Streptococcus Thermophilus UASt-09 Upregulates Goblet Cell Activity in Colonic Epithelial Cells to a Greater Degree than other Probiotic Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Origins
2.2. Cell Culture and Reagents
2.3. Treatments
2.4. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
2.5. Prime PCR Assay
2.6. Statistical Analysis
2.7. Expression Heatmap Analysis
3. Results
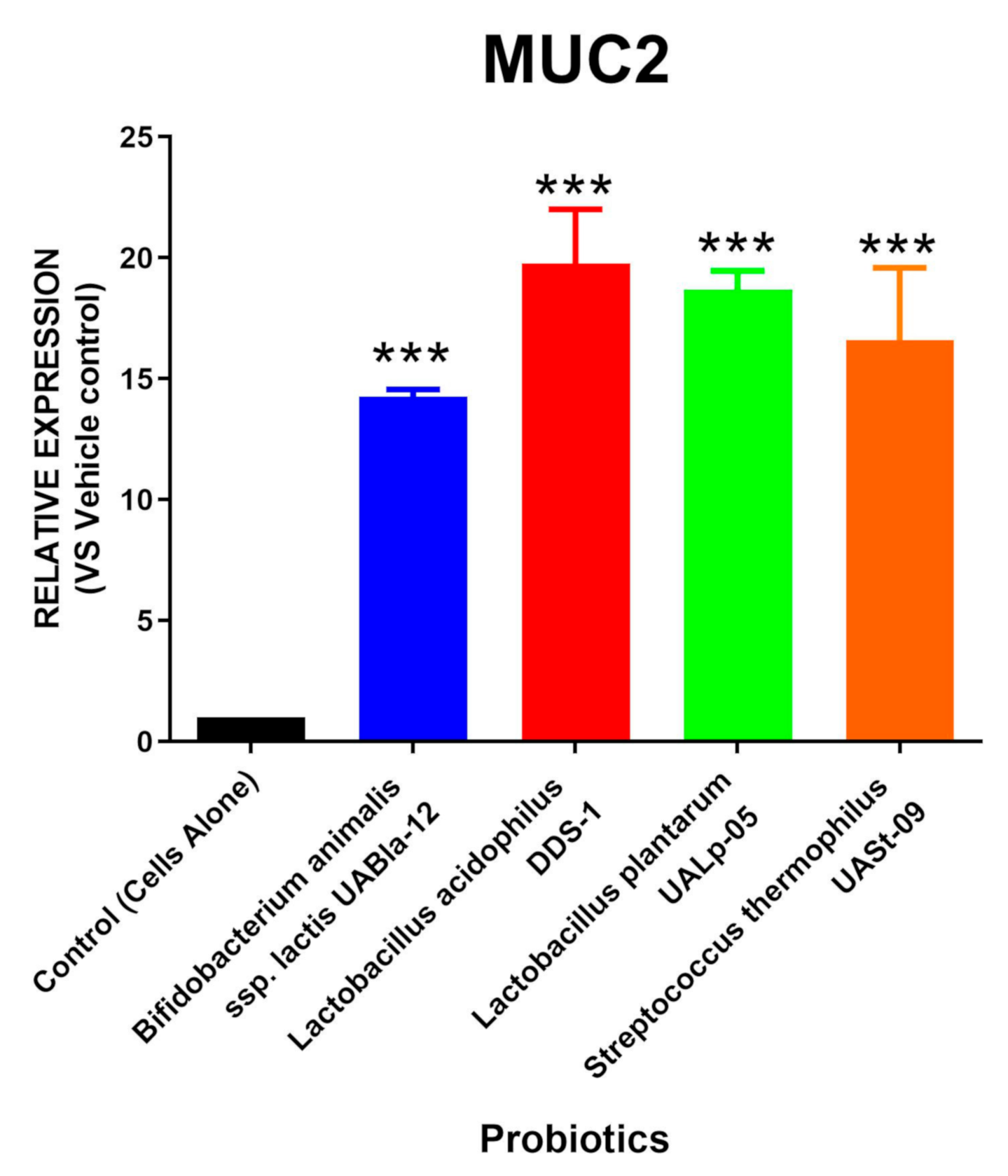
3.1. All Tested Probiotics Significantly Upregulate MUC2 Expression
3.2. Streptococcus Thermophilus UASt-09 Significantly Upregulates AGR2 Expression
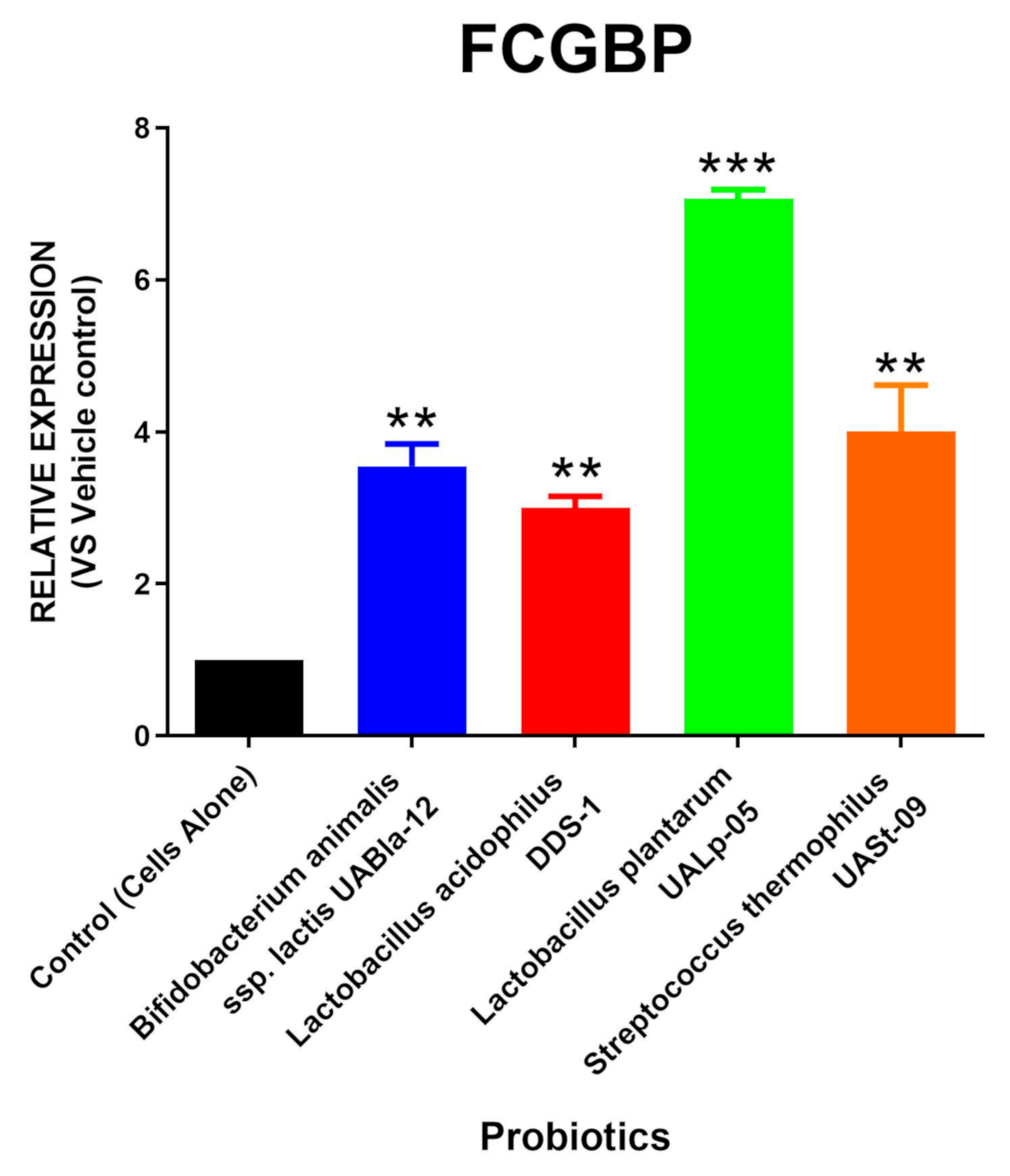
3.3. All Tested Probiotics Enhance FCGBP Expression
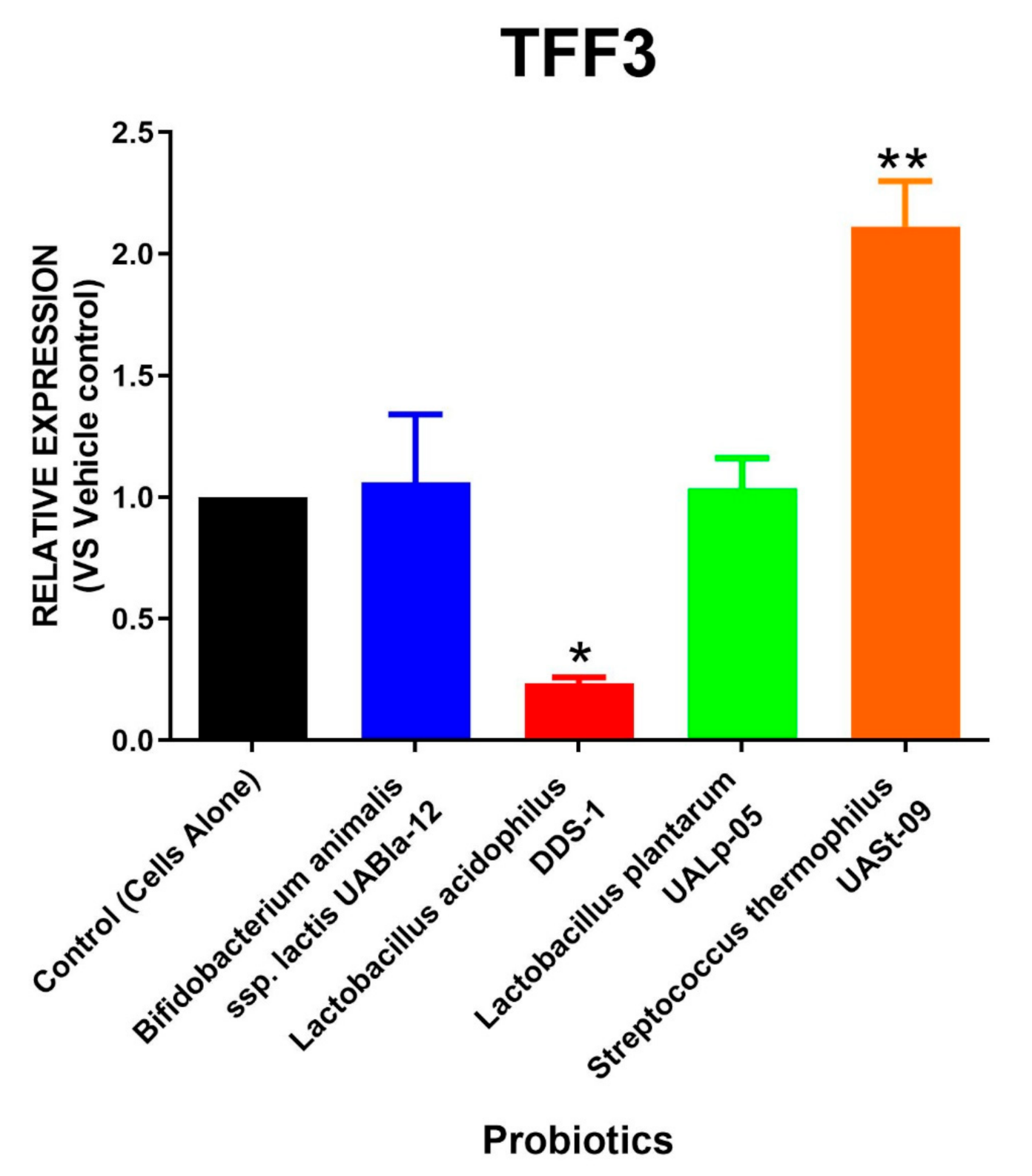
3.4. Streptococcus Thermophilus UASt-09 Elevates RELMβ and TFF3 Expression
3.5. The Role of Streptococcus Thermophilus UASt-09 in Regulating Immune Response in Human Colonic Cells
3.6. Heatmap Comparison of Probiotics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ficara, M.; Pietrella, E.; Spada, C.; Della Casa Muttini, E.; Lucaccioni, L.; Iughetti, L.; Berardi, A. Changes of intestinal microbiota in early life. J. Matern. Fetal Neonatal Med. 2020, 33, 1036–1043. [Google Scholar] [CrossRef]
- Kastl, A.J., Jr.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The structure and function of the human small intestinal microbiota: Current understanding and future directions. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Huang, J.; Luo, M.; Wang, Z.; Zhu, L.; Wang, S.; Zhu, D.; Liu, H. The influence of gut microbiota on the rheological characterization of soy hull polysaccharide and mucin interactions. RSC Adv. 2020, 10, 2830–2840. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.E.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [Green Version]
- Fyderek, K.; Strus, M.; Kowalska-Duplaga, K.; Gosiewski, T.; Wędrychowicz, A.; Jedynak-Wąsowicz, U.; Sładek, M.; Pieczarkowski, S.; Adamski, P.; Kochan, P.; et al. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J. Gastroenterol. 2009, 15, 5287–5294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, M.E.; Thomsson, K.A.; Hansson, G.C. Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J. Proteome Res. 2009, 8, 3549–3557. [Google Scholar] [CrossRef] [PubMed]
