Staphylococcus epidermidis and Cutibacterium acnes: Two Major Sentinels of Skin Microbiota and the Influence of Cosmetics
Abstract
1. Introduction
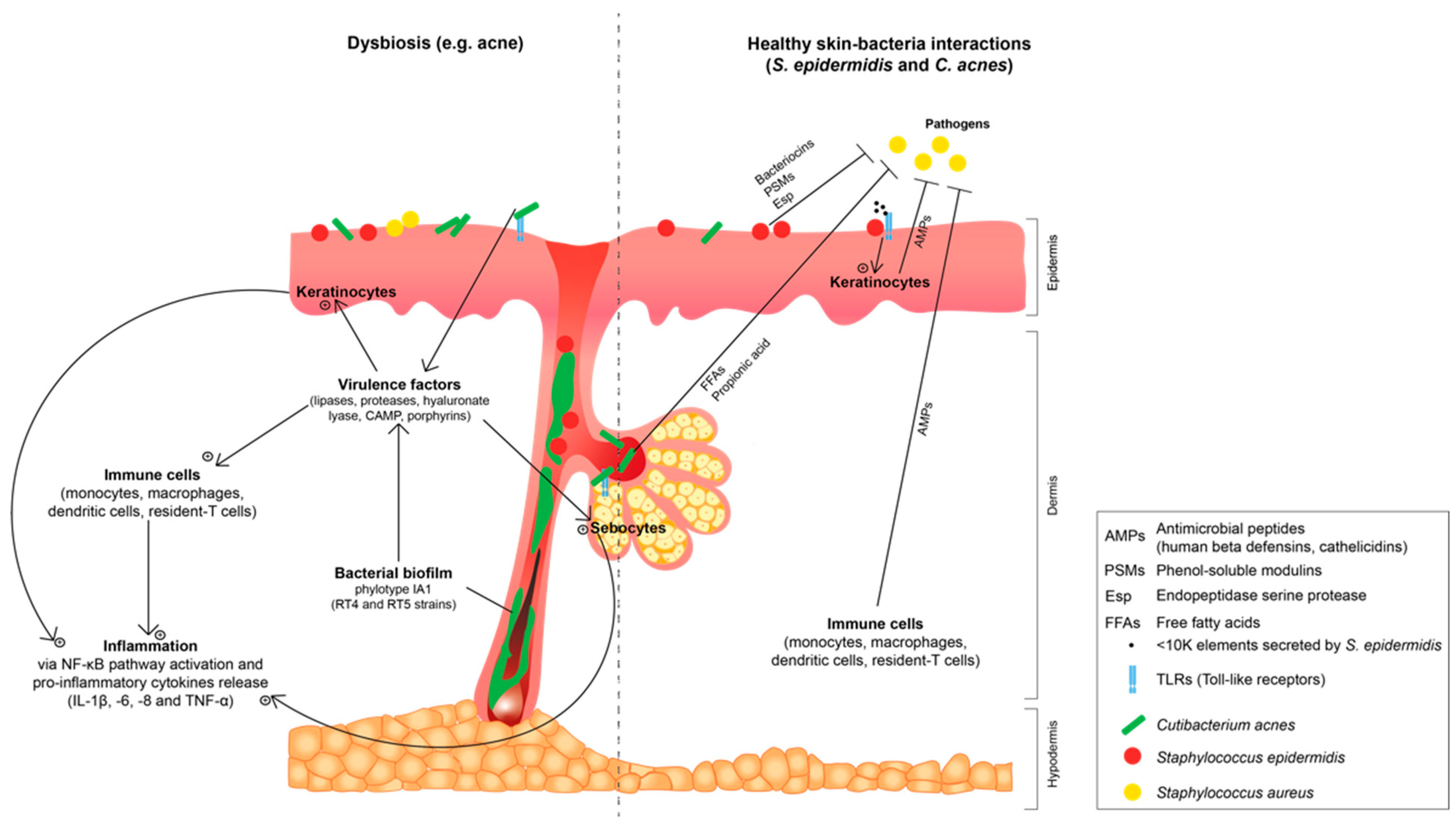
2. Interactions between Skin, Staphylococcus epidermidis and Cutibacterium acnes: A Possible Shift from Commensalism to Opportunistic Pathogenicity
2.1. Staphylococcus epidermidis and Cutibacterium acnes: Two Major Commensal Gram-Positive Bacteria of Skin Microbiota
2.2. Staphylococcus epidermidis and Cutibacterium acnes: Shift to an Opportunistic Pathogenicity and Correlation with Common Skin Dysbiosis
2.2.1. Staphylococcus epidermidis Biofilm and Loss in Staphylococcus aureus and Staphylococcus epidermidis Diversity: Involvement in Atopic Dermatitis
2.2.2. Cutibacterium acnes Biofilm and Loss in Phylotype Diversity: Involvement in Acne Dysbiosis
2.2.3. Influence of Skin Microenvironment on Staphylococcus epidermidis and Cutibacterium acnes Virulence and Biofilm Formation
2.3. Skin Ageing and Photoexposition Linked to Staphylococcus epidermidis and Cutibacterium acnes Dysbiosis
3. Evaluation Methods of Skin Microbiota Targeting Staphylococcus epidermidis and Cutibacterium acnes from a Cosmetics Perspective
3.1. Cutaneous Microbiome and Microbiota Evaluation
3.2. Skin–Microbiota Interactions and Cosmetic Active Ingredients Evaluation
4. Influence of Cosmetics on Skin Microbiota, Particularly on Staphylococcus epidermidis and Cutibacterium acnes
- active ingredients, algal- or plant-based, and thermal water-based, which are not a nutrient source for microorganism;
- prebiotics: nutrients that confer a health benefit with modulation of structure and functionality of the host microbiota in topical application for the cosmetic sector [26]. Cosmetic prebiotic approaches are to maintain healthy skin microbiota, or improve the skin microbiota composition by limiting or reducing pathogen growth and in the same time preserve or stimulate commensal bacteria growth [143,156,171,172];
- probiotics: fragmented bacteria that confer health benefits to the host. Cosmetic products with “probiotics” or “probiotic ingredients” often contain non-viable bacteria, products of bacterial fermentation or cell lysates, which do not require changes in the preservative ingredient system [26]. Nevertheless, cosmetic products containing fragments of microorganisms as probiotics require care regarding safe production. For now, a strict definition of a probiotic in cosmetic products has not been established and these products should only follow European Cosmetic Regulation 1223/2009 [149];
- post-biotics: bacterial metabolites and or cell wall components released by probiotic microorganisms [160].
