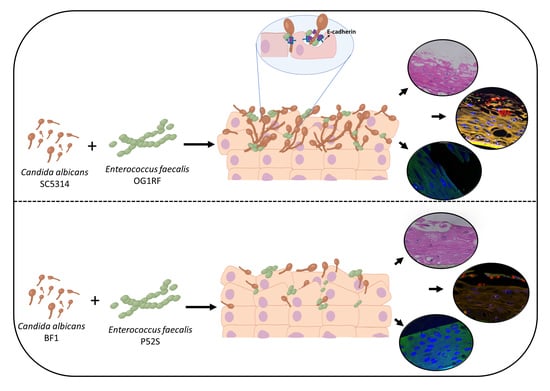
Interactions between Candida albicans and Enterococcus faecalis in an Organotypic Oral Epithelial Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Microbial Strains, Engineered Tissue, and Culture Conditions
2.2. Inoculation of Mucosal Tissues with C. albicans and E. faecalis
2.3. Analysis of Mucosal Biofilms and Microbial Invasion
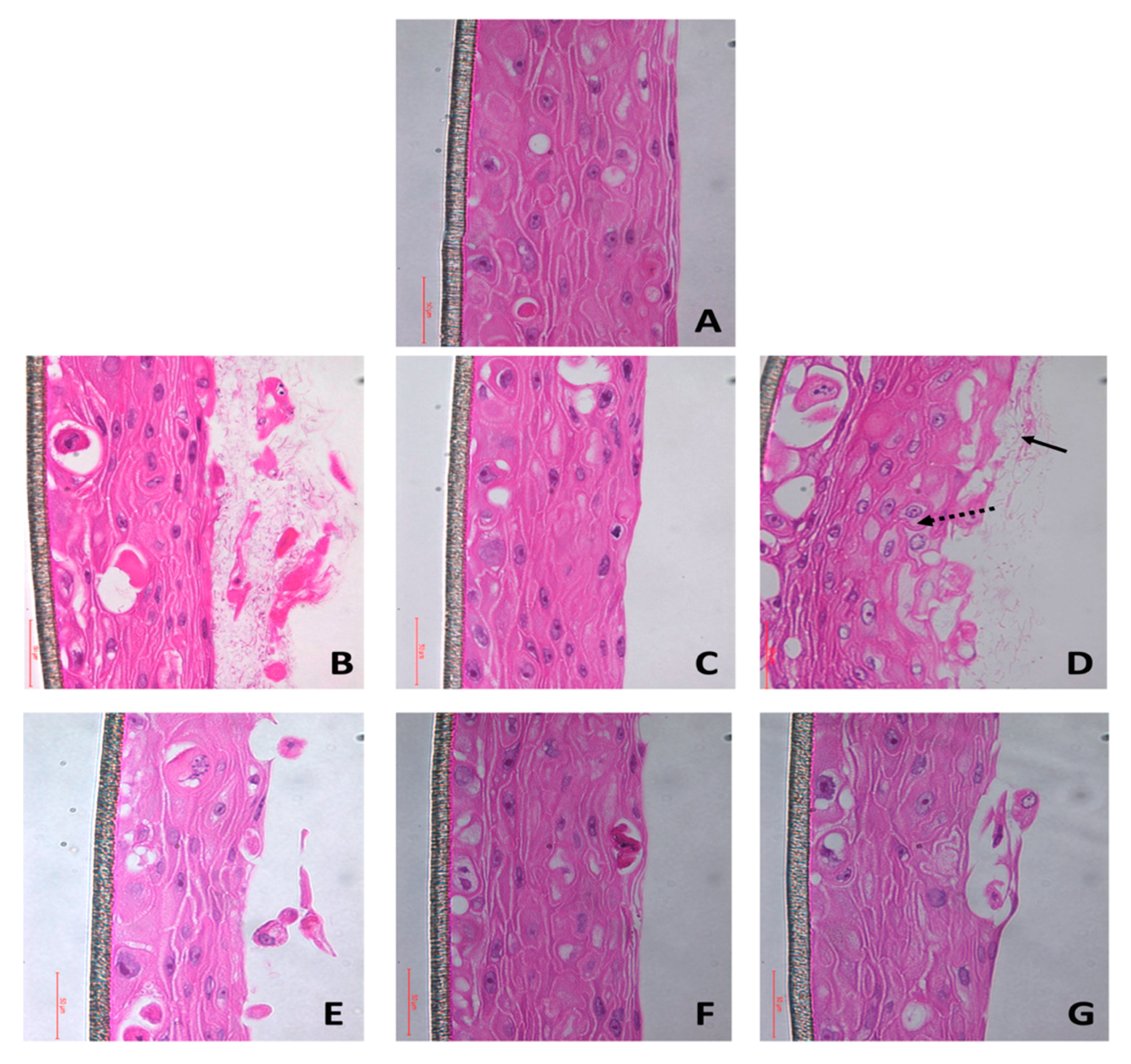
2.3.1. Hematoxylin and Eosin Staining
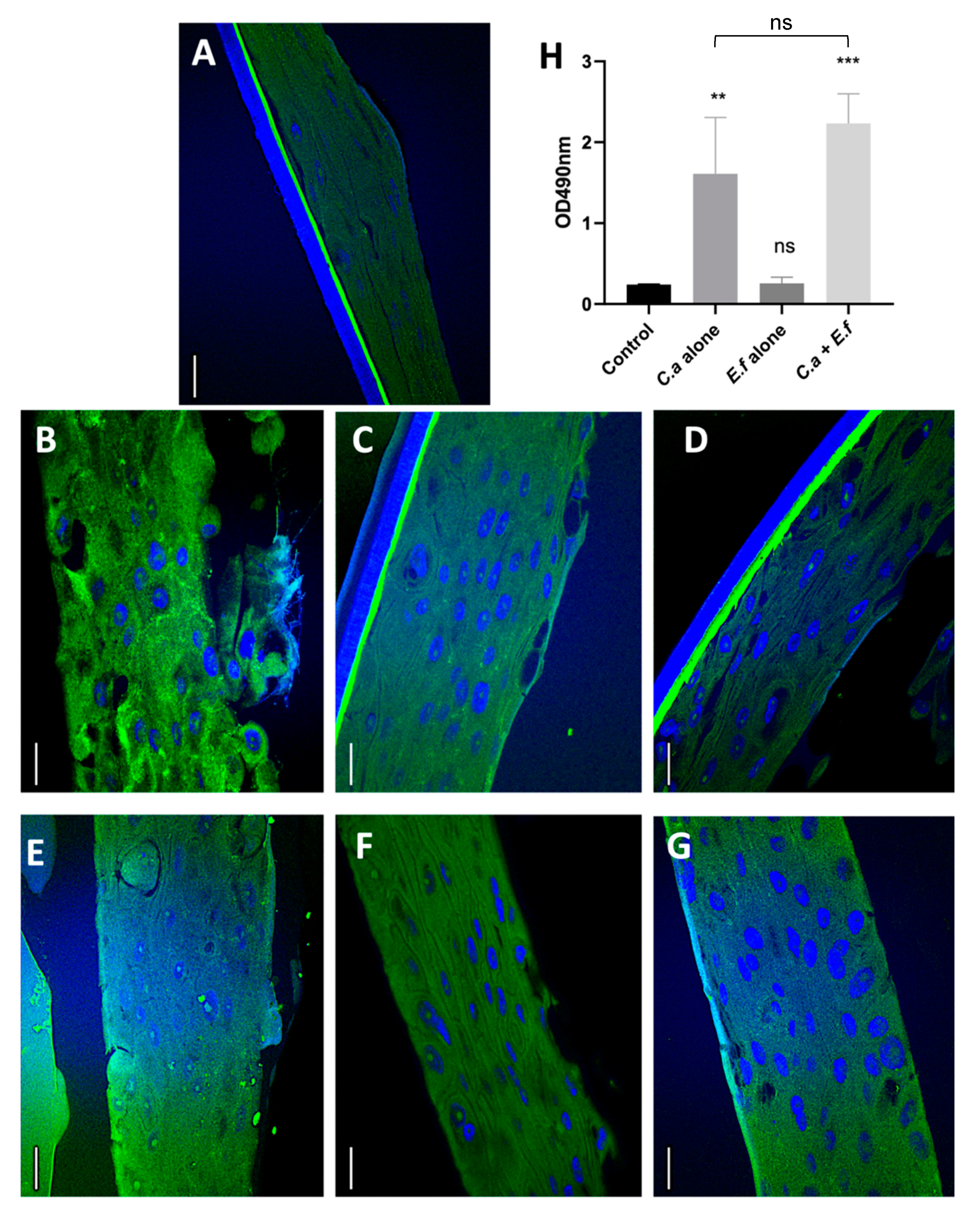
2.3.2. Fluorescent in situ Hybridisation (FISH) and Quantification of Tissue Invasion by Microbial Isolates
2.4. Evaluation of Tissue Integrity Using e-cadherin
2.5. Quantification of Tissue Destruction Using the Lactate Dehydrogenase (LDH) Assay
2.6. Gene Expression Studies
2.7. Statistical Analysis
3. Results and Discussion
3.1. Fungal Biofilms and Dual Species Biofilms cause Mucosal Tissue Erosion
3.2. Dual Species Biofilms Demonstrate Increased Microbial Invasion into Mucosal Compartments than Mono-Species Biofilms
3.3. E. faecalis Upregulates the Expression of Selected Virulence Genes of C. albicans
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dalle, F.; Wächtler, B.; L’Ollivier, C.; Holland, G.; Bannert, N.; Wilson, D.; Labruère, C.; Bonnin, A.; Hube, B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol. 2010, 12, 248–271. [Google Scholar] [CrossRef]
- Nett, J.E.; Marchillo, K.; Spiegel, C.A.; Andes, D.R. Development and validation of an in vivo Candida albicans biofilm denture model. Infect. Immun. 2010, 78, 3650–3659. [Google Scholar] [CrossRef]
- Bertolini, M.; Ranjan, A.; Thompson, A.; Diaz, P.I.; Sobue, T.; Maas, K.; Dongari-Bagtzoglou, A. Candida albicans induces mucosal bacterial dysbiosis that promotes invasive infection. PLoS Pathog. 2019, 15, e1007717. [Google Scholar] [CrossRef]
- Dongari-Bagtzoglou, A. Mucosal biofilms: Challenges and future directions. Expert Rev. Anti Infect. Ther. 2008, 6, 141–144. [Google Scholar] [CrossRef]
- Xu, H.; Sobue, T.; Thompson, A.; Xie, Z.; Poon, K.; Ricker, A.; Cervantes, J.; Diaz, P.I.; Dongari-Bagtzoglou, A. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014, 16, 214–231. [Google Scholar] [CrossRef]