- Bansil, R.; Turner, B.S. The biology of mucus: Composition, synthesis and organization. Adv. Drug Deliv. Rev. 2018, 124, 3–15. [Google Scholar] [CrossRef]
- Hansson, G.C. Mucins and the Microbiome. Annu. Rev. Biochem. 2020, 89, 769–793. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Rubin, B.K.; Voynow, J.A. Mucins, mucus, and goblet cells. Chest 2018, 154, 169–176. [Google Scholar] [CrossRef]
- Demouveaux, B.; Gouyer, V.; Gottrand, F.; Narita, T.; Desseyn, J.-L. Gel-forming mucin interactome drives mucus viscoelasticity. Adv. Colloid Interface Sci. 2018, 252, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Kamphuis, J.B.; Mercier-Bonin, M.; Eutamene, H.; Theodorou, V. Mucus organisation is shaped by colonic content; a new view. Sci. Rep. 2017, 7, 8527–8540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, S.; Araki, T.; Toiyama, Y.; Tanaka, K.; Kawamura, M.; Okugawa, Y.; Okita, Y.; Saigusa, S.; Inoue, Y.; Uchida, K.; et al. Downregulation of trefoil factor-3 expression in the rectum is associated with the development of ulcerative colitis-associated cancer. Oncol. Lett. 2018, 16, 3658–3664. [Google Scholar] [CrossRef] [Green Version]
- Batugedara, H.M.; Nair, M.G. Resistin-Like Molecules and Endocannabinoids Regulate Inflammation and Immunity during Helminth Infections. In Immunoregulatory Mechanisms in a Mouse Model of Hookworm Infection; University of California Riverside: Riverside, CA, USA, 2018; pp. 1–31. [Google Scholar]
- Propheter, D.C.; Chara, A.L.; Harris, T.A.; Ruhn, K.A.; Hooper, L.V. Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc. Natl. Acad. Sci. USA 2017, 114, 11027–11033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, S.P.; Seidu, L.; Blanchard, C.; Groschwitz, K.; Mishra, A.; Karow, M.L.; Ahrens, R.; Artis, D.; Murphy, A.J.; Valenzuela, D.M.; et al. Resistin-like molecule β regulates innate colonic function: Barrier integrity and inflammation susceptibility. J. Allergy Clin. Immunol. 2006, 118, 257–268. [Google Scholar] [CrossRef] [Green Version]