4.1. Promotion of Commensal Metabolism for Prevention of Pathogen Growth
| Active Names and Composition | Staphylococcus epidermidis | Cutibacterium acnes | References | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Growth | Virulence | Cytotoxicity on HaCat Keratinocytes | Biofilm Formation | Growth | Virulence | Cytotoxicity on HaCat Keratinocytes | Biofilm Formation | ||
| BioEcolia® Oligosaccharide with saccharose and maltose bond in α-1-2 and α-1-6 | commensal strain MFP04 | ns | [170,192,193] | ||||||
| + | ns | + | + | ||||||
| PS291® Polysaccharide rich in rhamnose | commensal strain MFP04 | normal skin strain and/or acneic strains (RT4: (HL045PA1/HM-516) and (RT5: HL043PA2/HM-514) | |||||||
| 0 | 0 | 0 | − | 0 | 0 | 0 | − | ||
| ExpoZen® Low molecular weight polysaccharides enriched in galactose produced by radical hydrolysis from Halymenia durvillei | + | ns | ns | [187,194] | |||||
| UriageTM Thermal Water (UTW) Enrich in natural minerals 11 g/L (sulfates, chloride, sodium, bicarbonate, calcium, magnesium, potassium and silicon) and trace elements (zinc, manganese, cupper and iron) | commensal strain MFP04 | acneic strains RT4 (HL045PA1/HM-516) and RT5 (HL043PA2/HM-514) | [103,170,192] | ||||||
| − (idd) | ns | + | + | − | 0 | 0 | − | ||
| Viniderm® Rich in polyphenol and δ-viniferine | commensal strain MFP04 | ns | [170] | ||||||
| + | ns | 0 | − | ||||||
| MPA-RegulTM Vegetal polysaccharide rich in gluconic acid (obtained from enzymatic process) with UTWTM | ns | acneic strains RT4 and RT5 | [103] | ||||||
| 0 | 0 | 0 | − (idd) | ||||||
| Myrtacine® Lipophilic extract from M. communis leaves | strain CIP 53117T | [195] | |||||||
| − | ns | ns | − | ||||||
| BGM Complex Bakuchiol, Gingko biloba extract, and mannitol | strain CIP A 179 | [196] | |||||||
| − | ns | ||||||||
| ACNILYS® Rhodomyrtus tomentosa berry extract | − | ns | [197,198] | ||||||
| DIOLÉNYL® Ester of diol and polyunsaturated fatty acid | strain ATCC6919 | [199] | |||||||
| − | ns | ||||||||
4.2. Reduction in Pathogen Growth, Biofilm Formation or Virulence: Example of Acne Dysbiosis, Frontier with Dermatology
4.2.1. Decrease in Virulence Factors
4.2.2. Antibacterial Activity
4.2.3. Promotion of Cutibacterium acnes Phylotype Diversity
4.2.4. Inhibition of Biofilm Formation and Maturation
4.3. Modulation of the Skin Microenvironment and Immune Responses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; NISC Comparative Sequencing Program; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; Fitzgerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on humang-microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef]
- Wilson, M.; Houpt, E.R. An introduction to the human–microbe symbiosis. In Microbial Inhabitants of Humans: Their Ecology and Role in Health and Disease; Cambridge University Press: Cambridge, UK, 2005; pp. 1–50. ISBN 9780511735080. [Google Scholar]
- Findley, K.; Oh, J.; Yang, J.; Conlan, S.; Deming, C.; Meyer, J.A.; Schoenfeld, D.; Nomicos, E.; Park, M.; Kong, H.H.; et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013, 498, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Perez-Perez, G.I.; Chen, Y.; Blaser, M.J. Quantitation of major human cutaneous bacterial and fungal populations. J. Clin. Microbiol. 2010, 48, 3575–3581. [Google Scholar] [CrossRef]
- Foulongne, V.; Sauvage, V.; Hebert, C.; Dereure, O.; Cheval, J.; Gouilh, M.A.; Pariente, K.; Segondy, M.; Burguière, A.; Manuguerra, J.-C.; et al. Human Skin Microbiota: High Diversity of DNA Viruses Identified on the Human Skin by High Throughput Sequencing. PLoS ONE 2012, 7, e38499. [Google Scholar] [CrossRef]
- Probst, A.J.; Auerbach, A.K.; Moissl-Eichinger, C. Archaea on Human Skin. PLoS ONE 2013, 8, e65388. [Google Scholar] [CrossRef]
- Lacey, N.; Kavanagh, K.; Tseng, S.C.G. Under the lash: Demodex mites in human diseases. Biochemist 2009, 31, 20–24. [Google Scholar] [CrossRef]
- Robinson, P.J. Skin. In Encyclopedia of Toxicology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 283–309. ISBN 9780123864543. [Google Scholar]
- Wilkes, G.L.; Brown, I.A.; Wildnauer, R.H. The biomechanical properties of skin. CRC Crit. Rev. Bioeng. 1973, 1, 453–495. [Google Scholar]
- Proksch, E.; Brandner, J.M.; Jensen, J.-M. The skin: An indispensable barrier. Exp. Dermatol. 2008, 17, 1063–1072. [Google Scholar] [CrossRef]
- Wilson, M. The Indigenous Microbiota of the Skin. In The Human Microbiota in Health and Disease: An Ecological and Community-Based Approach; Garland Science, Ed.; Taylor & Francis: Cambridge, UK, 2018; pp. 86–95. ISBN 978-0815345855. [Google Scholar]
- Karkman, A.; Lehtimäki, J.; Ruokolainen, L. The ecology of human microbiota: Dynamics and diversity in health and disease. Ann. N. Y. Acad. Sci. 2017, 1399, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Segre, J.A. Skin Microbiome: Looking Back to Move Forward. J. Investig. Dermatol. 2012, 132, 933–939. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science (80-.) 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Moeller Christensen, G.J.; Nakatsuji, T.; et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the human skin microbiome early in life. J. Investig. Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef]
- Ladizinski, B.; McLean, R.; Lee, K.C.; Elpern, D.J.; Eron, L. The human skin microbiome. Int. J. Dermatol. 2014, 53, 1177–1179. [Google Scholar] [CrossRef]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef]
- ALCIMED. Skin Microbiome—The Development of a Science that will Transform the Cosmetic Industry; ALCIMED: Paris, France, 2016. [Google Scholar]
- Sfriso, R.; Egert, M.; Gempeler, M.; Voegeli, R.; Campiche, R. Revealing the secret life of skin-with the microbiome you never walk alone. Int. J. Cosmet. Sci. 2019, 42, 116–126. [Google Scholar] [CrossRef]
- Grice, E.A. The intersection of microbiome and host at the skin interface: Genomic-and metagenomic-based insights. Genome Res. 2015, 25, 1514–1520. [Google Scholar] [CrossRef]
- Douglas, H.C.; Gunter, S.E. The Taxonomic Position of Corynebacterium acnes. J. Bacteriol. 1946, 52, 15–23. [Google Scholar] [CrossRef]
- Scholz, C.F.P.; Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef]
- Dagnelie, M.-A.; Corvec, S.; Saint-Jean, M.; Bourdès, V.; Nguyen, J.M.; Khammari, A.; Dréno, B. Decrease in diversity of Propionibacterium acnes phylotypes in patients with severe acne on the back. Acta Derm. Venereol. 2018, 98, 262–267. [Google Scholar] [CrossRef] [PubMed]
- McDowell, A. Over a Decade of recA and tly Gene Sequence Typing of the Skin Bacterium Propionibacterium acnes: What Have We Learnt? Microorganisms 2017, 6, 1. [Google Scholar] [CrossRef]
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.-H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Investig. Dermatol. 2013, 133, 2152–2160. [Google Scholar] [CrossRef]
- Corvec, S.; Dagnelie, M.-A.; Khammari, A.; Dréno, B. Taxonomy and phylogeny of Cutibacterium (formerly Propionibacterium) acnes in inflammatory skin diseases. Ann. Dermatol. Venereol. 2019, 146, 26–30. [Google Scholar] [CrossRef]
- Jahns, A.C.; Alexeyev, O.A. Three dimensional distribution of Propionibacterium acnes biofilms in human skin. Exp. Dermatol. 2014, 23, 687–689. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mitra, R.; Maitra, A.; Gupta, S.; Kumaran, S.; Chakrabortty, A.; Majumder, P.P. Sebum and Hydration Levels in Specific Regions of Human Face Significantly Predict the Nature and Diversity of Facial Skin Microbiome. Sci. Rep. 2016, 6, 36062. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chiang, H.-I.; Jiang, S.B.; Nagarajan, H.; Zengler, K.; Gallo, R.L. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 2013, 4, 1431. [Google Scholar] [CrossRef]
