1. Introduction
Colon microbes ferment dietary fiber to produce short chain fatty acids (SCFAs) that have many roles in promoting gut health [
1]. The ratio of SCFAs produced varies with the metabolism of each gut bacterial species and carbohydrate state [
2]. The SCFAs may reduce pH in the colon and results in shifting of the composition of gut microbiota, which is determined by the acid tolerance of gut bacterial species [
2]. Many species of Firmicutes, which are Gram-positive and characterized with a low percentage of G + C, and
Actinomycetes (high % G + C) show relatively greater acid tolerance, whereas the
Bacteroides spp. have less acid tolerance [
3].
Arabinogalactan (AG), which is a constituent of pectin, has been studied both in vivo and in vitro as a potential prebiotic in view of stimulating bifidobacteria in the gut microbiota [
4,
5,
6]. However, the enzyme functions for AG degradation in gut bacteria, even though predicted from genome analysis, are still poorly understood from a functional viewpoint. There are two structural types of AG, type I and type II [
7]. Type I AG consists of a β-1,3 and 1,4 linked galactan backbone, whereas type II AG has a more complex structure consisting of a β-1,3 linked galactan backbone with β-1,6 linked galactan side chains [
8,
9]. In addition, the backbone of type I AG and side chains of type II AG are substituted with α-arabinofuranose and/or, less frequently, β-arabinopyranose at the non-reducing terminus of side chains of both types of AG [
7,
8]. Gum arabic (GA) has a similar structure to type II AG, which includes a β-1,3 linked galactan backbone and β-1,6 linked galactan side chains with some arabinose substitutions [
10].
AG can be utilized by many
Bacteroides species, such as
Bacteroides thetaiotaomicron,
Bac. uniformis,
Bac. cellulosilyticus,
Bac. ovatus, and
Bac. caccae, while
Bac. distasonis,
Bac. eggerthii and
Bac. fragilis cannot [
11,
12].
Bifidobacterium species prefer fermentation of oligomers of relatively short degree of polymerization (DP), such as fructooligosaccharides (FOS) and galactooligosaccharides (GOS) [
13]. Furthermore, bifidobacteria also have the ability to degrade arabinoxylan oligosaccharides [(A)XOS], but this activity is strain-dependent [
14]. Many in vitro studies reported that some gut bacteria cannot degrade specific polysaccharides by themselves but take advantage of the metabolic products of other gut bacteria [
15]. These products may be carbohydrate fragments and/or fermentation products such as SCFAs [
16].
β-galactanases are the main enzymes for degradation of the backbone of AG, resulting in the release of galactose and/or GOS. Two kinds of galactanases, exo-β-1,3-galactanase and endo-β-1,4-galactanase, have been cloned from
Bifidobacterium longum and expressed in
Escherichia coli [
8,
17]. Both enzymes are extracellular but the degradation of the AG backbone by both enzymes is inhibited by arabinosyl side chains [
8,
17]. However, the study did not demonstrate which linkage type was inhibited. Furthermore, endo- and exo-β-1,3-galactanases from
Bac. thetaiotaomicron species have been characterized [
10,
18]. The exo-β-1,3-galactanase was also inhibited by arabinosyl side chains, but there is no information on whether endo-β-1,3-galactanases are inhibited in the same manner [
10].
Given that arabinose side chains of AG may inhibit cleavage of the backbone of AG by some β-galactanases, extracellular arabinosidases may have an important role in removing arabinosyl side chains for efficient AG degradation. Two of the extracellular α–arabinofuranosidase genes were induced in
B. longum subsp.
longum NCC 2705 when cocultured on (A)XOS with
Eubacteria rectale ATCC 33656 [
19]. However, these results represent gene expression, not enzyme activity. The β-L-arabinopyranosidases have been cloned from
B. longum and
Bac. thetaiotaomicron and characterized by heterologous expression in
E. coli, respectively [
10,
20]. The carbohydrate-active enzyme (CAZymes) database shows that the genome of most
Bacteroides spp. encode α-arabinosidase genes, and the enzyme activities were determined for
Bac. thetaiotaomicron,
Bac. plebeius,
Bac. coprocola, Bac. barnesiae,
Bac. salanitronis,
Bac. gallinarum and
Bac. intestinalis [
10,
21,
22,
23]. However, the cellular location and substrate specificity of the enzymes have not been demonstrated.