- van der Post, S.; Jabbar, K.S.; Birchenough, G.; Arike, L.; Akhtar, N.; Sjovall, H.; Johansson, M.E.; Hansson, G.C. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 2019, 68, 2142–2151. [Google Scholar] [CrossRef] [Green Version]
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Lopez Siles, M.; Enrich-Capó, N.; Aldeguer, X.; Sabat-Mir, M.; Duncan, S.; Garcia-Gil, J.; Martinez-Medina, M. Alterations in the abundance and co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the colonic mucosa of inflammatory bowel disease subjects. Front. Cell. Infect. Microbiol. 2018, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L.; Xia, Y.; Li, W.; Wang, K.; Wang, L.; Miao, Y.; Ma, S. The gut microbiota heterogeneity and assembly changes associated with the IBD. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Zhai, Z.; Zhang, F.; Cao, R.; Ni, X.; Xin, Z.; Deng, J.; Wu, G.; Ren, W.; Yin, Y.; Deng, B. Cecropin A alleviates inflammation through modulating the gut microbiota of C57BL/6 mice with DSS-induced IBD. Front. Microbiol. 2019, 10, 1595. [Google Scholar] [CrossRef]
- Burrello, C.; Pellegrino, G.; Giuffrè, M.R.; Lovati, G.; Magagna, I.; Bertocchi, A.; Cribiù, F.M.; Boggio, F.; Botti, F.; Trombetta, E.; et al. Mucosa-associated microbiota drives pathogenic functions in IBD-derived intestinal iNKT cells. Life Sci. Alliance 2019, 2, e201800229. [Google Scholar] [CrossRef]
- Vaughn, B.P.; Kaiser, T.; Staley, C.; Hamilton, M.J.; Reich, J.; Graiziger, C.; Singroy, S.; Kabage, A.J.; Sadowsky, M.J.; Khoruts, A. A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease and primary sclerosing cholangitis. Clin. Exp. Gastroenterol. 2019, 12, 9. [Google Scholar] [CrossRef] [Green Version]
- Ganji-Arjenaki, M.; Rafieian-Kopaei, M. Probiotics are a good choice in remission of inflammatory bowel diseases: A meta analysis and systematic review. J. Cell. Physiol. 2018, 233, 2091–2103. [Google Scholar] [CrossRef]