- Percoco, G. Methods and results in 3D skin biopsies. In Proceedings of the The Microbiome of the Skin-New Avenues of Research, in cosmetics, Amsterdam, The Netherlands, 17 April 2018. [Google Scholar]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Brüggemann, H. Skin: Acne and Propionibacterium acnes Genomics. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3215–3225. ISBN 978-3-540-77587-4. [Google Scholar]
- Arrecubieta, C.; Lee, M.-H.; Macey, A.; Foster, T.J.; Lowy, F.D. SdrF, a Staphylococcus epidermidis Surface Protein, Binds Type I Collagen. J. Biol. Chem. 2007, 282, 18767–18776. [Google Scholar] [CrossRef] [PubMed]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.L.; Nakatsuji, T. Microbial Symbiosis with the Innate Immune Defense System of the Skin. J. Investig. Dermatol. 2011, 131, 1974–1980. [Google Scholar] [CrossRef]
- Wiesner, J.; Vilcinskas, A. Antimicrobial peptides: The ancient arm of the human immune system. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef]
- Claudel, J.P.; Auffret, N.; Leccia, M.T.; Poli, F.; Corvec, S.; Dréno, B. Staphylococcus epidermidis: A Potential New Player in the Physiopathology of Acne? Dermatology 2019, 235, 287–294. [Google Scholar] [CrossRef]
- O’Neill, A.M.; Gallo, R.L. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 2018, 6, 177. [Google Scholar] [CrossRef]
- Christensen, G.J.M.; Brüggemann, H. Bacterial skin commensals and their role as host guardians. Benef. Microbes 2014, 5, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Kies, S.; Vuong, C.; Hille, M.; Peschel, A.; Meyer, C.; Götz, F.; Otto, M. Control of antimicrobial peptide synthesis by the agr quorum sensing system in Staphylococcus epidermidis: Activity of the lantibiotic epidermin is regulated at the level of precursor peptide processing. Peptides 2003, 24, 329–338. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Vandecandelaere, I.; Depuydt, P.; Nelis, H.J.; Coenye, T. Protease production by Staphylococcus epidermidis and its effect on Staphylococcus aureus biofilms. Pathog. Dis. 2014, 70, 321–331. [Google Scholar] [CrossRef]
- Cogen, A.L.; Yamasaki, K.; Muto, J.; Sanchez, K.M.; Crotty Alexander, L.; Tanios, J.; Lai, Y.; Kim, J.E.; Nizet, V.; Gallo, R.L. Staphylococcus epidermidis Antimicrobial δ-Toxin (Phenol-Soluble Modulin-γ) Cooperates with Host Antimicrobial Peptides to Kill Group A Streptococcus. PLoS ONE 2010, 5, e8557. [Google Scholar] [CrossRef]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; MacLeod, D.T.; Leichtle, A.; Ryan, A.F.; Di Nardo, A.; Gallo, R.L. Activation of TLR2 by a Small Molecule Produced by Staphylococcus epidermidis Increases Antimicrobial Defense against Bacterial Skin Infections. J. Investig. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef]
- Shu, M.; Wang, Y.; Yu, J.; Kuo, S.; Coda, A.; Jiang, Y.; Gallo, R.L.; Huang, C.M. Fermentation of Propionibacterium acnes, a Commensal Bacterium in the Human Skin Microbiome, as Skin Probiotics against Methicillin-Resistant Staphylococcus aureus. PLoS ONE 2013, 8, e55380. [Google Scholar] [CrossRef]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.-M. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: Implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Götz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin Commensals Amplify the Innate Immune Response to Pathogens by Activation of Distinct Signaling Pathways. J. Investig. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef]
- Percoco, G.; Merle, C.; Jaouen, T.; Ramdani, Y.; Bénard, M.; Hillion, M.; Mijouin, L.; Lati, E.; Feuilloley, M.G.J.; Lefeuvre, L.; et al. Antimicrobial peptides and pro-inflammatory cytokines are differentially regulated across epidermal layers following bacterial stimuli. Exp. Dermatol. 2013, 22, 800–806. [Google Scholar] [CrossRef]
- Yuki, T.; Yoshida, H.; Akazawa, Y.; Komiya, A.; Sugiyama, Y.; Inoue, S. Activation of TLR2 enhances tight junction barrier in epidermal keratinocytes. J. Immunol. 2011, 187, 3230–3237. [Google Scholar] [CrossRef]
- Lai, Y.; Di Nardo, A.; Nakatsuji, T.; Leichtle, A.; Yang, Y.; Cogen, A.L.; Wu, Z.R.; Hooper, L.V.; Schmidt, R.R.; Von Aulock, S.; et al. Commensal bacteria regulate toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 2009, 15, 1377–1382. [Google Scholar] [CrossRef]
- Wang, Z.; MacLeod, D.T.; Di Nardo, A. Commensal Bacteria Lipoteichoic Acid Increases Skin Mast Cell Antimicrobial Activity against Vaccinia Viruses. J. Immunol. 2012, 189, 1551–1558. [Google Scholar] [CrossRef]
- Xia, X.; Li, Z.; Liu, K.; Wu, Y.; Jiang, D.; Lai, Y. Staphylococcal LTA-induced miR-143 inhibits Propionibacterium acnes-Mediated inflammatory response in skin. J. Investig. Dermatol. 2016, 136, 621–630. [Google Scholar] [CrossRef]
- Wang, Y.; Kao, M.S.; Yu, J.; Huang, S.; Marito, S.; Gallo, R.L.; Huang, C.M. A precision microbiome approach using sucrose for selective augmentation of Staphylococcus epidermidis fermentation against Propionibacterium acnes. Int. J. Mol. Sci. 2016, 17, 1870. [Google Scholar] [CrossRef]
- Skabytska, Y.; Biedermann, T. Staphylococcus epidermidis Sets Things Right Again. J. Investig. Dermatol. 2016, 136, 559–560. [Google Scholar] [CrossRef]
- Dréno, B.; Martin, R.; Moyal, D.; Henley, J.B.; Khammari, A.; Seité, S. Skin microbiome and acne vulgaris: Staphylococcus, a new actor in acne. Exp. Dermatol. 2017, 26, 798–803. [Google Scholar] [CrossRef]
- Brandwein, M.; Steinberg, D.; Meshner, S. Microbial biofilms and the human skin microbiome. NPJ Biofilms Microbiomes 2016, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, T.; Biagini Myers, J.M.; Herr, A.B.; Khurana Hershey, G.K. Staphylococcal Biofilms in Atopic Dermatitis. Curr. Allergy Asthma Rep. 2017, 17, 81. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.B.; Mueller, J.L. A novel finding in atopic dermatitis: Film-producing Staphylococcus epidermidis as an etiology. Int. J. Dermatol. 2011, 50, 992–993. [Google Scholar] [CrossRef]
- Clausen, M.L.; Agner, T.; Lilje, B.; Edslev, S.M.; Johannesen, T.B.; Andersen, P.S. Association of disease severity with skin microbiome and filaggrin gene mutations in adult atopic dermatitis. JAMA Dermatol. 2018, 154, 293–300. [Google Scholar] [CrossRef]
- David Boothe, W.; Tarbox, J.A.; Tarbox, M.B. Atopic Dermatitis: Pathophysiology. Adv. Exp. Med. Biol. 2017, 1027, 21–37. [Google Scholar]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.I.; Conlan, S.; Belkaid, Y.; Segre, J.A.; Kong, H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017, 9, eaal4651. [Google Scholar] [CrossRef]
- Costerton, W.; Veeh, R.; Shirtliff, M.; Pasmore, M.; Post, C.; Ehrlich, G. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 2003, 112, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.B.; Vaze, N.D.; Choi, C.; Hailu, T.; Tulbert, B.H.; Cusack, C.A.; Joshi, S.G. The presence and impact of biofilm-producing staphylococci in atopic dermatitis. JAMA Dermatol. 2014, 150, 260–265. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar]
- Cheung, G.Y.C.; Joo, H.S.; Chatterjee, S.S.; Otto, M. Phenol-soluble modulins-critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [Google Scholar] [CrossRef]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef]
- O’Gara, J.P.; Humphreys, H. Staphylococcus epidermidis biofilms: Importance and implications. J. Med. Microbiol. 2001, 50, 582–587. [Google Scholar] [CrossRef]
- Mekni, M.A.; Bouchami, O.; Achour, W.; Ben Hassen, A. Strong biofilm production but not adhesion virulence factors can discriminate between invasive and commensal Staphylococcus epidermidis strains. APMIS 2012, 120, 605–611. [Google Scholar] [CrossRef]