- Hogan, D.A.; Vik, Å.; Kolter, R.A. Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterialg-fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef]
- Graham, C.E.; Cruz, M.R.; Garsin, D.A.; Lorenz, M.C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 2017, 114, 4507–4512. [Google Scholar] [CrossRef]
- Dahlén, G.; Blomqvist, S.; Almståhl, A.; Carlén, A. Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections. J. Oral Microbiol. 2012, 4, 10855. [Google Scholar] [CrossRef]
- Hermann, C.; Hermann, J.; Munzel, U.; Rüchel, R. Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 1999, 42, 619–627. [Google Scholar] [CrossRef]
- Abusrewil, S.; Alshanta, O.A.; Albashaireh, K.; Alqahtani, S.; Nile, C.J.; Scott, J.A.; McLean, W. Detection, treatment and prevention of endodontic biofilm infections: What’s new in 2020? Crit Rev. Microbiol. 2020, 46, 194–212. [Google Scholar] [CrossRef]
- Komiyama, E.Y.; Lepesqueur, L.S.; Yassuda, C.G.; Samaranayake, L.P.; Parahitiyawa, N.B.; Balducci, I.; Koga-Ito, C.Y. Enterococcus Species in the oral cavity: Prevalence, virulence factors and antimicrobial susceptibility. PLoS ONE 2016, 11, e0163001. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, Q.; Zhang, C.; Cheung, G.S.; Shen, Y. Prevalence, phenotype, and genotype of Enterococcus faecalis isolated from saliva and root canals in patients with persistent apical periodontitis. J. Endod. 2010, 36, 1950–1955. [Google Scholar] [CrossRef]
- Jin, Y.; Yip, H.K.; Samaranayake, Y.H.; Yau, J.Y.; Samaranayake, L.P. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J. Clin. Microbiol. 2003, 41, 2961–2967. [Google Scholar] [CrossRef]
- Seneviratne, C.J.; Silva, W.J.; Jin, L.J.; Samaranayake, Y.H.; Samaranayake, L.P. Architectural analysis, viability assessment and growth kinetics of Candida albicans and Candida glabrata biofilms. Arch. Oral Biol. 2009, 54, 1052–1060. [Google Scholar] [CrossRef]