- Alard, J.; Peucelle, V.; Boutillier, D.; Breton, J.; Kuylle, S.; Pot, B.; Holowacz, S.; Grangette, C. New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches. Benef. Microbes. 2018, 9, 317–331. [Google Scholar] [CrossRef]
- Ballini, A.; Santacroce, L.; Cantore, S.; Bottalico, L.; Dipalma, G.; Topi, S.; Saini, R.; De Vito, D.; Inchingolo, F. Probiotics efficacy on oxidative stress values in inflammatory bowel disease: A randomized double-blinded placebo-controlled pilot study. Endocr. Metab. Immune Disord. Drug Targets 2019, 19, 373–381. [Google Scholar] [CrossRef]
- Mao, N.; Cubillos-Ruiz, A.; Cameron, D.E.; Collins, J.J. Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 2018, 10, eaao2586. [Google Scholar] [CrossRef] [Green Version]
- Sassone-Corsi, M.; Nuccio, S.-P.; Liu, H.; Hernandez, D.; Vu, C.T.; Takahashi, A.A.; Edwards, R.A.; Raffatellu, M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016, 540, 280–283. [Google Scholar] [CrossRef]
- White, R.; Atherly, T.; Guard, B.; Rossi, G.; Wang, C.; Mosher, C.; Webb, C.; Hill, S.; Ackermann, M.; Sciabarra, P.; et al. Randomized, controlled trial evaluating the effect of multi-strain probiotic on the mucosal microbiota in canine idiopathic inflammatory bowel disease. Gut Microbes 2017, 8, 451–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo-Bolívar, J.F.; Pardo, R.Y.R.; Hume, M.E.; Nisbet, D.J.; Rodríguez-Villamizar, F.; Alzate, J.F.; Junca, H.; Diaz, L.M.V. Establishment and characterization of a competitive exclusion bacterial culture derived from Nile tilapia (Oreochromis niloticus) gut microbiomes showing antibacterial activity against pathogenic Streptococcus agalactiae. PLoS ONE 2019, 14, e0215375. [Google Scholar] [CrossRef]
- Freedman, S.; Williamson-Urquhart, S.; Farion, K.; Gouin, S.; Willan, A.; Poonai, N.; Hurley, K.; Sherman, P.; Finkelstein, Y.; Lee, B.; et al. PL02: Probiotic regimen for outpatient gastroenteritis utility of treatment (PROGUT) study: A multicenter randomized controlled trial. Can. J. Emerg. Med. 2018, 20, S5. [Google Scholar] [CrossRef] [Green Version]
