Integrated into Environmental Biofilm Chromobacterium vaccinii Survives Winter with Support of Bacterial Community
Abstract
:1. Introduction
2. Materials and Methods
3. Results
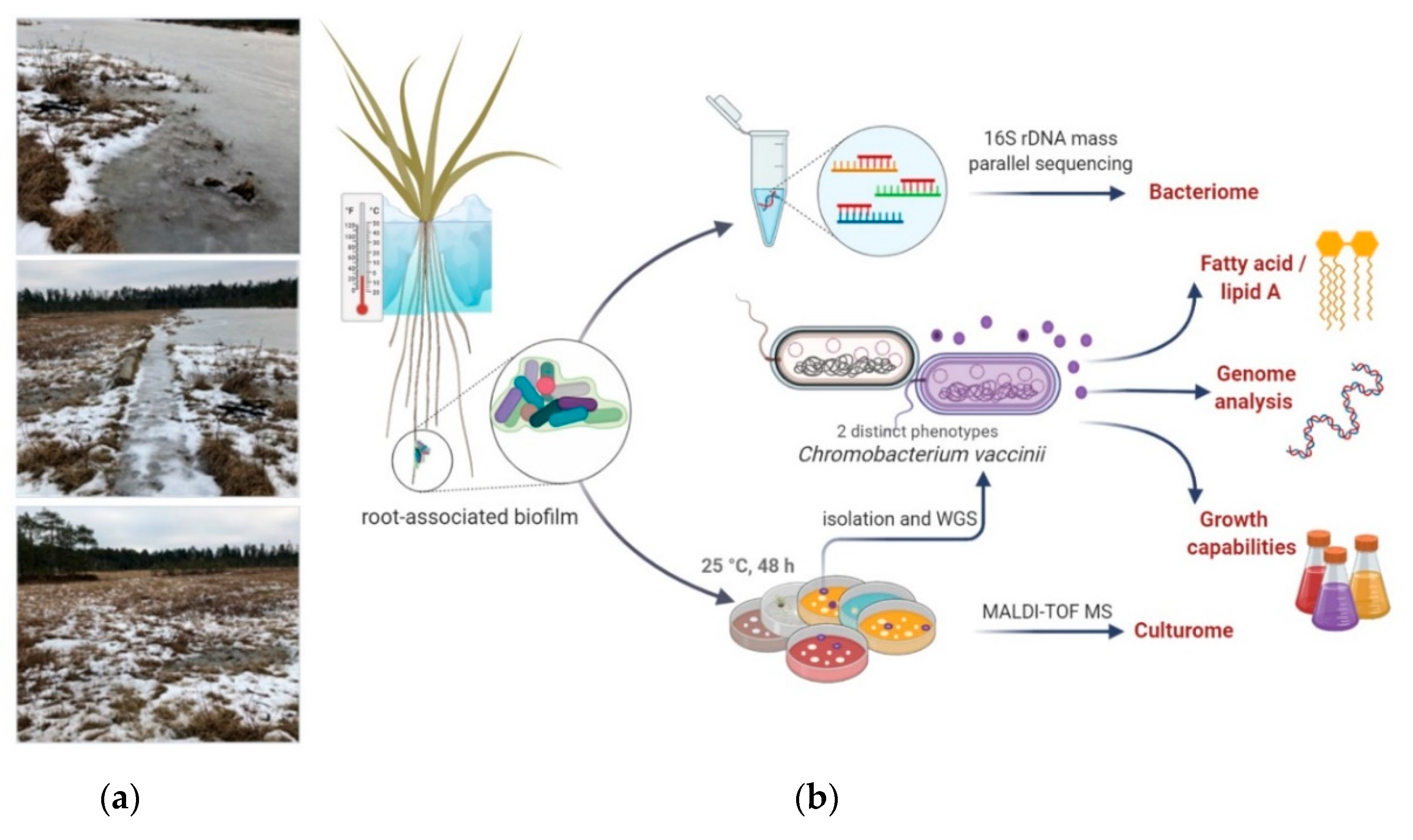
3.1. Quaking Bog Description
3.2. Individual Properties of Chromobacterium vaccinii
3.2.1. Isolation and Species-Level Identification
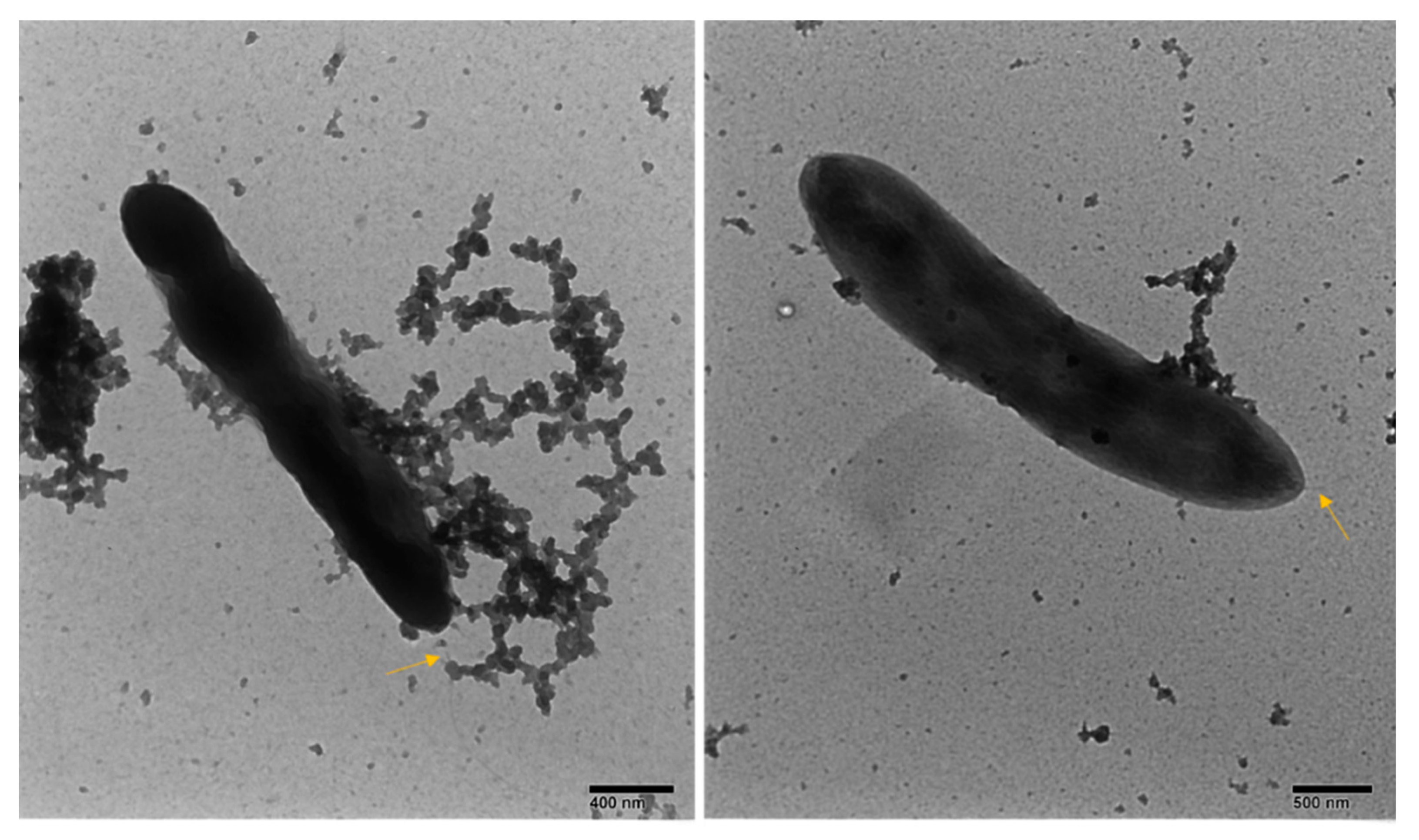
3.2.2. General Phenotype Characterization
3.2.3. Genome Characterization and Comparison with Known Strains of C. vaccinii.
3.3. Regulatory Nature of Unpigmented Isolate
3.4. Adaptability to Low Temperatures
3.5. Surrounding Bacterial Community
3.5.1. Culturome Analysis
3.5.2. Microbiome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hutchins, D.; Jansson, J.; Remais, J.; Rich, V.; Singh, B.; Trivedi, P. Climate change microbiology—Problems and perspectives. Nat. Rev. Microbiol. 2019, 17, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Antwis, R.; Griffiths; Harrison, X.; Aranega-Bou, P.; Arce, A.; Bettridge, A.; Brailsford, F.; Menezes, A.; Devaynes, A.; Forbes, K.; et al. Fifty important research questions in microbial ecology. FEMS Microbiol. Ecol. 2017, 93, 93. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaya, T.; Golovchenko, A.; Yurchenko, E.; Yakushev, A.; Manucharova, N.; Lysak, L.; Kostina, N. Bacterial Communities of Regressive Spots in Ombrotrophic Bogs: Structure and Functions. Microbiology 2020, 89, 107–114. [Google Scholar] [CrossRef]
- Stojek, N.; Dutkiewicz, J. Studies on the occurrence of Gram-negative bacteria in ticks: Ixodes ricinus as a potential vector of Pasteurella. Ann. Agric. Environ. Med. 2004, 11, 319–322. [Google Scholar] [PubMed]
- Jędruszczak, A.; Bąk, M.W.; Nosal, R.B.; Maciejewski, M.; Marczewski, K. Sepsis caused by Chromobacterium violaceum—Probably the first case in Europe, or Macbeth read anew. Ann. Agric. Environ. Med. 2019, 26, 508–510. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, R., Jr.; Sparks, M.; Kuhar, D.; Mitchell, A.; Gundersen-Rindal, D. Chromobacterium sphagni sp. nov., an insecticidal bacterium isolated from Sphagnum bogs. Int. J. Syst. Evol. Microbiol. 2017, 67, 3417–3422. [Google Scholar] [CrossRef] [PubMed]
- Soby, S.; Gadagkar, S.; Contreras, C.; Caruso, F. Chromobacterium vaccinii sp. nov., isolated from native and cultivated cranberry (Vaccinium macrocarpon Ait.) bogs and irrigation ponds. Int. J. Syst. Evol. Microbiol. 2013, 63, 1840–1846. [Google Scholar] [CrossRef]
- Zhou, S.; Guo, X.; Wang, H.; Kong, D.; Wang, Y.; Zhu, J.; Dong, W.; He, M.; Hu, G.; Zhao, B.; et al. Chromobacterium rhizoryzae sp. nov., isolated from rice roots. Int. J. Syst. Evol. Microbiol. 2016, 66, 3890–3896. [Google Scholar] [CrossRef]