The GOS released from AG can be transported into the bacteria or degraded to galactose outside the cell. A three-gene cluster encoding an ATP-binding cassette (ABC)-type uptake system for GOS has been shown in
B. breve UCC 2003, which includes one GOS-binding protein and two permease proteins [
24]. In addition, a total of nine β-galactosidase genes from
B. bifidum,
B. infantis and
B. breve have been isolated and characterized [
24,
25,
26]. Two of them are predicted to be extracellular enzymes, and seven of them are intracellular enzymes. Furthermore, β-galactosidase activities have also been reported from
Bac. polypragmatus,
Bac. fragilis,
Bac. thetaiotaomicron among
Bacteroides species [
27,
28,
29].
Compared to oligosaccharides, AG has longer DP and more complex monomer saccharide units, which may exceed the substrate specificity of bifidobacterial enzymes [
30]. One study reported that nine
B. longum strains were incapable of significant growth on AG in pure culture [
24]. In general,
B. longum,
B. breve, and
B. bifidum are dominant in infants, while
B. catenulatum,
B. adolescentis and
B. longum are isolated from adult fecal samples [
31,
32]. In 42 fecal samples from healthy Belgian adults, among bifidobacterial species,
B. longum, B. adolescentis, and
B. catenulatum were present in 90%, 79% and 38% of samples, respectively [
33]. Therefore, the observed bifidogenic effect of AG in vivo and in vitro (fecal inoculation) studies is likely due to the cross-feeding of products released by other AG-degrading bacteria, such as
Bacteroides species.
The aim of this study was to investigate the specificity of these enzymes from gut bacteria strains in order to understand their contribution to AG degradation. Furthermore, a better understanding of the metabolic interaction between bifidobacteria and
Bacteroides spp. in the presence of AG is required. Both
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 strains were isolated from human feces, and the whole genome sequences of both strains have been published [
34]. However, the enzyme activities for AG degradation from both strains have not been characterized. In this study, the characteristics of AG degradation enzymes from
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 were determined, which included quantification of activity, cellular location, and substrate specificity. The possible metabolic cooperation between
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 during AG fermentation was investigated.
2. Materials and Methods
2.1. Carbohydrate Substrates
Larch wood AG (average molecular weight (MW): around 20 kDa (after autoclaving)), GA from acacia tree and p-nitrophenyl (pNP) substrates were purchased from Sigma-Aldrich (Oakville, Canada). Potato galactan, de-arabinosylated potato galactan, and other small MW substrates (β-1,3-galactobiose, β-1,4-galactobiose, β-1,6-galactobiose, α-1,5-arabinobiose, α-1,5-arabinotriose, α-1,5-arabinotetraose, 23-α-L-arabinofuranosyl-xylotriose (A2XX) and 32-α-L-arabinofuranosyl-xylobiose (A3X)) were obtained from Megazyme (Burlington, Canada).