- Lai, H.-H.; Chiu, C.-H.; Kong, M.-S.; Chang, C.-J.; Chen, C.-C. Probiotic Lactobacillus casei: Effective for Managing Childhood Diarrhea by Altering Gut Microbiota and Attenuating Fecal Inflammatory Markers. Nutrients 2019, 11, 1150–1165. [Google Scholar] [CrossRef] [Green Version]
- Martoni, C.J.; Srivastava, S.; Leyer, G.J. Lactobacillus acidophilus DDS-1 and Bifidobacterium lactis UABla-12 Improve Abdominal Pain Severity and Symptomology in Irritable Bowel Syndrome: Randomized Controlled Trial. Nutrients 2020, 12, 363–378. [Google Scholar] [CrossRef] [Green Version]
- Bu, X.-D.; Li, N.; Tian, X.-Q.; Huang, P.-L. Caco-2 and LS174T cell lines provide different models for studying mucin expression in colon cancer. Tissue Cell 2011, 43, 201–206. [Google Scholar] [CrossRef]
- Bergstrom, K.S.; Morampudi, V.; Chan, J.M.; Bhinder, G.; Lau, J.; Yang, H.; Ma, C.; Huang, T.; Ryz, N.; Sham, H.P.; et al. Goblet cell derived RELM-β recruits CD4+ T cells during infectious colitis to promote protective intestinal epithelial cell proliferation. PLoS Pathog. 2015, 11, e1005108. [Google Scholar] [CrossRef]
- Raja, S.B.; Murali, M.R.; Devaraj, H.; Devaraj, S.N. Differential expression of gastric MUC5AC in colonic epithelial cells: TFF3-wired IL1 β/Akt crosstalk-induced mucosal immune response against Shigella dysenteriae infection. J. Cell Sci. 2012, 125, 703–713. [Google Scholar] [CrossRef] [Green Version]
- Heitkemper, M.; Cain, K.; Shulman, R.; Burr, R.; Ko, C.; Hollister, E.; Callen, N.; Zia, J.; Han, C.; Jarrett, M. Stool and urine trefoil factor 3 levels: Associations with symptoms, intestinal permeability, and microbial diversity in irritable bowel syndrome. Benef. Microbes. 2018, 9, 345–355. [Google Scholar] [CrossRef]
- Lu, P.; Burger-van Paassen, N.; van der Sluis, M.; Witte-Bouma, J.; Kerckaert, J.P.; van Goudoever, J.B.; Van Seuningen, I.; Renes, I.B. Colonic gene expression patterns of mucin Muc2 knockout mice reveal various phases in colitis development. Inflamm. Bowel Dis. 2011, 17, 2047–2057. [Google Scholar] [CrossRef]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.P.; Van Goudoever, J.B.; Büller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef]
