Isolation and Characterization of Cold-Tolerant Hyper-ACC-Degrading Bacteria from the Rhizosphere, Endosphere, and Phyllosphere of Antarctic Vascular Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Isolation of Culturable Heterotrophic Bacteria
2.3. Selection of Putative ACC-Degrading Bacteria
2.4. Screening of Putative ACC-Degrading Bacteria for Cold Tolerance
2.5. Quantification of ACC Deaminase Activity
2.6. Taxonomic Analysis of Selected Cold-Tolerant ACC-Degrading Bacteria
2.7. Statistical Analysis
3. Results
3.1. Culturable Bacterial Counts and Isolation of Putative ACC-Degrading Bacteria
3.2. Screening of Cold-Tolerant ACC-Degrading Bacteria and Detection of IRI Activity
3.3. ACC Deaminase Activity of Cold-Tolerant Bacterial Isolates
3.4. Identification of Cold-Tolerant Hyper-ACC-Degrading Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- FAO. Agriculture and Climate Change—Challenges and Opportunities at the Global and Local Level–Collaboration on Climate-Smart Agriculture; Licence: CC BY-NC-SA 3.0 IGO; Food and Agriculture Organization of the United Nations: Rome, Italy, 2019; p. 52. Available online: http://www.fao.org/3/CA3204EN/ca3204en.pdf (accessed on 27 July 2020).
- Becklin, M.K.; Anderson, J.T.; Gerhart, L.M.; Wadgymar, S.M.; Wessinger, C.A.; Ward, J.K. Examining plant physiological responses to climate change through an evolutionary lens. Plant Physiol. 2016, 172, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Classen, A.T.; Sundqvist, M.K.; Henning, J.A.; Newman, G.S.; Moore, J.A.M.; Cregger, M.A.; Moorhead, L.C.; Patterson, C.M. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere 2015, 6, 130. [Google Scholar] [CrossRef]
- Compant, S.; Van Der Heijden, M.; Sessitsch, A. Climate change effects on beneficial plant–microorganism interactions. FEMS Microbiol. Ecol. 2010, 73, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Sapre, S.; Gontia-Mishra, I.; Tiwari, S. ACC Deaminase-Producing Bacteria: A Key Player in Alleviating Abiotic Stresses in Plants. In Plant Growth Promoting Rhizobacteria for Agricultural Sustainability; Kumar, A., Meena, V., Eds.; Springer: Singapore, 2019; Volume 1, pp. 267–291. [Google Scholar]
- Barra, P.J.; Inostroza, N.G.; Acuña, J.J.; Mora, M.L.; Crowley, D.E.; Jorquera, M.A. Formulation of bacterial consortia from avocado (Persea americana Mill.) and their effect on growth, biomass and superoxide dismutase activity of wheat seedlings under salt stress. Appl. Soil. Ecol. 2016, 102, 80–91. [Google Scholar] [CrossRef] [Green Version]
- Barra, P.J.; Inostroza, N.G.; Mora, M.L.; Crowley, D.E.; Jorquera, M.A. Bacterial consortia inoculation mitigates the water shortage and salt stress in an avocado (Persea americana Mill.) nursery. Appl. Soil. Ecol. 2017, 111, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, G.; Duraivadivel, P.; Sharma, S.; Hariprasad, P. 1-Aminocyclopropane-1-carboxylic acid deaminase producing beneficial rhizobacteria ameliorate the biomass characters of Panicum maximumJacq. by mitigating drought and salt stress. Sci. Rep. 2018, 8, 17513. [Google Scholar] [CrossRef] [PubMed]