De-arabinosylated AG (de-AG) and de-arabinosylated GA (de-GA) were prepared by mild acid degradation, as described previously with minor modifications [
35]. Briefly, AG (90 g) or GA (90 g) were dissolved in water to a final volume of 450 mL, respectively. After heating to 95–98 °C with continuous stirring, 50 mL of trifluoroacetic acid (2.5 M) (Sigma-Aldrich, Oakville, Canada) were added to the solutions and stirred continuously at 90–95 °C for another 2 h. After incubation, the solutions were rapidly cooled on ice for 5 min, and the pH was adjusted to 7.0 using 5 M NaOH. The supernatant was collected by centrifugation at 15,000×
g for 5 min at room temperature, followed by filtration through a binder-free glass microfiber filter (GF/A grade; Whatman, Maidstone, UK). The de-AG and de-GA were precipitated with 2 volumes of absolute ethanol overnight at 4 °C and washed twice with absolute ethanol. The washed de-AG and de-GA precipitates were dried in the vacuum oven at 50 °C overnight. To remove fragments of sugar units, the de-AG, de-GA and de-arabinosylated potato galactan were dissolved in water and dialyzed in 3500 Da cut-off dialysis cassettes (Thermo Scientific, Mississauga, Canada) against water for 3 days at 4 °C, followed by drying in the vacuum oven at 50 °C overnight.
2.2. Media, Strains and Growth Conditions
The MRS medium (Oxoid, Nepean, Canada) was supplemented with 0.05% (w/v) L-cysteine·HCl for growth of B. longum subsp. longum NCC 2705. The yeast extract-casitone medium supplemented with short chain fatty acid (YCFA) contained (per liter): 10 g casitone, 2.5 g yeast extract, 4 g NaHCO3, 1 g L-cysteine·HCl, 0.45 g K2HPO4, 0.45 g KH2PO4, 0.9 g NaCl, 0.9 g (NH4)2SO4, 0.09 g MgSO4·7H2O, 0.09 g CaCl2, 0.001 g resazurin, 0.01 g hemin, 0.00001 g biotin, 0.00001 g cobalamin, 0.00003 g p-aminobenzoic acid, 0.00005 g folic acid, and 0.00015 g pyridoxamine. The following SCFAs were added (final concentrations): acetate (33 mM); propionate (9 mM); isobutyrate, isovalerate, and valerate (1 mM each). The SCFAs were excluded when appropriate, which was referred to as YC medium. The carbon sources were added at a final concentration of 0.5% (w/v) to YCFA or YC medium. The final pH of the medium was adjusted to 6.3 ± 0.1 by using 1 M NaOH before sterilization at 121 °C for 15 min. Sterilized medium was cooled and filter-sterilized solutions of thiamine and riboflavin were added to give a final concentration of 0.05 μg/mL of each.
B. longum subsp. longum NCC 2705 was obtained from the Nestlé Research Centre (Lausanne, Switzerland); Bac. caccae ATCC 43185 was purchased from the American Type Culture Collection (Manassas, VA, USA). B. longum subsp. longum NCC 2705 was stored in MRS-cysteine while Bac. caccae ATCC 43185 was stored in YCFA-glucose, both at −80 °C with 24% (v/v) glycerol. B. longum subsp. longum NCC 2705 and Bac. caccae ATCC 43185 were resuscitated from frozen stocks in MRS-cysteine and YCFA-glucose, respectively. Subsequently, the cultures were inoculated at 3% (v/v) into YC medium supplemented with the carbon source and YC medium without carbon source as a control. For coculture, 3% (v/v) of each strain culture was inoculated in YC with the carbon source. All cultures were incubated anaerobically in an anaerobic chamber (Model UM-041, Ruskinn Technology, Sanford, USA) containing an atmosphere of 80% N2, 10% H2, and 10% CO2 at 37 °C. At selected fermentation times, optical density (OD) at 600 nm and pH of harvested culture broth were measured by using a spectrophotometer (Model DU530, Beckman, Brea, USA) and a pH meter (Fisher Scientific, Nepean, Canada), respectively. All growth curves were repeated three independent times.
