 152, 153–160. [Google Scholar] [CrossRef]
- Lehmann, R.; Totsche, K.U. Multi-directional flow dynamics shape groundwater quality in sloping bedrock strata. J. Hydrol. 2020, 580, 124291. [Google Scholar] [CrossRef]
- Schwab, V.F.; Herrmann, M.; Roth, V.-N.; Gleixner, G.; Lehmann, R.; Pohnert, G.; Trumbore, S.; Küsel, K.; Totsche, K.U. Functional diversity of microbial communities in pristine aquifers inferred by PLFA- and sequencing-based approaches. Biogeosciences 2017, 14, 2697–2714. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, T.; Koch, B.; Hertkorn, N.; Kattner, G. A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol. Oceanogr. Methods 2008, 6, 230–235. [Google Scholar] [CrossRef]
- Simon, C.; Roth, V.-N.; Dittmar, T.; Gleixner, G. Molecular Signals of Heterogeneous Terrestrial Environments Identified in Dissolved Organic Matter: A Comparative Analysis of Orbitrap and Ion Cyclotron Resonance Mass Spectrometers. Front. Earth Sci. 2018, 6. [Google Scholar] [CrossRef]
- Hillebrand, H.; Bennett, D.M.; Cadotte, M.W. Consequences of dominance: A review of evenness effects on local and regional ecosystem processes. Ecology 2008, 89, 1510–1520. [Google Scholar] [CrossRef]
- Fuchs, G. Alternative Pathways of Carbon Dioxide Fixation: Insights into the Early Evolution of Life? Annu. Rev. Microbiol. 2011, 65, 631–658. [Google Scholar] [CrossRef]
- Gibson, J.L.; Tabita, F.R. The molecular regulation of the reductive pentose phosphate pathway in Proteobacteria and Cyanobacteria. Arch. Microbiol. 1996, 166, 141–150. [Google Scholar] [CrossRef]
- Koranda, M.; Kaiser, C.; Fuchslueger, L.; Kitzler, B.; Sessitsch, A.; Zechmeister-Boltenstern, S.; Richter, A. Seasonal variation in functional properties of microbial communities in beech forest soil. Soil Biol. Biochem. 2013, 60, 95–104. [Google Scholar] [CrossRef]
- Zhou, Y.; Kellermann, C.; Griebler, C. Spatio-temporal patterns of microbial communities in a hydrologically dynamic pristine aquifer. FEMS Microbiol. Ecol. 2012, 81, 230–242. [Google Scholar] [CrossRef]
- Yan, L.; Herrmann, M.; Kampe, B.; Lehmann, R.; Totsche, K.U.; Küsel, K. Environmental selection shapes the formation of near-surface groundwater microbiomes. Water Res. 2020, 170, 115341. [Google Scholar] [CrossRef]
- Emagnabosco, C.; Etekere, M.; Lau, M.C.Y.; Elinage, B.; Ekuloyo, O.; Eerasmus, M.; Ecason, E.; Heerden, E.E.; Eborgonie, G.; Kieft, T.L.; et al. Comparisons of the composition and biogeographic distribution of the bacterial communities occupying South African thermal springs with those inhabiting deep subsurface fracture water. Front. Microbiol. 2014, 5, 679. [Google Scholar] [CrossRef]
- Xie, K.; Deng, Y.; Zhang, S.; Zhang, W.; Liu, J.; Xie, Y.; Zhang, X.; Huang, H. Prokaryotic Community Distribution along an Ecological Gradient of Salinity in Surface and Subsurface Saline Soils. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Tran, P.Q.; Kieft, K.; Anantharaman, K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060–2077. [Google Scholar] [CrossRef] [PubMed]