- Inostroza, N.G.; Barra, P.J.; Wick, L.Y.; Mora, M.L.; Jorquera, M.A. Effect of rhizobacterial consortia from undisturbed arid- and agro-ecosystems on wheat growth under different conditions. Lett. Appl. Microbiol. 2017, 64, 158–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acuña, J.J.; Jaisi, D.; Campos, M.; Mora, M.L.; Jorquera, M.A. ACCD-producing rhizobacteria from an Andean Altiplano native plant (Parastrephia quadrangularis) and their potential to alleviate salt stress in wheat seedling. Appl. Soil. Ecol. 2019, 136, 184–190. [Google Scholar] [CrossRef]
- Gupta, S.; Pandey, S. ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French Bean (Phaseolus vulgaris) pants. Front. Microbiol. 2019, 10, 1506. [Google Scholar] [CrossRef]
- El-Tarabily, K.A.; AlKhajeh, A.S.; Ayyash, M.M.; Alnuaimi, L.H.; Sham, A.; ElBaghdady, K.Z.; Tariq, S.; AbuQamar, S.F. Growth promotion of Salicornia bigelovii by Micromonospora chalcea UAE1, an endophytic 1-aminocyclopropane-1-carboxylic acid deaminase-producing actinobacterial isolate. Front. Microbiol. 2019, 10, 1694. [Google Scholar] [CrossRef]
- Mathew, B.T.; Torky, Y.; Amin, A.; Mourad, A.H.I.; Ayyash, M.M.; El-Keblawy, A.; Hilal-Alnaqbi, A.; AbuQamar, S.F.; El-Tarabily, K.A. Halotolerant marine rhizosphere-competent actinobacteria promoteSalicornia bigeloviigrowth and seed production using seawater irrigation. Front. Microbiol. 2020, 11, 552. [Google Scholar] [CrossRef] [PubMed]
- Papagiannaki, K.; Lagouvardos, K.; Kotroni, V.; Papagiannakis, G. Agricultural losses related to frost events: Use of the 850 hPa level temperature as an explanatory variable of the damage cost. Nat. Hazards Earth Syst. Sci. 2014, 14, 2375–2381. [Google Scholar] [CrossRef] [Green Version]
- Jorquera, M.A.; Graether, S.P.; Maruyama, F. Editorial: Bioprospecting and Biotechnology of Extremophiles. Front. Bioeng. Biotechnol. 2019, 7, 204. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Montero, K.; Barrientos, L. Advances in Antarctic Research for Antimicrobial Discovery: A Comprehensive Narrative Review of Bacteria from Antarctic Environments as Potential Sources of Novel Antibiotic Compounds Against Human Pathogens and Microorganisms of Industrial Importance. Antibiotics 2018, 7, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cid, F.P.; Rilling, J.I.; Graether, S.P.; Bravo, L.A.; Mora, M.L.; Jorquera, M.A. Properties and biotechnological applications of ice-binding proteins in bacteria. FEMS Microbiol. Lett. 2016, 363, fnw099. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Goel, R. Principles, Applications and Future Aspects of Cold-Adapted PGPR. In Plant-Bacteria Interactions Strategies and Techniques to Promote Plant Growth; Ahmad, I., Pichtel, J., Hayat, S., Eds.; Wiley Online Library: Weinheim, Germany, 2008; Volume 1, pp. 195–212. [Google Scholar]