2.3. Analysis of Carbohydrate Degradation
The MW profile of carbohydrates in the spent culture media were analyzed by high-performance size exclusion chromatography (HPSEC). After 48-h fermentation, 1 mL of cell-free supernatants obtained through centrifugation at 14,000×
g for 15 min were deproteinated with Carrez clarification reagent kit (Sigma-Aldrich, Oakville, Canada) following the manufacturer’s protocol. The protein-free supernatant or dextran standards (Sigma-Aldrich, Oakville, Canada) were mixed with the same volume of mobile phase (0.1 M NaNO
3 and 0.01 M NaH
2PO
4; pH: 7.7), followed by centrifugation at 14,000×
g for 15 min. The supernatant of the mixture was filtered through a 0.45-μm filter (Fisher Scientific, Nepean, Canada). Forty microliters of sample or standard were analyzed on a Shimadzu LC-10A HPSEC system (Shimadzu Corporation, Kyoto, Japan) equipped with a PL Aquagel-OH-Mixed M column (7.5 × 300 mm, 8 µm, Agilent Technologies Canada Inc., Mississauga, Canada) and refractive index (RI) detector. The temperature of the column and detector were set at 40 °C, and mobile phase at a flow rate of 0.3 mL/min. All instrument control, analysis, and data processing were performed using the Labsolutions platform (Shimadzu corporation. Version 5.82). Known MW (1 kDa, 5 kDa, 12 kDa, 25 kDa, 50 kDa and 80 kDa) dextran standards were used for mass approximation of AG present in the culture. The percentage of degradation was quantified by peak area at retention time between 27.8 and 32.2 min (50 kDa-5 kDa).
2.4. Analysis of Enzyme Activity and Substrate Specificity
A volume of 1 mL of YC-AG broth culture was harvested after 16 h of fermentation and centrifuged at 14,000× g for 5 min at 4 °C. The cell pellets were washed twice with 500 μL of 0.2 M phosphate buffer (PB) (pH 6.3), disrupted by Bead Mill 24 (Fisher Scientific, Nepean, Canada) in 500 μL of the same buffer with 0.6 g of glass beads (0.1 mm, BioSpec Products, Inc., Bartlesville, USA) for 10 min, followed by centrifugation at 14,000× g for 5 min at 4 °C. The supernatant containing crude cytoplasmic enzymes was collected. The precipitate containing crude cell wall-associated enzymes was suspended in 500 μL of the same buffer after washing twice.
To quantify cell wall-associated and cytoplasmic enzymes, the
pNP test was performed as described previously [
36]. Briefly, 10 μL of the crude enzyme was mixed with 10 μL of 5 mM
pNP substrates and 80 μL of PB buffer (0.2 M; pH 6.3), followed by incubation at 37 °C for 30 min. The reaction was stopped by the addition of 100 μL of 1 M Na
2CO
3, and liberated
pNP was measured at 405 nm using a spectrophotometer (Multiskan
TM GO Microplate Spectrophotometer, Thermo Scientific, Mississauga, Canada). One unit (U) of enzyme activity was defined as the amount of enzyme that released 1 μmol of
pNP per min at 37 °C. The enzymatic assays were conducted in triplicate on each extract.