- Heilmann, C. Adhesion Mechanisms of Staphylococci. Adv. Exp. Med. Biol. 2011, 715, 105–123. [Google Scholar] [PubMed]
- Vuong, C.; Gerke, C.; Somerville, G.A.; Fischer, E.R.; Otto, M. Quorum-Sensing Control of Biofilm Factors in Staphylococcus epidermidis. J. Infect. Dis. 2003, 188, 706–718. [Google Scholar] [CrossRef]
- Mack, D.; Davies, A.P.; Harris, L.G.; Rohde, H.; Horstkotte, M.A.; Knobloch, J.K.-M. Microbial interactions in Staphylococcus epidermidis biofilms. Anal. Bioanal. Chem. 2007, 387, 399–408. [Google Scholar] [CrossRef]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci—An overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef]
- Downing, D.T.; Stewart, M.E.; Wertz, P.W.; Strauss, J.S. Essential fatty acids and acne. J. Am. Acad. Dermatol. 1986, 14, 221–225. [Google Scholar] [CrossRef]
- Zouboulis, C.C. Acne and sebaceous gland function. Clin. Dermatol. 2004, 22, 360–366. [Google Scholar] [CrossRef]
- Ottaviani, M.; Alestas, T.; Flori, E.; Mastrofrancesco, A.; Zouboulis, C.C.; Picardo, M. Peroxidated squalene induces the production of inflammatory mediators in HaCaT keratinocytes: A possible role in acne vulgaris. J. Investig. Dermatol. 2006, 126, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Jahns, A.C.; Lundskog, B.; Ganceviciene, R.; Palmer, R.H.; Golovleva, I.; Zouboulis, C.C.; McDowell, A.; Patrick, S.; Alexeyev, O.A. An increased incidence of Propionibacterium acnes biofilms in acne vulgaris: A case-control study. Br. J. Dermatol. 2012, 167, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B. What is new in the pathophysiology of acne, an overview. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 8–12. [Google Scholar] [CrossRef]
- Dagnelie, M.-A.; Corvec, S.; Saint-Jean, M.; Bourdès, V.; Nguyen, J.-M.; Khammari, A.; Dréno, B. La sévérité de l’acné est associée à une perte de la diversité des souches de Propionibacterium acnes en peau acnéique. Ann. Dermatol. Venereol. 2017, 144, S130. [Google Scholar] [CrossRef]
- Dagnelie, M.-A.; Corvec, S.; Saint-Jean, M.; Nguyen, J.M.; Khammari, A.; Dréno, B. Cutibacterium acnes phylotypes diversity loss: A trigger for skin inflammatory process. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 2340–2348. [Google Scholar] [CrossRef] [PubMed]
- Paugam, C.; Corvec, S.; Saint-Jean, M.; Le Moigne, M.; Khammari, A.; Boisrobert, A.; Nguyen, J.M.; Gaultier, A.; Dréno, B. Propionibacterium acnes phylotypes and acne severity: An observational prospective study. J. Eur. Acad. Dermatol. Venereol. 2017, 31, e398–e399. [Google Scholar] [CrossRef]
- Higaki, S.; Kitagawa, T.; Kagoura, M.; Morohashi, M.; Yamagishi, T. Correlation between Propionibacterium acnes Biotypes, Lipase Activity and Rash Degree in Acne Patients. J. Dermatol. 2000, 27, 519–522. [Google Scholar] [CrossRef]
- Oyewole, A.O.; Birch-Machin, M.A. Sebum, inflammasomes and the skin: Current concepts and future perspective. Exp. Dermatol. 2015, 24, 651–654. [Google Scholar] [CrossRef]
- Shu, M.; Kuo, S.; Wang, Y.; Jiang, Y.; Liu, Y.-T.; Gallo, R.L.; Huang, C.-M. Porphyrin Metabolisms in Human Skin Commensal Propionibacterium acnes Bacteria: Potential Application to Monitor Human Radiation Risk. Curr. Med. Chem. 2013, 20, 562–568. [Google Scholar]
- Johnson, T.; Kang, D.; Barnard, E.; Li, H. Strain-Level Differences in Porphyrin Production and Regulation in Propionibacterium acnes Elucidate Disease Associations. mSphere 2016, 1, e00023-15. [Google Scholar] [CrossRef] [PubMed]
- Nazipi, S.; Stødkilde, K.; Scavenius, C.; Brüggemann, H. The Skin Bacterium Propionibacterium acnes Employs Two Variants of Hyaluronate Lyase with Distinct Properties. Microorganisms 2017, 5, 57. [Google Scholar] [CrossRef]
- Dréno, B.; Pécastaings, S.; Corvec, S.; Veraldi, S.; Khammari, A.; Roques, C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: A brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 5–14. [Google Scholar] [CrossRef]
- Liu, P.F.; Nakatsuji, T.; Zhu, W.; Gallo, R.L.; Huang, C.M. Passive immunoprotection targeting a secreted CAMP factor of Propionibacterium acnes as a novel immunotherapeutic for acne vulgaris. Vaccine 2011, 29, 3230–3238. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Tang, D.C.; Zhang, L.; Gallo, R.L.; Huang, C.M. Propionibacterium acnes CAMP factor and host acid sphingomyelinase contribute to bacterial virulence: Potential targets for inflammatory acne treatment. PLoS ONE 2011, 6, e14797. [Google Scholar] [CrossRef]
- Beylot, C.; Auffret, N.; Poli, F.; Claudel, J.P.; Leccia, M.T.; Del Giudice, P.; Dréno, B. Propionibacterium acnes: An update on its role in the pathogenesis of acne. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 271–278. [Google Scholar] [CrossRef]
- Borrel, V.; Gannesen, A.V.; Barreau, M.; Gaviard, C.; Duclairoir-Poc, C.; Hardouin, J.; Konto-Ghiorghi, Y.; Lefeuvre, L.; Feuilloley, M.G.J. Adaptation of acneic and non acneic strains of Cutibacterium acnes to sebum-like environment. Microbiologyopen 2019, 8, e00841. [Google Scholar] [CrossRef]
- Coenye, T.; Peeters, E.; Nelis, H.J. Biofilm formation by Propionibacterium acnes is associated with increased resistance to antimicrobial agents and increased production of putative virulence factors. Res. Microbiol. 2007, 158, 386–392. [Google Scholar] [CrossRef]
- Gannesen, A.V.; Lesouhaitier, O.; Racine, P.-J.; Barreau, M.; Netrusov, A.I.; Plakunov, V.K.; Feuilloley, M.G.J. Regulation of Monospecies and Mixed Biofilms Formation of Skin Staphylococcus aureus and Cutibacterium acnes by Human Natriuretic Peptides. Front. Microbiol. 2018, 9, 2912. [Google Scholar] [CrossRef]
- Achermann, Y.; Goldstein, E.J.C.; Coenye, T.; Shirtliffa, M.E. Propionibacterium acnes: From Commensal to Opportunistic Biofilm-Associated Implant Pathogen. Clin. Microbiol. Rev. 2014, 27, 419–440. [Google Scholar] [CrossRef]
- Holmberg, A.; Lood, R.; Mörgelin, M.; Söderquist, B.; Holst, E.; Collin, M.; Christensson, B.; Rasmussen, M. Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin. Microbiol. Infect. 2009, 15, 787–795. [Google Scholar] [CrossRef]
- Ionescu, M.-A.A.; Feuilloley, M.G.J.; Enault, J.; Saguet, T.; Robert, G.; Lefeuvre, L. La modulation du microbiofilm et du P. acnes ribotypes 4 et 5 dans l’acné polymorphe: Étude microbiologique in vitro et essai clinique dans une série de 70 cas. Ann. Dermatol. Venereol. 2015, 142, S434–S435. [Google Scholar] [CrossRef]
- Kuehnast, T.; Cakar, F.; Weinhäupl, T.; Pilz, A.; Selak, S.; Schmidt, M.A.; Rüter, C.; Schild, S. Comparative analyses of biofilm formation among different Cutibacterium acnes isolates. Int. J. Med. Microbiol. 2018, 308, 1027–1035. [Google Scholar] [CrossRef]
- Burkhart, C.G.; Burkhart, C.N. Expanding the microcomedone theory and acne therapeutics: Propionibacterium acnes biofilm produces biological glue that holds corneocytes together to form plug. J. Am. Acad. Dermatol. 2007, 57, 722–724. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.B.; Byun, E.J.; Kim, H.S. Potential Role of the Microbiome in Acne: A Comprehensive Review. J. Clin. Med. 2019, 8, 987. [Google Scholar] [CrossRef]
- Choi, J.Y.; Piao, M.S.; Lee, J.B.; Oh, J.S.; Kim, I.G.; Lee, S.C. Propionibacterium acnes stimulates pro-matrix metalloproteinase-2 expression through tumor necrosis factor-α in human dermal fibroblasts. J. Investig. Dermatol. 2008, 128, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Jugeau, S.; Tenaud, I.; Knol, A.C.; Jarrousse, V.; Quereux, G.; Khammari, A.; Dréno, B. Induction of toll-like receptors by Propionibacterium acnes. Br. J. Dermatol. 2005, 153, 1105–1113. [Google Scholar] [CrossRef]