- Bertolini, M.M.; Xu, H.; Sobue, T.; Nobile, C.J.; Del Bel Cury, A.A.; Dongari-Bagtzoglou, A. Candida-streptococcal mucosal biofilms display distinct structural and virulence characteristics depending on growth conditions and hyphal morphotypes. Mol. Oral Microbiol. 2015, 30, 307–322. [Google Scholar] [CrossRef]
- Kempf, V.A.J.; Trebesius, K.; Autenrieth, I.B. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 2000, 38, 830–838. [Google Scholar] [CrossRef]
- Wang, Z.; Qi, Y.; Wang, J.; Pei, Y. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. World J. Microbiol Biotechnol. 2012, 28, 1919–1925. [Google Scholar] [CrossRef]
- Pernthaler, J.; Glöckner, F.O.; Schönhuber, W.; Amann, R. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 2001, 30, 1–31. [Google Scholar] [CrossRef]
- Valm, A.M.; Mark Welch, J.L.; Rieken, C.W.; Hasegawa, Y.; Sogin, M.L.; Oldenbourg, R. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. USA 2011, 108, 4152–4157. [Google Scholar] [CrossRef]
- Diaz, P.I.; Xie, Z.; Sobue, T.; Thompson, A.; Biyikoglu, B.; Ricker, A.; Ikonomou, L.; Dongari-Bagtzoglou, A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 2012, 80, 620–632. [Google Scholar] [CrossRef]
- Villar, C.C.; Kashleva, H.; Nobile, C.J.; Mitchell, A.P.; Dongari-Bagtzoglou, A. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect. Immun. 2007, 75, 2126–2135. [Google Scholar] [CrossRef]
- Cruz, M.R.; Graham, C.E.; Gagliano, B.C.; Lorenz, M.C.; Garsin, D.A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 2013, 81, 189–200. [Google Scholar] [CrossRef]
- Naglik, J.R.; Richardson, J.P.; Moyes, D.L. Candida albicans Pathogenicity and Epithelial Immunity. PLoS Pathog. 2014, 10, e1004257. [Google Scholar] [CrossRef]
- Barnes, A.M.T.; Dale, J.L.; Chen, Y.; Manias, D.A.; Greenwood-Quaintance, K.E.; Karau, M.K.; Kashyap, P.C.; Patel, R.; Wells, C.L.; Dunny, G.M. Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence 2017, 8, 282–296. [Google Scholar] [CrossRef]
- Yang, W.; Yan, L.; Wu, C.; Zhao, X.; Tang, J. Fungal invasion of epithelial cells. Microbiol. Res. 2014, 169, 803–810. [Google Scholar] [CrossRef]