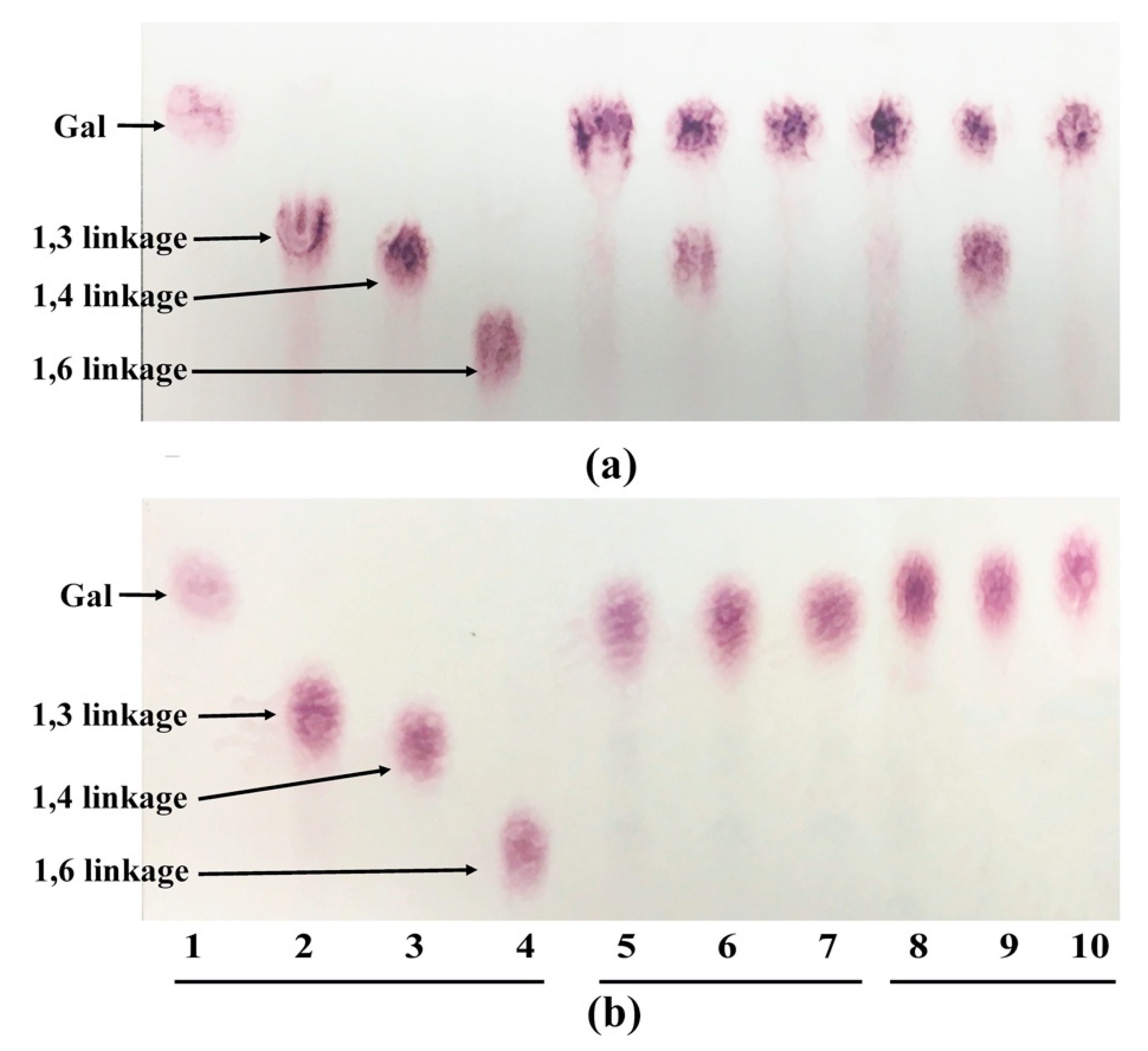
The enzymatic substrate specificity was analyzed by thin-layer chromatography (TLC) as described previously [
20]. Eighty μL of the crude enzyme was incubated with 20 μL of substrate (0.2 mg/mL in water) at 37 °C for 16 h. Ten microliters of the reaction products and standards were loaded on a TLC silica gel 60 aluminum sheet (Merck, Darmstadt, Germany) using a 7:1:2 (
v/
v/
v)
n-propanol-ethanol-water as mobile phase. The carbohydrate was visualized by spraying the plate with 180 mg orcinol in H
2SO
4 solution (water (5 mL): ethanol (75 mL): H
2SO
4 (10 mL)) and heating.