- Nagy, I.; Pivarcsi, A.; Kis, K.; Koreck, A.; Bodai, L.; McDowell, A.; Seltmann, H.; Patrick, S.; Zouboulis, C.C.; Kemény, L. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 2006, 8, 2195–2205. [Google Scholar] [CrossRef]
- Jarrousse, V.; Castex-Rizzi, N.; Khammari, A.; Charveron, M.; Dréno, B. Modulation of integrins and filaggrin expression by Propionibacterium acnes extracts on keratinocytes. Arch. Dermatol. Res. 2007, 299, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Akaza, N.; Akamatsu, H.; Kishi, M.; Mizutani, H.; Ishii, I.; Nakata, S.; Matsunaga, K. Effects of Propionibacterium acnes on various mRNA expression levels in normal human epidermal keratinocytes in vitro. J. Dermatol. 2009, 36, 213–223. [Google Scholar] [CrossRef]
- Feuilloley, M.G.J. Antidromic neurogenic activity and cutaneous bacterial flora. Semin. Immunopathol. 2018, 50, 281–289. [Google Scholar] [CrossRef]
- Roosterman, D.; Goerge, T.; Schneider, S.W.; Bunnett, N.W.; Steinhoff, M. Neuronal Control of Skin Function: The Skin as a Neuroimmunoendocrine Organ. Physiol. Rev. 2006, 86, 1309–1379. [Google Scholar] [CrossRef]
- Racine, P.; Janvier, X.; Clabaut, M.; Catovic, C.; Souak, D.; Boukerb, A.M.; Groboillot, A.; Konto-Ghiorghi, Y.; Duclairoir-Poc, C.; Lesouhaitier, O.; et al. Dialog between skin and its microbiota: Emergence of “Cutaneous Bacterial Endocrinology”. Exp. Dermatol. 2020, 29, exd.14158. [Google Scholar] [CrossRef]
- Choi, J.E.; Di Nardo, A. Skin Neurogenic inflammation. Semin. Immunopathol. 2018, 40, 249–259. [Google Scholar] [CrossRef]
- Lesouhaitier, O.; Clamens, T.; Rosay, T.; Desriac, F.; Louis, M.; Rodrigues, S.; Gannesen, A.V.; Plakunov, V.K.; Bouffartigues, E.; Tahrioui, A.; et al. Host Peptidic Hormones Affecting Bacterial Biofilm Formation and Virulence. J. Innate Immun. 2019, 11, 227–241. [Google Scholar] [CrossRef]
- Maksimovic, S.; Baba, Y.; Lumpkin, E.A. Neurotransmitters and synaptic components in the Merkel cell-neurite complex, a gentle-touch receptor. Ann. N. Y. Acad. Sci. 2013, 1279, 13–21. [Google Scholar] [CrossRef]
- Kähler, C.M.; Sitte, B.A.; Reinisch, N.; Wiedermann, C.J. Stimulation of the chemotactic migration of human fibroblasts by substance P. Eur. J. Pharmacol. 1993, 249, 281–286. [Google Scholar] [CrossRef]
- Mijouin, L.; Hillion, M.; Ramdani, Y.; Jaouen, T.; Duclairoir-Poc, C.; Follet-Gueye, M.-L.; Lati, E.; Yvergnaux, F.; Driouich, A.; Lefeuvre, L.; et al. Effects of a Skin Neuropeptide (Substance P) on Cutaneous Microflora. PLoS ONE 2013, 8, e78773. [Google Scholar] [CrossRef]
- N’Diaye, A.; Mijouin, L.; Hillion, M.; Diaz, S.; Konto-Ghiorghi, Y.; Percoco, G.; Chevalier, S.; Lefeuvre, L.; Harmer, N.J.; Lesouhaitier, O.; et al. Effect of Substance P in Staphylococcus aureus and Staphylococcus epidermidis virulence: Implication for skin homeostasis. Front. Microbiol. 2016, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- N’Diaye, A.; Borrel, V.; Racine, P.J.; Clamens, T.; Depayras, S.; Maillot, O.; Schaack, B.; Chevalier, S.; Lesouhaitier, O.; Feuilloley, M.G.J. Mechanism of action of the moonlighting protein EfTu as a Substance P sensor in Bacillus cereus. Sci. Rep. 2019, 9, 1304. [Google Scholar] [CrossRef]
- N’Diaye, A.; Leclerc, C.; Kentache, T.; Hardouin, J.; Poc, C.D.; Konto-Ghiorghi, Y.; Chevalier, S.; Lesouhaitier, O.; Feuilloley, M.G.J. Skin-bacteria communication: Involvement of the neurohormone Calcitonin Gene Related Peptide (CGRP) in the regulation of Staphylococcus epidermidis virulence. Sci. Rep. 2016, 6, 35379. [Google Scholar] [CrossRef]
- N’Diaye, A.; Gannesen, A.V.; Borrel, V.; Maillot, O.; Enault, J.; Racine, P.J.; Plakunov, V.; Chevalier, S.; Lesouhaitier, O.; Feuilloley, M.G.J. Substance P and calcitonin gene-related peptide: Key regulators of cutaneous microbiota homeostasis. Front. Endocrinol. 2017, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Borrel, V.; Thomas, P.; Catovic, C.; Racine, P.-J.; Konto-Ghiorghi, Y.; Lefeuvre, L.; Duclairoir-Poc, C.; Zouboulis, C.C.; Feuilloley, M.G.J. Acne and Stress: Impact of Catecholamines on Cutibacterium acnes. Front. Med. 2019, 6, 155. [Google Scholar] [CrossRef]
- Potter, L.R.; Yoder, A.R.; Flora, D.R.; Antos, L.K.; Dickey, D.M. Natriuretic peptides: Their structures, receptors, physiologic functions and therapeutic applications. In Handbook of Experimental Pharmacology; Schmidt, H.H.H.W., Hofmann, F., Stasch, J.-P., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 191, pp. 341–366. ISBN 9783540689607. [Google Scholar]
- Gannesen, A.V.; Lesouhaitier, O.; Netrusov, A.I.; Plakunov, V.K.; Feuilloley, M.G.J. Regulation of Formation of Monospecies and Binary Biofilms by Human Skin Microbiota Components, Staphylococcus epidermidis and Staphylococcus aureus, by Human Natriuretic Peptides. Microbiology 2018, 87, 597–609. [Google Scholar] [CrossRef]
- Rouaud-Tinguely, P.; Jugé, R.; Mainzer, C.; Boudier, D.; Roth, M.; Coppin, H.; Closs, B. Analysis of cutaneous microbiota between two age-group of Caucasian women. J. Investig. Dermatol. 2018, 138, S171. [Google Scholar] [CrossRef]
- Shibagaki, N.; Suda, W.; Clavaud, C.; Bastien, P.; Takayasu, L.; Iioka, E.; Kurokawa, R.; Yamashita, N.; Hattori, Y.; Shindo, C.; et al. Aging-related changes in the diversity of women’s skin microbiomes associated with oral bacteria. Sci. Rep. 2017, 7, 10567. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, W.; Shu, M.; Jiang, Y.; Gallo, R.L.; Liu, Y.-T.; Huang, C.-M. The response of human skin commensal bacteria as a reflection of UV radiation: UV-B decreases porphyrin production. PLoS ONE 2012, 7, e47798. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Choi, J.E.; Wu, C.C.; Di Nardo, A. Skin commensal bacteria Staphylococcus epidermidis promote survival of melanocytes bearing UVB-induced DNA damage, while bacteria Propionibacterium acnes inhibit survival of melanocytes by increasing apoptosis. Photodermatol. Photoimmunol. Photomed. 2018, 34, 405–414. [Google Scholar] [CrossRef]
- Christensen, G.J.M.; Scholz, C.F.P.; Enghild, J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genom. 2016, 17, 152. [Google Scholar] [CrossRef]
- Jo, J.H.; Kennedy, E.A.; Kong, H.H. Research Techniques Made Simple: Bacterial 16S Ribosomal RNA Gene Sequencing in Cutaneous Research. J. Investig. Dermatol. 2016, 136, e23–e27. [Google Scholar] [CrossRef]
- Kong, H.H.; Andersson, B.; Clavel, T.; Common, J.E.; Jackson, S.A.; Olson, N.D.; Segre, J.A.; Traidl-Hoffmann, C. Performing Skin Microbiome Research: A Method to the Madness. J. Investig. Dermatol. 2017, 137, 561–568. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Van Horn, K.G.; Audette, C.D.; Tucker, K.A.; Sebeck, D. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diagn. Microbiol. Infect. Dis. 2008, 62, 471–473. [Google Scholar] [CrossRef]
- Lange-Asschenfeldt, B.; Marenbach, D.; Lang, C.; Patzelt, A.; Ulrich, M.; Maltusch, A.; Terhorst, D.; Stockfleth, E.; Sterry, W.; Lademann, J. Distribution of bacteria in the epidermal layers and hair follicles of the human skin. Skin Pharmacol. Physiol. 2011, 24, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Ogai, K.; Nagase, S.; Mukai, K.; Iuchi, T.; Mori, Y.; Matsue, M.; Sugitani, K.; Sugama, J.; Okamoto, S. A Comparison of Techniques for Collecting Skin Microbiome Samples: Swabbing Versus Tape-Stripping. Front. Microbiol. 2018, 9, 2362. [Google Scholar] [CrossRef]
- Prast-Nielsen, S.; Tobin, A.-M.; Adamzik, K.; Powles, A.; Hugerth, L.W.; Sweeney, C.; Kirby, B.; Engstrand, L.; Fry, L. Investigation of the skin microbiome: Swabs vs. biopsies. Br. J. Dermatol. 2019, 181, 572–579. [Google Scholar]