- Heazlewood, C.K.; Cook, M.C.; Eri, R.; Price, G.R.; Tauro, S.B.; Taupin, D.; Thornton, D.J.; Png, C.W.; Crockford, T.L.; Cornall, R.J.; et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008, 5, e54. [Google Scholar] [CrossRef] [Green Version]
- Eri, R.; Adams, R.; Tran, T.; Tong, H.; Das, I.; Roche, D.; Oancea, I.; Png, C.W.; Jeffery, P.; Radford-Smith, G.L.; et al. An intestinal epithelial defect conferring ER stress results in inflammation involving both innate and adaptive immunity. Mucosal. Immunol. 2011, 4, 354–364. [Google Scholar] [CrossRef]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131S–1141S. [Google Scholar] [CrossRef] [Green Version]
- Albert, T.K.; Laubinger, W.; Müller, S.; Hanisch, F.-G.; Kalinski, T.; Meyer, F.; Hoffmann, W. Human intestinal TFF3 forms disulfide-linked heteromers with the mucus-associated FCGBP protein and is released by hydrogen sulfide. J. Proteome Res. 2010, 9, 3108–3117. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Klasson, S.; Larsson, E.; Johansson, M.E.; Hansson, G.C.; Samuelsson, T. Searching the evolutionary origin of epithelial mucus protein components—mucins and FCGBP. Mol. Biol. Evol. 2016, 33, 1921–1936. [Google Scholar] [CrossRef]
- Podolsky, D.K.; Gerken, G.; Eyking, A.; Cario, E. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology 2009, 137, 209–220. [Google Scholar] [CrossRef]
- zum Büschenfelde, D.M.; Tauber, R.; Huber, O. TFF3-peptide increases transepithelial resistance in epithelial cells by modulating claudin-1 and-2 expression. Peptides 2006, 27, 3383–3390. [Google Scholar] [CrossRef] [PubMed]
- Morampudi, V.; Dalwadi, U.; Bhinder, G.; Sham, H.; Gill, S.; Chan, J.; Bergstrom, K.; Huang, T.; Ma, C.; Jacobson, K.; et al. The goblet cell-derived mediator RELM-β drives spontaneous colitis in Muc2-deficient mice by promoting commensal microbial dysbiosis. Mucosal. Immunol. 2016, 9, 1218–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De’Broski, R.H.; Yang, J.-Q.; Hogan, S.P.; Groschwitz, K.; Khodoun, M.; Munitz, A.; Orekov, T.; Perkins, C.; Wang, Q.; Brombacher, F.; et al. Intestinal epithelial cell secretion of RELM-β protects against gastrointestinal worm infection. J. Exp. Med. 2009, 206, 2947–2957. [Google Scholar]