- Bolívar-Anillo, H.J.; Orozco-Sanchez, C.J.; Lima, G.S.; dos Santos, G.F. Endophytic Microorganisms Isolated of Plants Grown in Colombia: A Short Review. J. Microb. Biochem. Technol. 2016, 8, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Dall’Agnol, L.; Martins; Vallinoto, A.; Ribeiro, K. Diversity of Chromobacterium violaceum isolates from aquatic environments of state of Pará, Brazilian Amazon. Memórias Instituto Oswaldo Cruz 2008, 103, 678–682. [Google Scholar] [CrossRef] [Green Version]
- Barreto, E.; Torres, A.; Barreto, M.; Vasconcelos, A.; Astolfi-Filho, S.; Hungria, M. Diversity in antifungal activity of strains of Chromobacterium violaceum from the Brazilian Amazon. J. Ind. Microbiol. Biotechnol. 2008, 35, 783–790. [Google Scholar] [CrossRef]
- Dodou, H.; Batista, A.; Medeiros, S.; Sales, G.; Rodrigues, M.; Pereira, P.; Nogueira, P.; Silveira, E.; Grangeiro, T.; Nogueira, N. Violacein antimicrobial activity on Staphylococcus epidermidis biofilm. Nat. Prod. Res. 2019, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Andrighetti-Fröhner, C.; Antonio, R.; Creczynski-Pasa, T.; Barardi, C.; Simões, C. Cytotoxicity and potential antiviral evaluation of violacein produced by Chromobacterium violaceum. Memórias Instituto Oswaldo Cruz 2003, 98, 843–848. [Google Scholar] [CrossRef]
- Bilsland, E.; Tavella, T.A.; Krogh, R.; Stokes, J.E.; Roberts, A.; Ajioka, J.; Spring, D.R.; Andricopulo, A.D.; Costa, F.T.; Oliver, S.G. Antiplasmodial and trypanocidal activity of violacein and deoxyviolacein produced from synthetic operons. BMC Biotechnol. 2018, 18, 22. [Google Scholar] [CrossRef] [Green Version]
- Lozano, G.L.; Guan, C.; Cao, Y.; Borlee, B.R.; Broderick, N.A.; Stabb, E.V.; Handelsman, J. A chemical counterpunch: Chromobacterium violaceum ATCC31532 produces violacein in response to translation-inhibiting antibiotics. BioRxiv 2019, 589192. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Choi, H.S.; Yang, S.Y.; Kim, I.S.; Yamaguchi, T.; Sohng, J.K.; Park, S.K.; Kim, J.-C.; Lee, C.H.; Gardener, B.M.; et al. Both extracellular chitinase and a new cyclic lipopeptide, chromobactomycin, contribute to the biocontrol activity of Chromobacterium sp. C61. Mol. Plant Pathol. 2013, 15, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Mart’yanov, S.V.; Letarov, A.V.; Ivanov, P.A.; Plakunov, V.K. Stimulation of Violacein Biosynthesis in Chromobacterium violaceum Biofilms in the Presence of Dimethyl Sulfoxide. Microbiology 2018, 87, 437–440. [Google Scholar] [CrossRef]
- McClean, K.; Winson, M.; Fish, L.; Taylor, A.; Chhabra, S.; Camara, M.; Daykin, M.; Lamb, J.; Swift, S.; Bycroft, B.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, K.; Heilmann, S.; Ditmarsch, D.; Xavier, J. Exploiting social evolution in biofilms. Curr. Opin. Microbiol. 2013, 16, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Freiwald, A.; Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 2009, 4, 732–742. [Google Scholar] [CrossRef]