- Cangelosi, G.A.; Meschke, J.S. Dead or alive: Molecular assessment of microbial viability. Appl. Environ. Microbiol. 2014, 80, 5884–5891. [Google Scholar] [CrossRef]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of metatranscriptomics in microbiome research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef]
- Meisel, J.S.; Hannigan, G.D.; Tyldsley, A.S.; SanMiguel, A.J.; Hodkinson, B.P.; Zheng, Q.; Grice, E.A. Skin microbiome surveys are strongly influenced by experimental design. J. Investig. Dermatol. 2016, 136, 947–956. [Google Scholar] [CrossRef]
- Feuilloley, M.G.J. Skin microbiota: Variability and new perspectives. In Proceedings of the The Microbiome of the Skin-New Avenues of Research, in Cosmetics, Amsterdam, The Netherlands, 17 April 2018. [Google Scholar]
- Krutmann, J. Pre- and probiotics for human skin. J. Dermatol. Sci. 2009, 54, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef]
- Jahns, A.C.; Oprica, C.; Vassilaki, I.; Golovleva, I.; Palmer, R.H.; Alexeyev, O.A. Simultaneous visualization of Propionibacterium acnes and Propionibacterium granulosum with immunofluorescence and fluorescence in situ hybridization. Anaerobe 2013, 23, 48–54. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef]
- Coenye, T.; Nelis, H.J. In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Methods 2010, 83, 89–105. [Google Scholar] [CrossRef]
- Gannesen, A.V.; Zdorovenko, E.L.; Botchkova, E.A.; Hardouin, J.; Massier, S.; Kopitsyn, D.S.; Gorbachevskii, M.V.; Kadykova, A.A.; Shashkov, A.S.; Zhurina, M.V.; et al. Composition of the biofilm matrix of Cutibacterium acnes acneic strain RT5. Front. Microbiol. 2019, 10, 1284. [Google Scholar] [CrossRef]
- Parlement Européen et du Conseil. Règlement (CE) No 1223/2009 du PARLEMENT Européen et du Conseil du 30 Novembre 2009 Relatif Aux Produits Cosmétiques; Office des publications de l’Union européenne: Luxembourg, 2009. [Google Scholar]
- Vogelgesang, B. (BASF Beauty Care Solutions France) Recent advances and new strategies to substantiate microbiota-related cosmetic claims. In Proceedings of the The Microbiome of the Skin-New Avenues of Research, in Cosmetics, Amsterdam, The Netherlands, 17 April 2018. [Google Scholar]
- Krieken, D.; Ederveen, T.; Hijum, S.; Jansen, P.; Melchers, W.; Scheepers, P.; Schalkwijk, J.; Zeeuwen, P. An in vitro model for bacterial growth on human stratum corneum. Acta Derm. Venereol. 2016, 96, 873–879. [Google Scholar] [CrossRef]
- Niehues, H.; Bouwstra, J.A.; El Ghalbzouri, A.; Brandner, J.M.; Zeeuwen, P.L.J.M.; van den Bogaard, E.H. 3D skin models for 3R research: The potential of 3D reconstructed skin models to study skin barrier function. Exp. Dermatol. 2018, 27, 501–511. [Google Scholar] [CrossRef]
- Cadau, S.; Valla-Dury, L.; Cenizo, V.; Le-Beux, C.; Rival, D.; Gault, M.; Vianney, A.; Andre-Frei, V. Studying Microbiote Competition and Skin Interaction Using Organotypic 3D Skin Models. Adv. Tissue Eng. Regen. Med. Open Access 2017, 2, 5. [Google Scholar]
- Holland, D.B.; Bojar, R.A.; Jeremy, A.H.T.; Ingham, E.; Holland, K.T. Microbial colonization of an in vitro model of a tissue engineered human skin equivalent—A novel approach. FEMS Microbiol. Lett. 2008, 279, 110–115. [Google Scholar] [CrossRef]
- Duckney, P.; Wong, H.K.; Serrano, J.; Yaradou, D.; Oddos, T.; Stamatas, G.N. The role of the skin barrier in modulating the effects of common skin microbial species on the inflammation, differentiation and proliferation status of epidermal keratinocytes. BMC Res. Notes 2013, 6, 474. [Google Scholar] [CrossRef]
- Bojar, R.A. Studying the human skin microbiome using 3D in vitro skin models. Appl. In Vitro Toxicol. 2015, 1, 165–171. [Google Scholar] [CrossRef]
- Landemaine, L.; Cenizo, V.; Lemaire, G.; Portes, P. Colonization of a 3D skin model with a complete microbiota is more beneficial to the skin barrier than with Staphylococcus epidermidis alone. J. Investig. Dermatol. 2018, 138, S163. [Google Scholar] [CrossRef]
- Rademacher, F.; Simanski, M.; Gläser, R.; Harder, J. Skin microbiota and human 3D skin models. Exp. Dermatol. 2018, 27, 489–494. [Google Scholar] [CrossRef]
- Kuhbacher, A.; Henkel, H.; Stevens, P.; Grumaz, C.; Finkelmeier, D.; Burger-Kentischer, A.; Sohn, K.; Rupp, S. Dermal Fibroblasts Play a Central Role in Skin Model Protection against Candida albicans Invasion. J. Infect. Dis. 2017, 215, 1742–1752. [Google Scholar] [CrossRef]
- Holland, K.T.; Bojar, R.A. Cosmetics: What is their influence on the skin microflora? Am. J. Clin. Dermatol. 2002, 3, 445–449. [Google Scholar] [CrossRef]
- Wang, Q.; Cui, S.; Zhou, L.; He, K.; Song, L.; Liang, H.; He, C. Effect of cosmetic chemical preservatives on resident flora isolated from healthy facial skin. J. Cosmet. Dermatol. 2019, 18, 652–658. [Google Scholar] [CrossRef]
- Staudinger, T.; Pipal, A.; Redl, B. Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make-up. J. Appl. Microbiol. 2011, 110, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jeong, S.E.; Lee, S.; Kim, S.; Han, H.; Jeon, C.O. Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. Microbiologyopen 2017, 7, e00557. [Google Scholar] [CrossRef]
- Dobler, D.; Schmidts, T.; Wildenhain, S.; Seewald, I.; Merzhäuser, M.; Runkel, F. Impact of Selected Cosmetic Ingredients on Common Microorganisms of Healthy Human Skin. Cosmetics 2019, 6, 45. [Google Scholar] [CrossRef]
- Callewaert, C.; Hutapea, P.; Van de Wiele, T.; Boon, N. Deodorants and antiperspirants affect the axillary bacterial community. Arch. Dermatol. Res. 2014, 306, 701–710. [Google Scholar] [CrossRef]
- Wallen-Russell, C. The Role of Every-Day Cosmetics in Altering the Skin Microbiome: A Study Using Biodiversity. Cosmetics 2018, 6, 2. [Google Scholar] [CrossRef]
- Lopes, E.G.; Moreira, D.A.; Gullón, P.; Gullón, B.; Cardelle-Cobas, A.; Tavaria, F.K. Topical application of probiotics in skin: Adhesion, antimicrobial and antibiofilm in vitro assays. J. Appl. Microbiol. 2017, 122, 450–461. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Faseleh Jahromi, M.; Ho, Y.W. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. BioMed Res. Int. Int. 2014, 2014, 927268. [Google Scholar] [CrossRef]
- Khmaladze, I.; Butler, É.; Fabre, S.; Gillbro, J.M. Lactobacillus reuteri DSM 17938—A comparative study on the effect of probiotics and lysates on human skin. Exp. Dermatol. 2019, 28, 822–828. [Google Scholar] [CrossRef]
- Hillion, M. Interactions peau/microbiote cutané: Étude du microbiote cutané cultivable et influence de produits cosmétiques sur la virulence bactérienne. Apports de la technique de spectrométrie de masse MALDI-TOF. Ph.D. Thesis, Thèse de l’Université de Rouen, Évreux, France, 2013. [Google Scholar]
- Baldwin, H.E.; Bhatia, N.D.; Friedman, A.; Eng, R.M.; Seite, S. The role of cutaneous microbiota harmony in maintaining a functional skin barrier. J. Drugs Dermatol. 2017, 16, 12–18. [Google Scholar] [CrossRef]
- Seite, S.; Misery, L. Skin sensitivity and skin microbiota: Is there a link? Exp. Dermatol. 2018, 27, 1061–1064. [Google Scholar] [CrossRef]
- Cinque, B.; Palumbo, P.; La Torre, C.; Melchiorre, E.; Corridoni, D.; Miconi, G.; Di Marzio, L.; Cifone, M.G.; Giuliani, M. Probiotics in aging skin. In Textbook of Aging Skin; Farage, M.A., Miller, K.W., Howard I, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 811–820. ISBN 9783540896555. [Google Scholar]
- BASF Beauty Care Solutions France. RELIPIDIUM® BC10096. Available online: https://www.carecreations.basf.com/product-formulations/products/products-detail/RELIPIDIUMBC10096/307139450 (accessed on 23 March 2020).