- Park, S.-W.; Zhen, G.; Verhaeghe, C.; Nakagami, Y.; Nguyenvu, L.T.; Barczak, A.J.; Killeen, N.; Erle, D.J. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. USA 2009, 106, 6950–6955. [Google Scholar] [CrossRef] [Green Version]
- Bergström, J.H.; Berg, K.A.; Rodríguez-Piñeiro, A.M.; Stecher, B.; Johansson, M.E.; Hansson, G.C. AGR2, an endoplasmic reticulum protein, is secreted into the gastrointestinal mucus. PLoS ONE 2014, 9, e104186. [Google Scholar] [CrossRef]
- Plaza-Díaz, J.; Robles-Sánchez, C.; Abadía-Molina, F.; Morón-Calvente, V.; Sáez-Lara, M.J.; Ruiz-Bravo, A.; Jiménez-Valera, M.; Gil, Á.; Gómez-Llorente, C.; Fontana, L. Adamdec1, Ednrb and Ptgs1/Cox1, inflammation genes upregulated in the intestinal mucosa of obese rats, are downregulated by three probiotic strains. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- De Marco, S.; Sichetti, M.; Muradyan, D.; Piccioni, M.; Traina, G.; Pagiotti, R.; Pietrella, D. Probiotic cell-free supernatants exhibited anti-inflammatory and antioxidant activity on human gut epithelial cells and macrophages stimulated with LPS. Evid. Based Complementary Altern. Med. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Bhatia, S.; Prabhu, P.N.; Benefiel, A.C.; Miller, M.J.; Chow, J.; Davis, S.R.; Gaskins, H.R. Galacto-oligosaccharides may directly enhance intestinal barrier function through the modulation of goblet cells. Mol. Nutr. Food Res. 2015, 59, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Byrd, J.C.; Koo, J.S.; Bresalier, R.S. Bile acids induce MUC2 overexpression in human colon carcinoma cells. Cancer 2005, 103, 1606–1614. [Google Scholar] [CrossRef]




| Goblet Cell Secreted Components | Functions | Reference |
|---|---|---|
| Mucin 2 (MUC2) | Major component involved in the formation of mucus layer that acts as the first host-defense barrier | [4] |
| Fc-Gamma Binding Protein (FCGBP) | Stabilization and crosslinking of the MUC2 mucin barrier to the inner mucin layer | [6] |