- Schumann, P.; Maier, T. Chapter 13 MALDI-TOF Mass Spectrometry Applied to Classification and Identification of Bacteria. Method Microbiol. 2014, 41, 275–306. [Google Scholar] [CrossRef]
- Clark, C.M.; Costa, M.S.; Conley, E.; Li, E.; Sanchez, L.M.; Murphy, B.T. Using the Open-Source MALDI TOF-MS IDBac Pipeline for Analysis of Microbial Protein and Specialized Metabolite Data. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987. [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.-Y.; Cohoon, M.; de Crécy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R.; et al. The Subsystems Approach to Genome Annotation and its Use in the Project to Annotate 1000 Genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Sasser, M.; Kunitsky, C.; Jackoway, G.; Ezzell, J.; Teska, J.; Harper, B.; Parker, S.; Cheek, W.; Ezzell, J.; Hopkins, K.; et al. Identification of Bacillus anthracis from Culture Using Gas Chromatographic Analysis of Fatty Acid Methyl Esters. J. Aoac. Int. 2019, 88, 178–181. [Google Scholar] [CrossRef] [Green Version]
- Christie, W. Equivalent chain-lengths of methyl ester derivatives of fatty acids on gas chromatography a reappraisal. J. Chromatogr. A 1988, 447, 305–314. [Google Scholar] [CrossRef]
- Gavrilov, M.; Zhmyleva, A.; Shakhparonov, V.; Zakharchenko, D. Flora and Fauna of the Western Part of Moscow Region; Selected Student Research Works Conducted at Summer Field Practice at S. N. Skadovsky Zvenigorod Biological Station; KMK Scientific Press: Moscow, Russia, 2020; Volume 10, ISBN 978-5-907099-05-0. [Google Scholar]
- Dhar, S. The oxidase activity of chromobacterium. J. Clin. Pathol. 1973, 26, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivendra, R.; Lo, H.S. Identification of Chromobacterium violaceum: Pigmented and non-pigmented strains. J. Gen. Microbiol. 1975, 90, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Sneath, P.H.A. Identification methods applied to Chromobacterium. In Identification Methods for Microbiologists; Gibbs, B.M., Skinner, F.A., Eds.; Academic Press: London, UK; New York, NY, USA, 1966; part A; pp. 15–20. [Google Scholar]
- Niven, D.F.; Collins, P.A.; Knowles, C.J. The respiratory system of Chromobacterium violaceum grown under conditions of high and low cyanide evolution. J. Gen. Microbiol. 1975, 90, 271–285. [Google Scholar] [CrossRef] [Green Version]
- Forte, E.; Borisov, V.; Vicente, J.; Giuffrè, A. Chapter Four Cytochrome bd and Gaseous Ligands in Bacterial Physiology. Adv. Microb. Physiol. 2017, 71, 171–234. [Google Scholar] [CrossRef]