- Paetzold, B.; Willis, J.R.; Pereira De Lima, J.; Knödlseder, N.; Brüggemann, H.; Quist, S.R.; Gabaldón, T.; Güell, M. Skin microbiome modulation induced by probiotic solutions. Microbiome 2019, 7, 95. [Google Scholar] [CrossRef]
- Solabia Group. Ecoskin. Available online: http://www.solabia.com/Produto_29 (accessed on 8 March 2020).
- Maquart, F.; Bellon, G.; Marchal, C.; Ducatel, H.; Dupuis, O.; Picton, L.; Lecerf, D.; Forbice, R. Method for Producing a Mixture of Neutral Oligosaccharides Extracted from Flaxseed. Patent WO2014174221A1, 13 June 2017. [Google Scholar]
- BASF Beauty Care Solutions France. OLIGOLIN® BC10028. Available online: https://www.carecreations.basf.com/product-formulations/products/products-detail/OLIGOLIN-BC10028/30666770 (accessed on 29 April 2020).
- BASF Beauty Care Solutions France. PROTEASYL® PW PSE LS 8951. Available online: https://www.carecreations.basf.com/product-formulations/product-highlights/product-highlights-detail/PROTEASYLPWPSELS8951/30531031 (accessed on 29 April 2020).
- De Jesus Raposo, M.F.; De Morais, A.M.M.B.; De Morais, R.M.S.C. Emergent sources of prebiotics: Seaweeds and microalgae. Mar. Drugs 2016, 14, 27. [Google Scholar] [CrossRef]
- Morvan, P.-Y.; Vallee, R. Evaluation of the Effects of Stressful Life on Human Skin Microbiota. Appl. Microbiol. 2018, 4, 1000140. [Google Scholar]
- CODIF International. EPS SEAPUR. Available online: http://www.codif-tn.com/principesactifs/eps-seapur/ (accessed on 27 February 2020).
- Misery, L.; Myon, E.; Martin, N.; Verrière, F.; Nocera, T.; Taieb, C. Sensitive skin in France: An epidemiological approach. Ann. Dermatol. Venereol. 2005, 132, 425–429. [Google Scholar] [CrossRef]
- Berardesca, E.; Farage, M.; Maibach, H. Sensitive skin: An overview. Int. J. Cosmet. Sci. 2013, 35, 2–8. [Google Scholar] [CrossRef]
- Farage, M.A.; Katsarou, A.; Maibach, H.I. Sensory, clinical and physiological factors in sensitive skin: A review. Contact Dermat. 2006, 55, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hillion, M.; Mijouin, L.; Jaouen, T.; Barreau, M.; Meunier, P.; Lefeuvre, L.; Lati, E.; Chevalier, S.; Feuilloley, M.G.J. Comparative study of normal and sensitive skin aerobic bacterial populations. Microbiologyopen 2013, 2, 953–961. [Google Scholar] [CrossRef]
- Filaire, E.; Vialleix, C.; Cadoret, J.-P.; Dreux, A.; Berthon, J.-Y. ExpoZen®: An Active Ingredient Modulating Reactive and Sensitive Skin Microbiota; Euro Cosmetics: Buchs, Switzerland, 2019. [Google Scholar]
- Nodake, Y.; Matsumoto, S.; Miura, R.; Honda, H.; Ishibashi, G.; Matsumoto, S.; Dekio, I.; Sakakibara, R. Pilot study on novel skin care method by augmentation with Staphylococcus epidermidis, an autologous skin microbe—A blinded randomized clinical trial. J. Dermatol. Sci. 2015, 79, 119–126. [Google Scholar] [CrossRef]
- Lew, L.-C.; Liong, M.-T. Bioactives from probiotics for dermal health: Functions and benefits. J. Appl. Microbiol. 2013, 114, 1241–1253. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Knobloch, J.K.M.; Bartscht, K.; Sabottke, A.; Rohde, H.; Feucht, H.H.; Mack, D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: Differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 2001, 183, 2624–2633. [Google Scholar] [CrossRef]
- Gannesen, A.V.; Borrel, V.; Lefeuvre, L.; Netrusov, A.I.; Plakunov, V.K.; Feuilloley, M.G.J. Effect of two cosmetic compounds on the growth, biofilm formation activity, and surface properties of acneic strains of Cutibacterium acnes and Staphylococcus aureus. Microbiologyopen 2018, 8, e00659. [Google Scholar] [CrossRef]
- Enault, J.; Saguet, T.; Yvergnaux, F.; Feuilloley, M.G.J. PS291®, a rhamnose-rich polysaccharide obtained by fermentation, is reducing Propionibacterium acnes adhesion and biofilm formation activity. In Proceedings of the International Federation of Societies of Cosmetic Chemists, Paris, France, 30 October 2014; pp. 2801–2810. [Google Scholar]
- Filaire, E.; Vialleix, C.; Cadoret, J.-P.; Guénard, S.; Muller, C.; Dreux-Zigha, A.; Berthon, J.-Y. Characterization of Reactive and Sensitive Skin Microbiota: Effect of Halymenia durvillei (HD) Extract Treatment. Cosmetics 2019, 6, 69. [Google Scholar] [CrossRef]
- Feuillolay, C.; Pecastaings, S.; Gac, C.L.; Fiorini-Puybaret, C.; Luc, J.; Joulia, P.; Roques, C. A Myrtus communis extract enriched in myrtucummulones and ursolic acid reduces resistance of Propionibacterium acnes biofilms to antibiotics used in acne vulgaris. Phytomedicine 2016, 23, 307–315. [Google Scholar] [CrossRef]
- Trompezinski, S.; Weber, S.; Cadars, B.; Larue, F.; Ardiet, N.; Chavagnac-Bonneville, M.; Sayag, M.; Jourdan, E. Assessment of a new biological complex efficacy on dysseborrhea, inflammation, and Propionibacterium acnes proliferation. Clin. Cosmet. Investig. Dermatol. 2016, 9, 233–239. [Google Scholar]
- Saising, J.; Voravuthikunchai, S.P. Anti Propionibacterium acnes activity of rhodomyrtone, an effective compound from Rhodomyrtus tomentosa (Aiton) Hassk. leaves. Anaerobe 2012, 18, 400–404. [Google Scholar] [CrossRef]
- Greentech. Acnilys®. Available online: https://www.greentech.fr/en/acnilys-2/ (accessed on 20 March 2020).
- Redoules, D.; Daunes-Marion, S.; Aries, M.-F. Polyunsaturated Fatty Acid and Diol Ester as an Anti-Acne Agent. Patent WO2010072738A1, 1 July 2010. [Google Scholar]
- Petigny, L.; Périno-Issartier, S.; Wajsman, J.; Chemat, F. Batch and Continuous Ultrasound Assisted Extraction of Boldo Leaves (Peumus boldus Mol.). Int. J. Mol. Sci 2013, 14, 5750–5764. [Google Scholar] [CrossRef]
- BASF Beauty Care Solutions France. BIX’ACTIV® BC10050. Available online: https://www.carecreations.basf.com/product-formulations/product-highlights/product-highlights-detail/BIX’ACTIVBC10050/30704818 (accessed on 23 March 2020).