- Suyal, D.C.; Shukla, A.; Goel, R. Growth promotory potential of the cold adapted diazotroph Pseudomonas migulae S10724 against native green gram (Vigna radiate (L.) Wilczek). 3 Biotech 2014, 4, 665–668. [Google Scholar] [CrossRef] [Green Version]
- Cid, F.P.; Inostroza, N.G.; Graether, S.P.; Bravo, L.A.; Jorquera, M.L. Bacterial community structures and ice recrystallization inhibition activity of bacteria isolated from the phyllosphere of the Antarctic vascular plant Deschampsia antarctica. Polar Biol. 2017, 40, 1319–1331. [Google Scholar] [CrossRef]
- Cid, F.P.; Maruyama, F.; Murase, K.; Graether, S.P.; Larama, G.; Bravo, L.A.; Jorquera, M.A. Draft genome sequences of bacteria isolated from the Deschampsia Antarctica phyllosphere. Extremophiles 2018, 22, 537–552. [Google Scholar] [CrossRef]
- Acuña-Rodríguez, I.S.; Hansen, H.; Gallardo-Cerda, J.; Atala, C.; Molina-Montenegro, M.A. Antarctic Extremophiles: Biotechnological Alternative to Crop Productivity in Saline Soils. Front. Bioeng. Biotechnol. 2019, 7, 22. [Google Scholar] [CrossRef]
- Gallardo-Cerda, J.; Levihuan, J.; Lavín, P.; Oses, R.; Atala, C.; Torres-Díaz, C.; Molina-Montenegro, M.A. Antarctic rhizobacteria improve salt tolerance and physiological performance of the Antarctic vascular plants. Polar Biol. 2018, 41, 1973–1982. [Google Scholar] [CrossRef]
- Zhang, Q.; Acuña, J.J.; Inostroza, N.G.; Durán, P.; Mora, M.L.; Sadowsky, M.J.; Jorquera, M.A. Niche differentiation in the composition, predicted function, and co-occurrence networks in bacterial communities associated with Antarctic vascular plants. Front. Microbiol. 2020, 11, 1036. [Google Scholar] [CrossRef] [PubMed]
- Lagos, L.; Navarrete, O.; Maruyama, F.; Crowley, D.E.; Cid, F.; Mora, M.L.; Jorquera, M.A. Bacterial community structures in rhizosphere microsites of ryegrass (Lolium perennevar. Nui) as revealed by pyrosequencing. Biol. Fert. Soils 2014, 50, 1253–1266. [Google Scholar] [CrossRef]
- Jorquera, M.A.; Inostroza, N.G.; Lagos, L.M.; Barra, P.J.; Marileo, L.G.; Rilling, J.I.; Campos, D.C.; Crowley, D.E.; Richardson, A.E.; Mora, M.L. Bacterial community structure and detection of putative plant growth-promoting rhizobacteria associated with plants grown in Chilean agro-ecosystems and undisturbed ecosystems. Biol. Fertil. Soils 2014, 50, 1141–1153. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upson, R.; Read, D.J.; Newsham, K.K. Nitrogen form influences the response of Deschampsia antarctica to dark septate root endophytes. Mycorrhiza 2009, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Santiago, I.F.; Rosa, C.A.; Rosa, L.H. Endophytic symbiont yeasts associated with the Antarctic angiosperms Deschampsia Antarctica and Colobanthus quitensis. Polar Biol. 2017, 40, 177–183. [Google Scholar] [CrossRef]
- Vanderstraeten, L.; Van Der Straeten, D. Accumulation and transport of 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: Current status, considerations for future research and agronomic applications. Front. Plant. Sci. 2017, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Vanderstraeten, L.; Depaepe, T.; Bertrand, S.; Van Der Straeten, D. The ethylene precursor ACC affects early vegetative development independently of ethylene signaling. Front. Plant Sci. 2019, 10, 1591. [Google Scholar] [CrossRef] [Green Version]