- Martin, P.; Gundersen-Rindal, D.; Blackburn, M.; Buyer, J. Chromobacterium subtsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int. J. Syst. Evol. Microbiol. 2007, 57, 993–999. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Han, F.; Segal, J. Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. Int. J. Syst. Evol. Microbiol. 2008, 58, 1398–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, C.; Arun, A.; Lai, W.; Chen, W.; Chou, J.; Chao, J.; Shen, F.; Rekha, P.; Kämpfer, P. Chromobacterium aquaticum sp. nov., isolated from spring water samples. Int. J. Syst. Evol. Microbiol. 2008, 58, 877–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kämpfer, P.; Busse, H.-J.; Scholz, H.C. Chromobacterium piscinae sp. nov. and Chromobacterium pseudoviolaceum sp. nov., from environmental samples. Int. J. Syst. Evol. Microbiol. 2009, 59, 2486–2490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackburn, M.B.; Farrar, R.R., Jr.; Sparks, M.E.; Kuhar, D.; Mowery, J.D.; Mitchell, A.; Gundersen-Rindal, D.E. Chromobacterium phragmitis sp. nov., isolated from estuarine marshes. Int. J. Syst. Evol. Microbiol. 2019. [Google Scholar] [CrossRef]
- Menezes, C.; Tonin, M.; Corrêa, D.; Parma, M.; Melo, I.; Zucchi, T.; Destéfano, S.; Fantinatti-Garboggini, F. Chromobacterium amazonense sp. nov. isolated from water samples from the Rio Negro, Amazon, Brazil. Antonie Van Leeuwenhoek 2015, 107, 1057–1063. [Google Scholar] [CrossRef]
- Hase, S.; Rietschel, E. The Chemical Structure of the Lipid a Component of Lipopolysaccharides from Chromobacterium violaceum NCTC 9694. Eur. J. Biochem. 1977, 75, 23–34. [Google Scholar] [CrossRef]
- Vinogradov, E.; Brade, H.; Holst, O. The structure of the O-specific polysaccharide of the lipopolysaccharide from Chromobacterium violaceum NCTC 9694. Carbohydr. Res. 1994, 264, 313–317. [Google Scholar] [CrossRef]
- Lima, D.C.; Nyberg, L.K.; Westerlund, F.; de Medeiros, S.R.B. Identification and DNA annotation of a plasmid isolated from Chromobacterium violaceum. Sci. Rep. UK 2018, 8, 5327. [Google Scholar] [CrossRef]
- Batista, J.H.; Leal, F.C.; Fukuda, T.T.H.; Diniz, J.A.; Almeida, F.; Pupo, M.T.; Neto, J.F.S. Interplay between two quorum sensing-regulated pathways, violacein biosynthesis and VacJ/Yrb, dictates outer membrane vesicle biogenesis in Chromobacterium violaceum. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- Rooney, L.M.; Amos, W.B.; Hoskisson, P.A.; McConnell, G. Intra-colony channels in E. coli function as a nutrient uptake system. ISME J. 2020. [Google Scholar] [CrossRef]
- Efthimion, M.; Corpe, W. Effect of Cold Temperatures on the Viability of Chromobacterium violaceum. Appl. Microbiol. 1969, 17, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, Y.; Lee, H.; Lee, S.; Kim, S.; Choi, K. Membrane fluidity-related adaptive response mechanisms of foodborne bacterial pathogens under environmental stresses. Food Res. Int. 2015, 72, 25–36. [Google Scholar] [CrossRef]
- Clark, C.; Costa, M.S.; Sanchez, L.; Murphy, B. Coupling MALDI-TOF mass spectrometry protein and specialized metabolite analyses to rapidly discriminate bacterial function. Proc. National. Acad. Sci. USA 2018, 115, 201801247. [Google Scholar] [CrossRef] [Green Version]