- Leboulanger, C.; Agogué, H.; Bernard, C.; Bouvy, M.; Carré, C.; Cellamare, M.; Duval, C.; Fouilland, E.; Got, P.; Intertaglia, L.; et al. Microbial Diversity and Cyanobacterial Production in Dziani Dzaha Crater Lake, a Unique Tropical Thalassohaline Environment. PLoS ONE 2017, 12, e0168879. [Google Scholar] [CrossRef]
- Hu, B.-L.; Rush, D.; Van Der Biezen, E.; Zheng, P.; Van Mullekom, M.; Schouten, S.; Damsté, J.S.S.; Smolders, A.J.P.; Jetten, M.S.M.; Kartal, B. New Anaerobic, Ammonium-Oxidizing Community Enriched from Peat Soil. Appl. Environ. Microbiol. 2010, 77, 966–971. [Google Scholar] [CrossRef]
- Herrmann, M.; Wegner, C.-E.; Taubert, M.; Geesink, P.; Lehmann, K.; Yan, L.; Lehmann, R.; Totsche, K.U.; Küsel, K. Predominance of Cand. Patescibacteria in Groundwater Is Caused by Their Preferential Mobilization From Soils and Flourishing Under Oligotrophic Conditions. Front. Microbiol. 2019, 10, 1407. [Google Scholar] [CrossRef]
- Lang, M.; Bei, S.; Li, X.; Kuyper, T.W.; Zhang, J. Rhizoplane Bacteria and Plant Species Co-determine Phosphorus-Mediated Microbial Legacy Effect. Front. Microbiol. 2019, 10, 2856. [Google Scholar] [CrossRef] [PubMed]
- Sinkko, H.; Lukkari, K.; Sihvonen, L.M.; Sivonen, K.; Leivuori, M.; Rantanen, M.; Paulin, L.; Lyra, C. Bacteria Contribute to Sediment Nutrient Release and Reflect Progressed Eutrophication-Driven Hypoxia in an Organic-Rich Continental Sea. PLoS ONE 2013, 8, e67061. [Google Scholar] [CrossRef] [PubMed]
- Lau, M.C.Y.; Kieft, T.L.; Kuloyo, O.; Linage-Alvarez, B.; Van Heerden, E.; Lindsay, M.R.; Magnabosco, C.; Wang, W.; Wiggins, J.B.; Guo, L.; et al. An oligotrophic deep-subsurface community dependent on syntrophy is dominated by sulfur-driven autotrophic denitrifiers. Proc. Natl. Acad. Sci. USA 2016, 113, E7927–E7936. [Google Scholar] [CrossRef]
- Ehsani, E.; Hernandez-Sanabria, E.; Kerckhof, F.-M.; Props, R.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Boon, N. Initial evenness determines diversity and cell density dynamics in synthetic microbial ecosystems. Sci. Rep. 2018, 8, 340. [Google Scholar] [CrossRef]
- Schiebenhoefer, H.; Bossche, T.V.D.; Fuchs, S.; Renard, B.Y.; Muth, T.; Martens, L. Challenges and promise at the interface of metaproteomics and genomics: An overview of recent progress in metaproteogenomic data analysis. Expert Rev. Proteom. 2019, 16, 375–390. [Google Scholar] [CrossRef]
- Gutleben, J.; De Mares, M.C.; Van Elsas, J.D.; Smidt, H.; Overmann, J.; Sipkema, D. The multi-omics promise in context: From sequence to microbial isolate. Crit. Rev. Microbiol. 2017, 44, 212–229. [Google Scholar] [CrossRef]
- Mori, J.F.; Chen, L.-X.; Jessen, G.L.; Rudderham, S.B.; McBeth, J.M.; Lindsay, M.B.J.; Slater, G.F.; Banfield, J.F.; Warren, L.A. Putative Mixotrophic Nitrifying-Denitrifying Gammaproteobacteria Implicated in Nitrogen Cycling Within the Ammonia/Oxygen Transition Zone of an Oil Sands Pit Lake. Front. Microbiol. 2019, 10, 2435. [Google Scholar] [CrossRef]