2.5. PMA Treatment and Droplet Digital PCR (ddPCR) Analysis
To enumerate viable cells,
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 in mono and co-cultures were treated with propidium monoazide (PMA) (Biotium, Fremont, USA) using a previously described method with minor modifications [
19]. Briefly, 1-mL aliquots of culture were centrifuged at 14,000
g for 15 min at 4 °C. After removing the supernatant, 500 μL of sterile peptone water (1 g/L peptone, 0.5 g/L cysteine-HCL, pH 6.8) was added. The cell suspension was treated with 10 μL PMA (2.5 mM) and shaken in the dark for 5 min at room temperature. The PMA-treated cells were exposed in a UV light (PhAST Blue, GenIUL, Barcelona, Spain) for 15 min, followed by centrifugation at 14,000
g for 15 min at 4 °C. The PMA-treated cell pellets were washed by 800 μL of sterile physiological saline (0.85% NaCl), followed by centrifugation at 14,000
g for 15 min at 4 °C. The cell pellets were stored at −20 °C until DNA extraction. The DNA of both strains was extracted by using the DNeasy UltraClean Microbial Kit (Qiagen, Mississauga, Canada) following the manufacturer’s protocol. The DNA extracts were quantified and qualified using Qubit 4 (Invitrogen Canada Inc., Burlington, Canada) and stored at −20 °C until use.
The number of viable
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 in mono and co-culture were quantified by the ddPCR system following the manufacturer’s instructions. Briefly, a total of 25 μL of each reaction mixture contained 12.5 μL of QX200 ddPCR EvaGreen supermix, 5 μL of DNA, and 0.7 μL each of 10 μM forward and reverse primers (
Table 1). Droplets were generated using QX200 Droplet Generator with a disposable cartridge. The cartridge was filled by 20 μL of each reaction mixture in the sample well and 70 μL of droplet generator oil in the oil well. After droplet generation, 40 μL of droplet suspension were transferred to a 96-well ddPCR plate and the plate was sealed with foil in a PX1 PCR Plate sealer at 180 °C for 5 s. The sealed plate was inserted into a C1000 Touch Thermal Cycler with the following amplification program: 95.0 °C for 10 min, 40 cycles at 95.0 °C for 30 s, the appropriate annealing temperature and time for each primer set (
Table 1), and one cycle of 98 °C for 10 min with a 4 °C hold. The fluorescence was read using the QX200 Droplet Plate Reader, and data were analyzed using Quantasoft software. All of the equipment, materials and regents for ddPCR analysis were purchased from Bio-Rad (Mississauga, ON, Canada).
2.6. Quantification of SCFAs and Other Metabolites
The concentration of acetate, butyrate, lactate, formate, ethanol, propionate and succinate were determined through single-dimension proton nuclear magnetic resonance (1D 1H NMR) following a previously described protocol [
38]. Briefly, culture samples were prepared first by serial filtration (1 μm, 0.8 μm, 0.45 μm, and 0.22 μm) (Whatman GE, Maidstone, UK). Filtered samples (630 μL) were mixed with 70 μL of internal standard, IS-2 Chenomx internal standard-DSS-d6 solution (Chenomx Inc., Edmonton, Canada), on the day of the analysis. Sample mixtures were transferred to 5-mm glass NMR tubes (NE-UL5-7, New Enterprises Inc., Vineland, NJ, USA) and analyzed by a Bruker Avance 600.13 MHz spectrometer with a triple resonance probe (TXI 600). The TXI 600 for scanning was performed in a single batch using the first increment of a 1D NOESY pulse sequence that had t
mix of 100 ms, 5.25-s acquisition time, 1-s relaxation delay, and a spectral width of 14 ppm. The spectra were analyzed by Chenomx NMR Suite 7.0-7.7 (Chenomx Inc., Edmonton, Canada). The compounds were determined through comparison with the compound library. A known concentration of internal standard was used for the quantification of compounds.
2.7. Analysis of Nucleotide Sequences
The enzyme coding genes were analyzed through the Integrated Microbial Genomes and Microbiomes web server (
https://img.jgi.doe.gov). The IMG genome ID of
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC43185 are 637000031 and 640963023, respectively. Cellular localization of putative AG degradation genes was defined based on the PSORTb v3.0 web server (
https://www.psort.org/psortb/).