- Spring, S.; Merkhoffer, B.; Weiss, N.; Kroppenstedt, R.; Hippe, H.; Stackebrandt, E. Characterization of novel psychrophilic clostridia from an Antarctic microbial mat: Description of Clostridium frigoris sp. nov., Clostridium lacusfryxellense sp. nov., Clostridium bowmanii sp. nov. and Clostridium psychrophilum sp. nov. and reclassification of Clostridium laramiense as Clostridium estertheticum subsp. laramiense subsp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1019–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankratov, T.A.; Serkebaeva, Y.M.; Kulichevskaya, I.S.; Liesack, W.; Dedysh, S.N. Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J. 2008, 2, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.H.; Zhu, J.N.; Liu, Q.F.; Liu, Y.; Liu, M.; Liu, L.; Zhang, Q. Comparison of the diversity of root-associated bacteria in Phragmites australis and Typha angustifolia L. in artificial wetlands. World J. Microb. Biot. 2013, 29, 1499–1508. [Google Scholar] [CrossRef]
- Kurniawan, A.; Yamamoto, T. Accumulation of NH4+ and NO3− inside Biofilms of Natural Microbial Consortia: Implication on Nutrients Seasonal Dynamic in Aquatic Ecosystems. Int. J. Microbiol. 2019, 2019, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Prescott, R.; Decho, A. Flexibility and Adaptability of Quorum Sensing in Nature. Trends Microbiol. 2020, 28, 436–444. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [Green Version]
- Blanco, Y.; Rivas, L.; González-Toril, E.; Ruiz-Bermejo, M.; Moreno-Paz, M.; Parro, V.; Palacín, A.; Aguilera, Á.; Puente-Sánchez, F. Environmental parameters, and not phylogeny, determine the composition of extracellular polymeric substances in microbial mats from extreme environments. Sci. Total Environ. 2019, 650, 384–393. [Google Scholar] [CrossRef]
- Kulichevskaya, I.; Guzev, V.; Gorlenko, V.; Liesack, W.; Dedysh, S. Rhodoblastus sphagnicola sp. nov., a novel acidophilic purple non-sulfur bacterium from Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 2006, 56, 1397–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imhoff, J. Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–11. [Google Scholar] [CrossRef]
- Zheng, Y.; Dzakpasu, M.; Wang, X.; Zhang, L.; Ngo, H.H.; Guo, W.; Zhao, Y. Molecular characterization of long-term impacts of macrophytes harvest management in constructed wetlands. Bioresource Technol. 2018, 268, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Pankratov, T.A.; Tindall, B.J.; Liesack, W.; Dedysh, S.N. Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan- and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 2007, 57, 2349–2354. [Google Scholar] [CrossRef] [PubMed]
- Trappen, S.V.; Mergaert, J.; Eygen, S.V.; Dawyndt, P.; Cnockaert, M.C.; Swings, J. Diversity of 746 Heterotrophic Bacteria Isolated from Microbial Mats from Ten Antarctic Lakes. Syst. Appl. Microbiol. 2002, 25, 603–610. [Google Scholar] [CrossRef]