- BASF Beauty Care Solutions France. BETAPUR®. Available online: https://www.carecreations.basf.com/product-formulations/products/products-detail/BETAPURA00067/30459417 (accessed on 29 April 2020).
- Wunnoo, S.; Saising, J.; Voravuthikunchai, S.P. Rhodomyrtone inhibits lipase production, biofilm formation, and disorganizes established biofilm in Propionibacterium acnes. Anaerobe 2017, 43, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Borrel, V. Influence du microenvironnement sur la virulence et la formation de biofilm de Cutibacterium acnes. Ph.D. Thesis, Thèse de l’Université de Rouen, Évreux, France, 2019. [Google Scholar]
- Zeichner, J.A. The use of lipohydroxy acid in skin care and acne treatment. J. Clin. Aesthet. Dermatol. 2016, 9, 40–43. [Google Scholar] [CrossRef]
- Choi, J.S.; Bae, H.J.; Kim, S.J.; Choi, I.S. In vitro antibacterial and anti-inflammatory properties of seaweed extracts against acne inducing bacteria, Propionibacterium acnes. J. Environ. Biol. 2011, 32, 313–318. [Google Scholar] [PubMed]
- Kamei, Y.; Sueyoshi, M.; Hayashi, K.I.; Terada, R.; Nozaki, H. The novel anti-Propionibacterium acnes compound, Sargafuran, found in the marine brown alga Sargassum macrocarpum. J. Antibiot. 2009, 62, 259–263. [Google Scholar] [CrossRef]
- Katsuta, R.; Aoki, K.; Yajima, A.; Nukada, T. Synthesis of the core framework of the proposed structure of sargafuran. Tetrahedron Lett. 2013, 54, 347–350. [Google Scholar] [CrossRef]
- Amiguet, V.T.; Jewell, L.E.; Mao, H.; Sharma, M.; Hudson, J.B.; Durst, T.; Allard, M.; Rochefort, G.; Arnason, J.T. Antibacterial properties of a glycolipid-rich extract and active principle from Nunavik collections of the macroalgae Fucus evanescens C. Agardh (Fucaceae). Can. J. Microbiol. 2011, 57, 745–749. [Google Scholar] [CrossRef]
- Lee, J.-H.; Eom, S.-H.; Lee, E.-H.; Jung, Y.-J.; Kim, H.-J.; Jo, M.-R.; Son, K.-T.; Lee, H.-J.; Kim, J.H.; Lee, M.-S.; et al. In vitro antibacterial and synergistic effect of phlorotannins isolated from edible brown seaweed Eisenia bicyclis against acne-related bacteria. ALGAE 2014, 29, 47–55. [Google Scholar] [CrossRef]
- Bateni, E.; Tester, R.; Al-Ghazzewi, F.; Bateni, S.; Alvani, K.; Piggott, J. The Use of Konjac Glucomannan Hydrolysates (GMH) to Improve the Health of the Skin and Reduce Acne Vulgaris. Am. J. Dermatol. Venereol. 2013, 2013, 10–14. [Google Scholar]
- Al-Ghazzewi, F.H.; Khanna, S.; Tester, R.F.; Piggott, J. The potential use of hydrolysed konjac glucomannan as a prebiotic. J. Sci. Food Agric. 2007, 87, 1758–1766. [Google Scholar] [CrossRef]
- Al-Ghazzewi, F.H.; Tester, R.F. Effect of konjac glucomannan hydrolysates and probiotics on the growth of the skin bacterium Propionibacterium acnes in vitro. Int. J. Cosmet. Sci. 2010, 32, 139–142. [Google Scholar] [CrossRef]
- Yang, A.-J.; Marito, S.; Yang, J.-J.; Keshari, S.; Chew, C.-H.; Chen, C.-C.; Huang, C.-M. A Microtube Array Membrane (MTAM) Encapsulated Live Fermenting Staphylococcus epidermidis as a Skin Probiotic Patch against Cutibacterium acnes. Int. J. Mol. Sci. 2018, 20, 14. [Google Scholar] [CrossRef]
- Castillo, D.E.; Nanda, S.; Keri, J.E. Propionibacterium (Cutibacterium) acnes Bacteriophage Therapy in Acne: Current Evidence and Future Perspectives. Dermatol. Ther. 2019, 9, 19–31. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Fitz-Gibbon, S.; Hayes, C.; Bowman, C.; Inkeles, M.; Loncaric, A.; Russell, D.A.; Jacobs-Sera, D.; Cokus, S.; Pellegrini, M.; et al. Propionibacterium acnes Bacteriophages Display Limited Genetic Diversity and Broad Killing Activity against Bacterial Skin Isolates. mBio 2012, 3, e00279-12. [Google Scholar] [CrossRef] [PubMed]
- Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Żaczek, M.; Międzybrodzki, R.; Letkiewicz, S.; Łusiak-Szelchowska, M.; Górski, A. Prospects of Phage Application in the Treatment of Acne Caused by Propionibacterium acnes. Front. Microbiol. 2017, 8, 164. [Google Scholar] [CrossRef]
- Liu, J.; Yan, R.; Zhong, Q.; Ngo, S.; Bangayan, N.J.; Nguyen, L.; Lui, T.; Liu, M.; Erfe, M.C.; Craft, N.; et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 2015, 9, 2078–2093. [Google Scholar] [CrossRef]
- Gervason, S.; Metton, I.; Gemrot, E.; Ranouille, E.; Skorski, G.; Cabannes, M.; Berthon, J.-Y.; Filaire, E. Rhodomyrtus tomentosa Fruit Extract and Skin Microbiota: A Focus on C. acnes Phylotypes in Acne Subjects. Cosmetics 2020, 7, 53. [Google Scholar] [CrossRef]
- Cinque, B.; La Torre, C.; Melchiorre, E.; Marchesani, G.; Zoccali, G.; Palumbo, P.; Di Marzio, L.; Masci, A.; Mosca, L.; Mastromarino, P.; et al. Use of Probiotics for Dermal Applications. In Probiotics; Liong, M.-T., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 221–241. ISBN 978-3-642-20838-6. [Google Scholar]
- Bey, M.; Nachat-Kapes, R.; Ranouille, E.; Berthon, J. Targeting skin microbiome: Innovative approach for the development of cosmetic active ingredients. In Proceedings of the COSM’ING 2016, Saint Malo, France, 29 June 2016; p. 13. [Google Scholar]
- SILAB. LACTOBIOTYL®. Available online: https://www.silab.fr/produit-108-lactobiotyl_fra.html (accessed on 27 February 2020).
- Rouaud-Tinguely, P.; Laporte, D.; Bordes, S.; Roth, M.-P.; Coppin, H.; Closs, B. Microbiota: A Topic at The Crossroads of Many Fields of Expertise. In Proceedings of the COSM’ING 2018, Saint Malo, France, 4–6 July 2018; p. 4. [Google Scholar]
- Al-Ghazzewi, F.H.; Tester, R.F. Impact of prebiotics and probiotics on skin health. Benef. Microbes 2014, 5, 99–107. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fournière, M.; Latire, T.; Souak, D.; Feuilloley, M.G.J.; Bedoux, G. Staphylococcus epidermidis and Cutibacterium acnes: Two Major Sentinels of Skin Microbiota and the Influence of Cosmetics. Microorganisms 2020, 8, 1752. https://doi.org/10.3390/microorganisms8111752
Fournière M, Latire T, Souak D, Feuilloley MGJ, Bedoux G. Staphylococcus epidermidis and Cutibacterium acnes: Two Major Sentinels of Skin Microbiota and the Influence of Cosmetics. Microorganisms. 2020; 8(11):1752. https://doi.org/10.3390/microorganisms8111752
Chicago/Turabian StyleFournière, Mathilde, Thomas Latire, Djouhar Souak, Marc G. J. Feuilloley, and Gilles Bedoux. 2020. "Staphylococcus epidermidis and Cutibacterium acnes: Two Major Sentinels of Skin Microbiota and the Influence of Cosmetics" Microorganisms 8, no. 11: 1752. https://doi.org/10.3390/microorganisms8111752
APA StyleFournière, M., Latire, T., Souak, D., Feuilloley, M. G. J., & Bedoux, G. (2020). Staphylococcus epidermidis and Cutibacterium acnes: Two Major Sentinels of Skin Microbiota and the Influence of Cosmetics. Microorganisms, 8(11), 1752. https://doi.org/10.3390/microorganisms8111752