| Trefoil Factor 3 (TFF3) | Associated in mucosal protection and healing process | [12] |
| Resistin-like Molecule β (RELMβ) | Involved in mucosal immune defense against microbial infection. | [13] |
| Promote spatial segregation of the microbiota and the colonic epithelium | [14] | |
| Anterior Gradient Homolog 2 (AGR2) | Essential in mucin biosynthesis | [15] |
| Genus | Species | Strain | Origin |
|---|---|---|---|
| Bifidobacterium | animalis subsp. lactis | UABla-12 | Human |
| Lactobacillus | acidophilus | DDS-1 | Human |
| Lactobacillus | plantarum | UALp-05 | Plant |
| Streptococcus | thermophilus | UASt-09 | Dairy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shastri, M.D.; Chong, W.C.; Vemuri, R.; Martoni, C.J.; Adhikari, S.; Bhullar, H.; Kunde, D.; Tristram, S.G.; Eri, R.D. Streptococcus Thermophilus UASt-09 Upregulates Goblet Cell Activity in Colonic Epithelial Cells to a Greater Degree than other Probiotic Strains. Microorganisms 2020, 8, 1758. https://doi.org/10.3390/microorganisms8111758
Shastri MD, Chong WC, Vemuri R, Martoni CJ, Adhikari S, Bhullar H, Kunde D, Tristram SG, Eri RD. Streptococcus Thermophilus UASt-09 Upregulates Goblet Cell Activity in Colonic Epithelial Cells to a Greater Degree than other Probiotic Strains. Microorganisms. 2020; 8(11):1758. https://doi.org/10.3390/microorganisms8111758
Chicago/Turabian StyleShastri, Madhur D., Wai Chin Chong, Ravichandra Vemuri, Christopher J. Martoni, Santosh Adhikari, Harinder Bhullar, Dale Kunde, Stephen G. Tristram, and Rajaraman D. Eri. 2020. "Streptococcus Thermophilus UASt-09 Upregulates Goblet Cell Activity in Colonic Epithelial Cells to a Greater Degree than other Probiotic Strains" Microorganisms 8, no. 11: 1758. https://doi.org/10.3390/microorganisms8111758
APA StyleShastri, M. D., Chong, W. C., Vemuri, R., Martoni, C. J., Adhikari, S., Bhullar, H., Kunde, D., Tristram, S. G., & Eri, R. D. (2020). Streptococcus Thermophilus UASt-09 Upregulates Goblet Cell Activity in Colonic Epithelial Cells to a Greater Degree than other Probiotic Strains. Microorganisms, 8(11), 1758. https://doi.org/10.3390/microorganisms8111758