- Kevbrina, M.V.; Okhapkina, A.A.; Akhlynin, D.S.; Kravchenko, I.K.; Nozhevnikova, A.N.; Gal’chenko, V.F. Growth of Mesophilic Methanotrophs at Low Temperatures. Microbiology 2001, 70, 384–391. [Google Scholar] [CrossRef]
- Hahn, M.W.; Schmidt, J.; Koll, U.; Rohde, M.; Verbarg, S.; Pitt, A.; Nakai, R.; Naganuma, T.; Lang, E. Silvanigrella aquatica gen. nov., sp. nov., isolated from a freshwater lake, description of Silvanigrellaceae fam. nov. and Silvanigrellales ord. nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales ord. nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexales. Int. J. Syst. Evol. Microbiol. 2017, 67, 2555–2568. [Google Scholar] [CrossRef]
- Mishra, P.K.; Mishra, S.; Bisht, S.C.; Selvakumar, G.; Kundu, S.; Bisht, J.K.; Gupta, H.S. Isolation, molecular characterization and growth-promotion activities of a cold tolerant bacterium Pseudomonas sp. NARs9 (MTCC9002) from the Indian Himalayas. Biol. Res. 2009, 42, 305–313. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.C.Y.; Dunfield, P.F.; Stott, M.B. The Prokaryotes, Other Major Lineages of Bacteria and the Archaea; Springer: New York, NY, USA, 2014; pp. 447–458. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Panikov, N.S.; Tiedje, J.M. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl. Environ. Microb. 1998, 64, 922–929. [Google Scholar] [CrossRef] [Green Version]
- Stackebrandt, E. The Prokaryotes, Actinobacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 5–12. [Google Scholar] [CrossRef]
- Lv, X.; Yu, J.; Fu, Y.; Ma, B.; Qu, F.; Ning, K.; Wu, H. A meta-analysis of the bacterial and archaeal diversity observed in wetland soils. Sci. World J. 2014, 2014, 437684. [Google Scholar] [CrossRef] [Green Version]
- Oloo, F.; Valverde, A.; Quiroga, M.V.; Vikram, S.; Cowan, D.; Mataloni, G. Habitat heterogeneity and connectivity shape microbial communities in South American peatlands. Sci. Rep. UK 2016, 6, 25712. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Ganta, R.R. Laboratory Maintenance of Ehrlichia chaffeensis and Ehrlichia canis and Recovery of Organisms for Molecular Biology and Proteomics Studies. Curr. Protoc. Microbiol. 2008, 9, 3A.1.1–3A.1.21. [Google Scholar] [CrossRef] [PubMed]
- Zintl, A.; Moutailler, S.; Stuart, P.; Paredis, L.; Dutraive, J.; Gonzalez, E.; O’Connor, J.; Devillers, E.; Good, B.; OMuireagain, C.; et al. Ticks and Tick-borne diseases in Ireland. Ir. Vet J. 2017, 70, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, J.S.; Dautel, H.; Estrada-Peña, A.; Kahl, O.; Lindgren, E. Effects of climate change on ticks and tick-borne diseases in Europe. Interdiscip. Perspect. Infect. Dis. 2009, 2009, 593232. [Google Scholar] [CrossRef] [PubMed]




| General Characteristics | Cations | Anions | Heavy Metals | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Turbidity | 2.37 | OD530nm | Mg2+ | 0.26 | mg/L | [SO4]- | 1.21 | mg/L | Hg | <0.00001 | mg/L |