- Danish, S.; Zafar-ul-Hye, M. Co-application of ACC-deaminase producing PGPR and timber-waste biochar improves pigments formation, growth and yield of wheat under drought stress. Sci. Rep. 2019, 9, 5999. [Google Scholar] [CrossRef] [Green Version]
- Danish, S.; Zafar-ul-Hye, M.; Mohsin, F.; Hussain, M. ACC-deaminase producing plant growth promoting rhizobacteria and biochar mitigate adverse effects of drought stress on maize growth. PLoS ONE 2020, 15, e0230615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glick, B.R. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 2005, 251, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Rossi, M.J. The role of rhizobial ACC deaminase in the nodulation process of leguminous plants. Int. J. Agron. 2016, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Prashar, P.; Kapoor, N.; Sachdeva, S. Rhizosphere: Its structure, bacterial diversity and significance. Rev. Environ. Sci. Biotechnol. 2013, 13, 63–77. [Google Scholar] [CrossRef]
- Barrientos-Díaz, L.; Gidekel, M.; Gutiérrez-Moraga, A. Characterization of rhizospheric bacteria isolated from Deschampsia Antarctica Desv. World J. Microbiol, Biotechnol. 2008, 24, 2289. [Google Scholar] [CrossRef]
- Smith, J.J.; Ah Tow, L.; Stafford, W.; Cary, C.; Cowan, D. Bacterial diversity in three different Antarctic cold desert mineral soils. Microb. Ecol. 2006, 51, 413–442. [Google Scholar] [CrossRef]
- Teixeira, L.C.; Peixoto, R.S.; Cury, J.C.; Sul, W.J.; Pellizari, V.H.; Tiedje, J.; Rosado, A.S. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J. 2010, 4, 989–1001. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.H.; Crowley, D.E.; Borneman, J.; Keen, N.T. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 2001, 98, 3889–3894. [Google Scholar] [CrossRef] [Green Version]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef] [Green Version]
- Nissinen, R.M.; Männistö, M.K.; Elsas, J.D. Endophytic bacterial communities in three arctic plants from low arctic fell tundra are cold-adapted and host-plant specific. FEMS Microbiol. Ecol. 2012, 82, 510–522. [Google Scholar] [CrossRef]
- Kawahara, H.; Nakano, Y.; Omiya, K.; Muryoi, N.; Nishikawa, J.; Obata, H. Production of two types of ice crystal-controlling proteins in Antarctic bacterium. J. Biosci. Bioeng. 2004, 98, 220–222. [Google Scholar] [CrossRef]