2.8. Statistical Analysis
Statistical analyses were performed using Graph-Pad Prism 8. Means and standard deviations of three replicates were analyzed by Student’s two-tailed
t-test (Figure 4 and
Figure S1;
Table 2) or Dunnett’s multiple comparisons test (Figure 5); where
p < 0.05 was considered as significant.
4. Discussion
AG must be degraded by extracellular enzymes to release mono- or oligo-saccharides for further transport and use [
12,
39]. β-galactanase is the main enzyme for degrading the backbone of AG. The genomes of both
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 encode β-galactanases. The β-galactanase of
B. longum subsp.
longum NCC 2705 is predicted to be exported outside the cytoplasmic membrane, due to the presence of a signal peptide and cell wall anchor motif [
40]. The LPXTG membrane-anchoring motif has been found in some Gram-positive bacteria genomes that encode several surface proteins [
8]. Although the β-galactanase gene from
Bac. caccae ATCC 43185 does not have an LPXTG membrane-anchoring motif, it does have a signal peptide. Therefore, the β-galactanases from both
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 are predicted to be extracellular enzymes.
After the breakdown of the AG backbone, the oligosaccharides that are released from AG can then be either transported into the cell or hydrolyzed to monosaccharides by extracellular enzymes [
8].
Bacteroides spp. are generally not efficient at transporting carbohydrate fragments into the cells after extracellular degradation [
30]. However, the specific activity of β-galactosidase and α-arabinofuranosidase showed that both strains have extracellular oligosaccharide-degrading activities. In addition, analysis of substrate specificity showed that extracellular β-galactosidases and α-arabinofuranosidases of both strains are able to cleave all types of glycosidic linkages of AG. These results likely indicate that
Bac. caccae ATCC 43185 has the ability and prefers external degradation of oligosaccharides that are released from AG. In contrast, the intracellular β-galactosidase activity of
B. longum subsp.
longum NCC 2705 is around 10 times higher than extracellular activity. Moreover, a three-gene cluster encoded in the genome of
B. longum subsp.
longum NCC 2705 is highly similar to the gene cluster that encodes an ABC-GOS uptake system in
B. breve UCC 2003 [
24]. These results imply that for GOS,
B. longum subsp.
longum NCC 2705 prefers internal degradation to sequester substrate and compete effectively with other gut bacteria.
β-galactosidase is only able to release galactose from the non-reducing end of the substrate, and may be inhibited by substituted side chains [
41,
42]. As the substrates used in this study have limited non-reducing ends, just a small amount of galactose could be released by β-galactosidase. If the endo-β-galactanase internally cleaves the main chain of AG to release more carbohydrate fragments with non-reducing ends, more galactose could be released from the non-reducing ends by β-galactosidase. However, the extracellular endo-β-galactanase activities for cleaving the galactans at β-1,3/1,6 linkages from both strains were inhibited by the arabinose side chains. Furthermore, the ability of the extracellular endo-β-galactanase of
Bac. caccae ATCC 43185 to cleave the β-1,4 galactan was partially inhibited by the arabinosyl side chains. These findings suggest that without removing arabinose side chains from galactans, both strains are unable to degrade the β-1,3/1,6 linkages; only the β-1,4 linkages of the galactan can be cleaved. This means that type II AG is mostly unusable, and only part of type I AG can be cleaved at the β-1,4 linkage position by both strains without removing the arabinose side chains.
Bac. caccae ATCC 43185 had better growth on the de-arabinosylated GA compared to GA, which confirmed that the arabinose side chains had a negative effect on GA utilization by this strain. However, this growth stimulation was not observed for de-AG versus AG for
Bac. caccae ATCC 43185. This likely resulted from the remaining arabinosyl side chains that were not completely removed from AG by using mild acid degradation. This method has only been validated when preparing the de-arabinosylated from of GA, but not for AG [
35]. Furthermore, both de-AG and de-GA did not show any stimulation of the growth of
B. longum subsp.
longum NCC 2705. These results indicate that the DP of de-AG and de-GA is likely too long to be utilized effectively by
B. longum subsp.
longum NCC 2705. Therefore, AG utilization by
B. longum subsp.
longum NCC 2705 requires the contribution of other gut bacteria to provide lower DP oligosaccharides such as GOS.