| Chromaticity | 36.9 | ° | Ca2+ | 0.66 | mg/L | [Cl]- | 1.14 | mg/L | V | <0.0001 | mg/L |
| Odour | 0 | grade 0–5 | Fe2+ | 0.154 | mg/L | [NO]- | 0.419 | mg/L | Ba | 0.003 | mg/L |
| pH | 5.68 | pH units | K+ | 0.43 | mg/L | [HCO3]- | <6.1 | mg/L | Be | <0.0001 | mg/L |
| Hardness | <0.060 | mg-CaCO3/L | Na+ | 0.66 | mg/L | [CO3]2- | <6.0 | mg/L | B | <0.05 | mg/L |
| Chemical oxygen demand | 22.2 | mg/L | Al3+ | 0.051 | mg/L | [NO2]- | <0.1 | mg/L | Mo | <0.0001 | mg/L |
| H2S | <0.002 | mg/L | [NH4]+ | 0.38 | mg/L | [Br]- | <0.05 | mg/L | Co | <0.0001 | mg/L |
| Petroleum products | 0.048 | mg/L | Li+ | <0.001 | mg/L | [PO3]- | <0.1 | mg/L | Ag | <0.0001 | mg/L |
| Free alkalinity | <0.1 | mM/L | [F]- | 0.159 | mg/L | Zn | 0.01 | mg/L | |||
| Total alkalinity | <0.1 | mM/L | Ni | <0.0001 | mg/L | ||||||
| Sulfide minerals | <0.002 | mg/L | Si | 0.556 | mg/L | ||||||
| Dry weight | 7.37 | mg/L | Cr | <0.0001 | mg/L | ||||||
| Conductivity | 11 | mkS/sm | Sr | 0.003 | mg/L | ||||||
| Cd | <0.0001 | mg/L | |||||||||
| As | <0.0001 | mg/L | |||||||||
| Cu | 0.002 | mg/L | |||||||||
| Pb | <0.0001 | mg/L | |||||||||
| Characteristic | Fermentation or Number of +/− Isolates if Variable |
|---|---|
| oxidase | − |
| indole production | − |
| bGL | − |
| NAG | 16/18 |
| SCI | 16/18 |
| LAC | − |
| MAN | 1/18 |
| TRE | + |
| XYL | 5/18 |
| ARA | 3/18 |
| aGA | − |
| bGA | − |
| MAL | − |
| GAL | 1/18 |
| MLT | − |
| CEL | − |
| SUC | 2/18 |
| INO | − |
| GGT | + |
| PHS | + |
| ESL | − |
| H2S | − |
| MAL | − |
| ONP | − |
| SAL | − |
| SOR | − |
| MLB | − |
| GLP | − |
| DUL | − |
| ADO | − |
| ART | − |
| RAF | − |
| bXY | − |
| NaCl, % range | 0–3 |
| pH range | 4.0–8.0 |
| pigmentation | 3/18 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11:0 | − | − | 0.2 | − | − | − | − | − | − | 0.2 | − | − | − | − |
| 10:0 3OH | 1.8 | 3.2 | 3.4 | 4.3 | 4.7 | 2.9 | 3.7 | 1.5 | 3.2 | 4.6 | 3 | 2.2 | 5.1 | 2.4 |
| 12:0 | 2.4 | 3.8 | 3.8 | 5.0 | 3.9 | 4.0 | 4.2 | 9.7 | 3.3 | 8.8 | 3.2 | 3.1 | 4.9 | 3.3 |
| 11:0 3OH | − | − | − | − | − | − | − | − | − | − | − | − | 0.4 | − |
| 13:0 | − | − | − | − | − | − | − | − | − | − | − | − | 0.4 | − |
| 12:0 2OH | 1.8 | 1.9 | 2.0 | 2.9 | 2.4 | 1.4 | 2.3 | − | 1.9 | 0.2 | 1.9 | 1.6 | 3.3 | 1.7 |
| 12:0 3OH | 2.8 | 3.3 | 3.4 | 4.0 | 3.6 | 2.5 | 2.9 | 1.4 | 2.9 | 4.4 | 2.8 | 2.5 | 4.8 | 2.6 |
| 13:0 2OH | − | − | − | − | − | − | − | 0.4 | − | − | − | − | − | − |
| 14:1 w7c | 0.4 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 14:1 w5c | 0.1 | 0.2 | 0.2 | 0 | 0.4 | − | 0.3 | 0.4 | − | − | 0 | 0.2 | − | − |
| 14:0 | 2.2 | 2.3 | 2.1 | 3.2 | 2.5 | 2.3 | 3.3 | 4.0 | 2 | 2.6 | 3.1 | 2.5 | 3.5 | 2.0 |
| 15:0 iso | − | − | − | − | − | − | − | 0.5 | − | − | − | − | − | − |
| 15:0 iso G | − | − | − | − | − | − | − | 0.7 | − | − | − | − | − | − |