- Xu, H.; Sobue, T.; Bertolini, M.; Thompson, A.; Dongari-Bagtzoglou, A. Streptococcus oralis and Candida albicans synergistically activate μ-calpain to degrade e-cadherin from oral epithelial junctions. J. Infect. Dis. 2016, 214, 925–934. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Garsin, D.A.; Lorenz, M.C. Candida albicans and Enterococcus faecalis in the gut: Synergy in commensalism? Gut Microbes. 2013, 4, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell. 2011, 10, 168–173. [Google Scholar] [CrossRef]
- Ali, L.; Goraya, M.U.; Arafat, Y.; Ajmal, M.; Chen, J.L.; Yu, D. Molecular mechanism of quorum-sensing in Enterococcus faecalis: Its role in virulence and therapeutic approaches. Int J. Mol. Sci. 2017, 18, 960. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, N.; Varahan, S.; Gorman, M.J.; Palmer, K.L.; Zaidman-Remy, A.; Yokohata, R.; Nakayama, J.; Hancock, L.E.; Jacinto, A.; Gilmore, M.S.; et al. Drosophila host model reveals new Enterococcus faecalis quorum-sensing associated virulence factors. PLoS ONE 2013, 8, e64740. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnamoorthy, A.L.; Lemus, A.A.; Solomon, A.P.; Valm, A.M.; Neelakantan, P. Interactions between Candida albicans and Enterococcus faecalis in an Organotypic Oral Epithelial Model. Microorganisms 2020, 8, 1771. https://doi.org/10.3390/microorganisms8111771
Krishnamoorthy AL, Lemus AA, Solomon AP, Valm AM, Neelakantan P. Interactions between Candida albicans and Enterococcus faecalis in an Organotypic Oral Epithelial Model. Microorganisms. 2020; 8(11):1771. https://doi.org/10.3390/microorganisms8111771
Chicago/Turabian StyleKrishnamoorthy, Akshaya Lakshmi, Alex A. Lemus, Adline Princy Solomon, Alex M. Valm, and Prasanna Neelakantan. 2020. "Interactions between Candida albicans and Enterococcus faecalis in an Organotypic Oral Epithelial Model" Microorganisms 8, no. 11: 1771. https://doi.org/10.3390/microorganisms8111771
APA StyleKrishnamoorthy, A. L., Lemus, A. A., Solomon, A. P., Valm, A. M., & Neelakantan, P. (2020). Interactions between Candida albicans and Enterococcus faecalis in an Organotypic Oral Epithelial Model. Microorganisms, 8(11), 1771. https://doi.org/10.3390/microorganisms8111771