As shown above, extracellular arabinosidase is very important to AG degradation. Both strains have extracellular α-arabinofuranosidase and β-arabinopyranosidase that are able to cleave α-1,2; 1,3; 1,5 linkages, when tested with low DP substrates. However, the extracellular arabinosidases from both strains were unable to release arabinose from AG, GA and galactan. These results indicate that the extracellular arabinosidases from both strains were not able to cleave arabinose from large MW substrates. One reason may be that large MW substrates have more complex structure, which could hide the non-reducing ends that are the preferred enzyme target sites.
The mono- and co-culture experiments showed that the cross-feeding of the partial breakdown products from AG occurred between
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 at the early fermentation stage (before 12 h of fermentation). The degraded AG fragments produced by extracellular enzymes from
Bac. caccae ATCC 43185 may accumulate in the coculture and are likely to be utilized by
B. longum subsp.
longum NCC 2705 for growth. Growth of bifidobacteria can reduce the pH due to organic acid production, which results in restraining the growth of other less acid-tolerant bacteria [
43]. Therefore, the pH of coculture was lower than that of
Bac. caccae ATCC 43185 monoculture due to the metabolism of
B. longum subsp.
longum NCC 2705.
Bacteroides species have poor growth at pH 5.5 due to lower acid tolerance [
3]. The low pH condition inhibited the growth of
Bac. caccae ATCC 43185, which may result in no further breakdown of carbohydrates in the coculture for
B. longum subsp.
longum NCC 2705 utilization. Therefore, the coculture of
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185 showed a lower OD
600 value than that of monoculture of
Bac. caccae ATCC 43185. These results are consistent with one study that reported the abundance of
Bac. thetaiotaomicron was maintained in the presence of bifidobacteria with high dilution rates during AG fermentation [
30]. These results support the importance of pH in the gut environment for limiting the growth of
Bacteroides species. In the distal colon where the pH would be optimal for the growth of
Bacteroides, this species may release carbohydrate fragments of AG that can be utilized by other gut bacterial species, consequently influencing the composition of gut microbiota and their interactions.
Prevotella ruminicola represents another fiber-degrading species that may participate in AG degradation in the gut, as this species has been enriched from cecal contents fermented with gum arabic [
44], which has similar monosaccharide linkages to type II AG.
There is less information about the limitation of metabolic activity of
Bacteroides species in low pH conditions.
Bacteroides species ferment carbohydrates to mainly produce acetate, propionate and succinate [
2]. However, the succinate can be converted to propionate via succinyl-, methylmalonyl-, and propionyl-coenzyme A by
Bac. fragilis [
45,
46]. The concentration of succinate was significantly higher, whereas propionate was significantly lower in the coculture compared to
Bac. caccae ATCC 43185 monoculture. These results imply that the metabolic activity of
Bac. caccae ATCC 43185 was inhibited at low pH, which resulted in lower conversion of succinate to propionate. Therefore, the succinate accumulated in the coculture. The concentration of formate was higher than that of lactate in monoculture of
B. longum subsp.
longum NCC 2705, which correlates with nutrient limitation in the culture. Formate accumulates because extra ATP can be formed when bifidobacteria produce formate instead of lactate in a carbohydrate-limited environment [
30]. The lactate that was mainly produced by
B. longum subsp.
longum NCC 2705 was not observed in the coculture, which was not surprising since lactate can be utilized by
Bacteroides spp. [
47,
48]. The concentration of acetate was significantly higher in coculture compared to monoculture of
B. longum subsp.
longum NCC 2705 and
Bac. caccae ATCC 43185. However, more evidence is required to demonstrate which strain contributed predominantly to the production of acetate.