| 15:1 w8c | 0.1 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 15:1 w6c | 0.2 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 15:0 | 1.0 | − | − | 1.3 | − | − | 0.9 | 1 | 0.6 | 2.3 | − | 3.0 | − | |
| 16:1 w7c | 43.7 | 42.7 | 41.9 | 41.9 | 47.1 | 42.5 | 34.1 | 38.6 | 38.9 | 33.4 | 28.7 | 38.5 | 27.5 | 36.3 |
| 16:1 w5c | 0.4 | 0.3 | 0.3 | 0.3 | 0.5 | − | − | − | − | 0.3 | 0 | 0.2 | − | − |
| 16:0 | 28.4 | 28.4 | 29.6 | 25.0 | 24.0 | 27.3 | 26.1 | 29.7 | 30.2 | 25.8 | 32 | 31.5 | 26.6 | 28.5 |
| 17:1 w6c | − | − | − | 0.2 | − | − | 0 | − | 0.2 | − | − | 0.4 | − | |
| 17:0 CYCLO | 0.2 | 0.4 | − | − | 0.4 | − | 2.9 | − | − | − | 13.2 | 0.2 | 4.3 | 1.3 |
| 18:2 w6,9c | − | − | − | − | − | − | 4.2 | − | − | − | − | − | − | − |
| 18:1 w9c | − | − | − | − | − | − | 2 | − | − | − | − | − | − | − |
| 18:1 w7c/12t/9t | 12.5 | 13.1 | 12.6 | 10.6 | 10.3 | 12.0 | 12.3 | 5.5 | 15.7 | 18.8 | 8.7 | 15.9 | 14.8 | 19.3 |
| 18:0 | 1.8 | 0.4 | 0.5 | − | 0.2 | 0.6 | 0.4 | 1.6 | 0.5 | 0 | 0.6 | 0.5 | 0.3 | 0.4 |
| SFA/MUFA * | 0.6 | 0.6 | 0.7 | 0.7 | 0.5 | 0.6 | 0.7 | 1.0 | 0.7 | 0.7 | 1.1 | 0.7 | 0.8 | 0.6 |
| Hydroxy FA ** | 6.4 | 8.4 | 8.8 | 11.2 | 10.7 | 6.8 | 8.6 | 3.3 | 8 | 9.2 | 7.7 | 6.3 | 13.6 | 6.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Egorova, D.A.; Voronina, O.L.; Solovyev, A.I.; Kunda, M.S.; Aksenova, E.I.; Ryzhova, N.N.; Danilova, K.V.; Rykova, V.S.; Scherbakova, A.A.; Semenov, A.N.; et al. Integrated into Environmental Biofilm Chromobacterium vaccinii Survives Winter with Support of Bacterial Community. Microorganisms 2020, 8, 1696. https://doi.org/10.3390/microorganisms8111696
Egorova DA, Voronina OL, Solovyev AI, Kunda MS, Aksenova EI, Ryzhova NN, Danilova KV, Rykova VS, Scherbakova AA, Semenov AN, et al. Integrated into Environmental Biofilm Chromobacterium vaccinii Survives Winter with Support of Bacterial Community. Microorganisms. 2020; 8(11):1696. https://doi.org/10.3390/microorganisms8111696
Chicago/Turabian StyleEgorova, Daria A., Olga L. Voronina, Andrey I. Solovyev, Marina S. Kunda, Ekaterina I. Aksenova, Natalia N. Ryzhova, Ksenya V. Danilova, Valentina S. Rykova, Anastasya A. Scherbakova, Andrey N. Semenov, and et al. 2020. "Integrated into Environmental Biofilm Chromobacterium vaccinii Survives Winter with Support of Bacterial Community" Microorganisms 8, no. 11: 1696. https://doi.org/10.3390/microorganisms8111696
APA StyleEgorova, D. A., Voronina, O. L., Solovyev, A. I., Kunda, M. S., Aksenova, E. I., Ryzhova, N. N., Danilova, K. V., Rykova, V. S., Scherbakova, A. A., Semenov, A. N., Polyakov, N. B., Grumov, D. A., Shevlyagina, N. V., Dolzhikova, I. V., Romanova, Y. M., & Gintsburg, A. L. (2020). Integrated into Environmental Biofilm Chromobacterium vaccinii Survives Winter with Support of Bacterial Community. Microorganisms, 8(11), 1696. https://doi.org/10.3390/microorganisms8111696