- Wegner, C.-E.; Gaspar, M.; Geesink, P.; Herrmann, M.; Marz, M.; Küsel, K. Biogeochemical Regimes in Shallow Aquifers Reflect the Metabolic Coupling of the Elements Nitrogen, Sulfur, and Carbon. Appl. Environ. Microbiol. 2019, 85, 85. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nat. Cell Biol. 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Nawaz, A.; Purahong, W.; Lehmann, R.; Herrmann, M.; Totsche, K.U.; Küsel, K.; Wubet, T.; Buscot, F. First insights into the living groundwater mycobiome of the terrestrial biogeosphere. Water Res. 2018, 145, 50–61. [Google Scholar] [CrossRef]
- Opitz, S.; Küsel, K.; Spott, O.; Totsche, K.U.; Herrmann, M. Oxygen availability and distance to surface environments determine community composition and abundance of ammonia-oxidizing prokaroytes in two superimposed pristine limestone aquifers in the Hainich region, Germany. FEMS Microbiol. Ecol. 2014, 90, 39–53. [Google Scholar] [CrossRef]
- Humbert, S.; Tarnawski, S.; Fromin, N.; Mallet, M.-P.; Aragno, M.; Ezopfi, J. Molecular detection of anammox bacteria in terrestrial ecosystems: Distribution and diversity. ISME J. 2009, 4, 450–454. [Google Scholar] [CrossRef]
- Dodsworth, J.A.; Hungate, B.A.; Hedlund, B.P. Ammonia oxidation, denitrification and dissimilatory nitrate reduction to ammonium in two US Great Basin hot springs with abundant ammonia-oxidizing archaea. Environ. Microbiol. 2011, 13, 2371–2386. [Google Scholar] [CrossRef]
- Schouten, S.; Strous, M.; Kuypers, M.M.M.; Rijpstra, W.I.C.; Baas, M.; Schubert, C.J.; Jetten, M.S.M.; Damsté, J.S.S. Stable Carbon Isotopic Fractionations Associated with Inorganic Carbon Fixation by Anaerobic Ammonium-Oxidizing Bacteria. Appl. Environ. Microbiol. 2004, 70, 3785–3788. [Google Scholar] [CrossRef]
- Strous, M.; Pelletier, E.; Mangenot, S.; Rattei, T.; Lehner, A.; Taylor, M.W.; Horn, M.; Daims, H.; Bartol-Mavel, D.; Wincker, P.; et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nat. Cell Biol. 2006, 440, 790–794. [Google Scholar] [CrossRef]
- Mußmann, M.; Pjevac, P.; Krüger, K.; Dyksma, S. Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 2017, 11, 1276–1281. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lohmann, P.; Benk, S.; Gleixner, G.; Potthast, K.; Michalzik, B.; Jehmlich, N.; Bergen, M.v. Seasonal Patterns of Dominant Microbes Involved in Central Nutrient Cycles in the Subsurface. Microorganisms 2020, 8, 1694. https://doi.org/10.3390/microorganisms8111694
Lohmann P, Benk S, Gleixner G, Potthast K, Michalzik B, Jehmlich N, Bergen Mv. Seasonal Patterns of Dominant Microbes Involved in Central Nutrient Cycles in the Subsurface. Microorganisms. 2020; 8(11):1694. https://doi.org/10.3390/microorganisms8111694
Chicago/Turabian StyleLohmann, Patrick, Simon Benk, Gerd Gleixner, Karin Potthast, Beate Michalzik, Nico Jehmlich, and Martin von Bergen. 2020. "Seasonal Patterns of Dominant Microbes Involved in Central Nutrient Cycles in the Subsurface" Microorganisms 8, no. 11: 1694. https://doi.org/10.3390/microorganisms8111694
APA StyleLohmann, P., Benk, S., Gleixner, G., Potthast, K., Michalzik, B., Jehmlich, N., & Bergen, M. v. (2020). Seasonal Patterns of Dominant Microbes Involved in Central Nutrient Cycles in the Subsurface. Microorganisms, 8(11), 1694. https://doi.org/10.3390/microorganisms8111694