- Raymond, J. Dependence on epiphytic bacteria for freezing protection in an Antarctic moss, Bryum argenteum. Environ. Microbiol. Rep. 2015, 8, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.L. Antarctic moss is home to many epiphytic bacteria that secrete antifreeze proteins. Environ. Microbiol. Rep. 2016, 8, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.J.M.; Miranda, K.R.; Lobo, L.A.; Granato, A.; de Carvalho Maalouf, P.; de Jesus, H.E.; Rachid, C.T.C.C.; Moraes, S.R.; dos Santos, H.F.; Peixoto, R.S.; et al. Antarctic strict anaerobic microbiota from Deschampsia antarctica vascular plants rhizosphere reveals high ecology and biotechnology relevance. Extremophiles 2016, 20, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Molina-Montenegro, M.A.; Ballesteros, G.I.; Castro-Nallar, E.; Meneses, C.; Torres-Díaz, C.; Gallardo-Cerda, J. Metagenomic exploration of soils microbial communities associated to Antarctic vascular plants. PeerJ Preprints 2018, 6, e26508v1. [Google Scholar]
- Morgan-Kiss, R.M.; Priscu, J.C.; Pocock, T.; Gudynaite-Savitch, L.; Huner, N.P. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 2006, 70, 222–252. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, F.X.; Rossi, M.J.; Soares, C.R.F.S.; McConkey, B.J.; Glick, B.R. New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE 2014, 9, e99168. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, F.X.; Glick, B.R.; Rossi, M.J. Isolation and characterization of novel soil- and plant-associated bacteria with multiple phytohormone-degrading activities using a targeted methodology. Access Microbiol. 2019, 1, e000053. [Google Scholar] [CrossRef]
- Subramanian, P.; Kim, K.; Krishnamoorthy, R.; Mageswari, A.; Selvakumar, G.; Sa, T. Cold stress tolerance in psychrotolerant soil bacteria and their conferred chilling resistance in tomato (Solanum lycopersicum Mill.) under low temperatures. PLoS ONE 2016, 11, e0161592. [Google Scholar] [CrossRef]
- Berríos, G.; Cabrera, G.; Gidekel, M.; Gutiérrez-Moraga, A. Characterization of a novel Antarctic plant growth-promoting bacterial strain and its interaction with Antarctic hair grass (Deschampsia antarctica Desv). Polar Biol. 2013, 36, 349–362. [Google Scholar] [CrossRef]
- Orellana-Saez, M.; Pacheco, N.; Costa, J.I.; Mendez, K.N.; Miossec, M.J.; Meneses, C.; Castro-Nallar, E.; Marcoleta, A.E.; Poblete-Castro, I. In-depth genomic and phenotypic characterization of the Antarctic psychrotolerant strain Pseudomonassp. MPC6 reveals unique metabolic features, plasticity, and biotechnological potential. Front Microbiol. 2019, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.A.; Newsham, K.K.; Thorne, M.A.S.; Calvo-Bado, L.; Krsek, M.; Laskaris, P.; Hodson, A.; Wellington, E.M. Metagenomic analysis of a southern maritime Antarctic soil. Front. Microbiol. 2012, 3, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivaji, S.; Reddy, G.S.N.; Aduri, R.P.; Kutty, R.; Ravenschlag, K. Bacterial diversity of a soil sample from Schirmacher Oasis, Antarctica. Cell Mol. Biol. 2004, 50, 525–536. [Google Scholar] [PubMed]
- Da Silva, A.C.; Rachid, C.T.C.; de Jesus, H.E.; Rosado, A.S.; Peixoto, R.S. Predicting the biotechnological potential of bacteria isolated from Antarctic soils, including the rhizosphere of vascular plants. Polar Biol. 2017, 40, 1393–1407. [Google Scholar] [CrossRef]


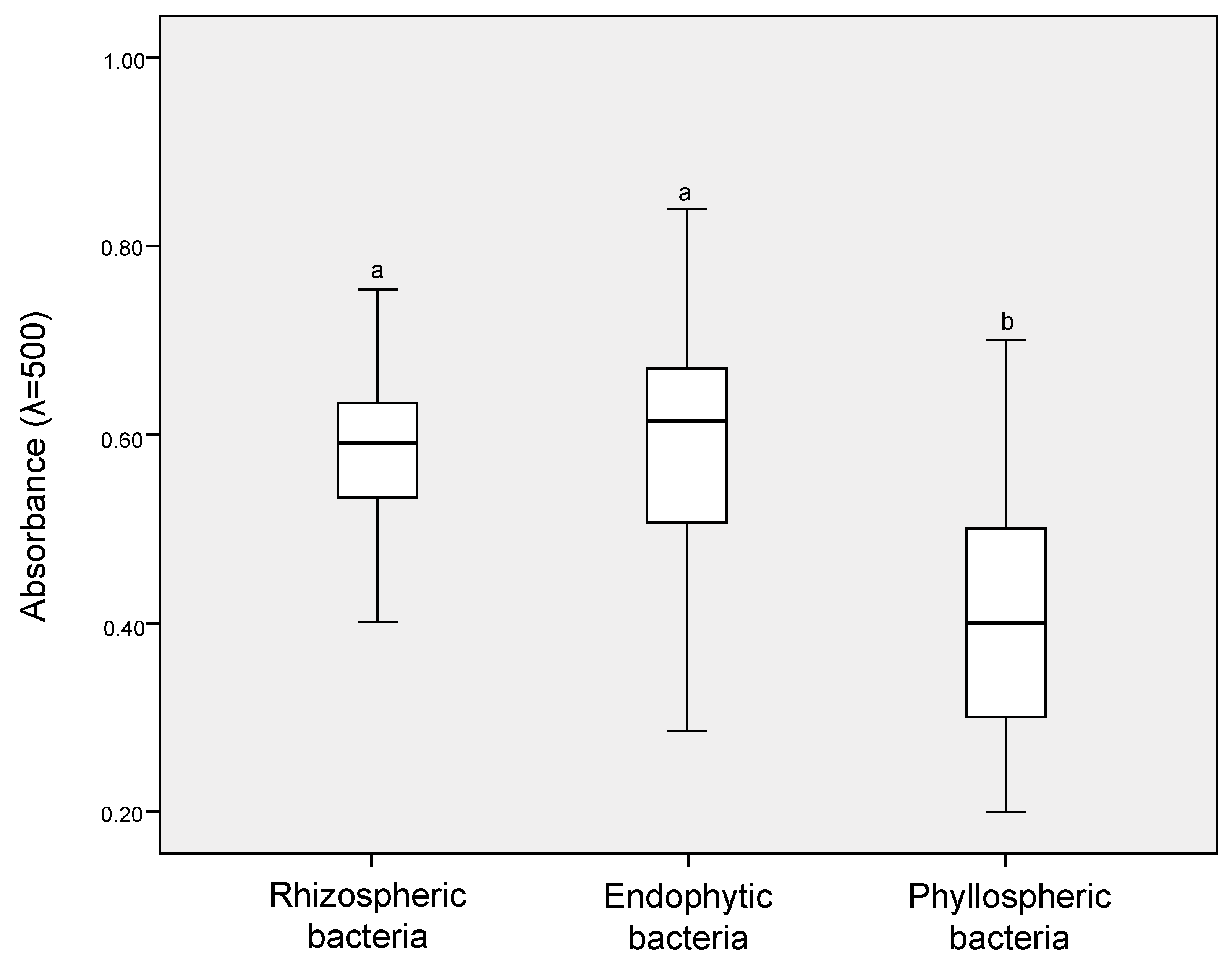
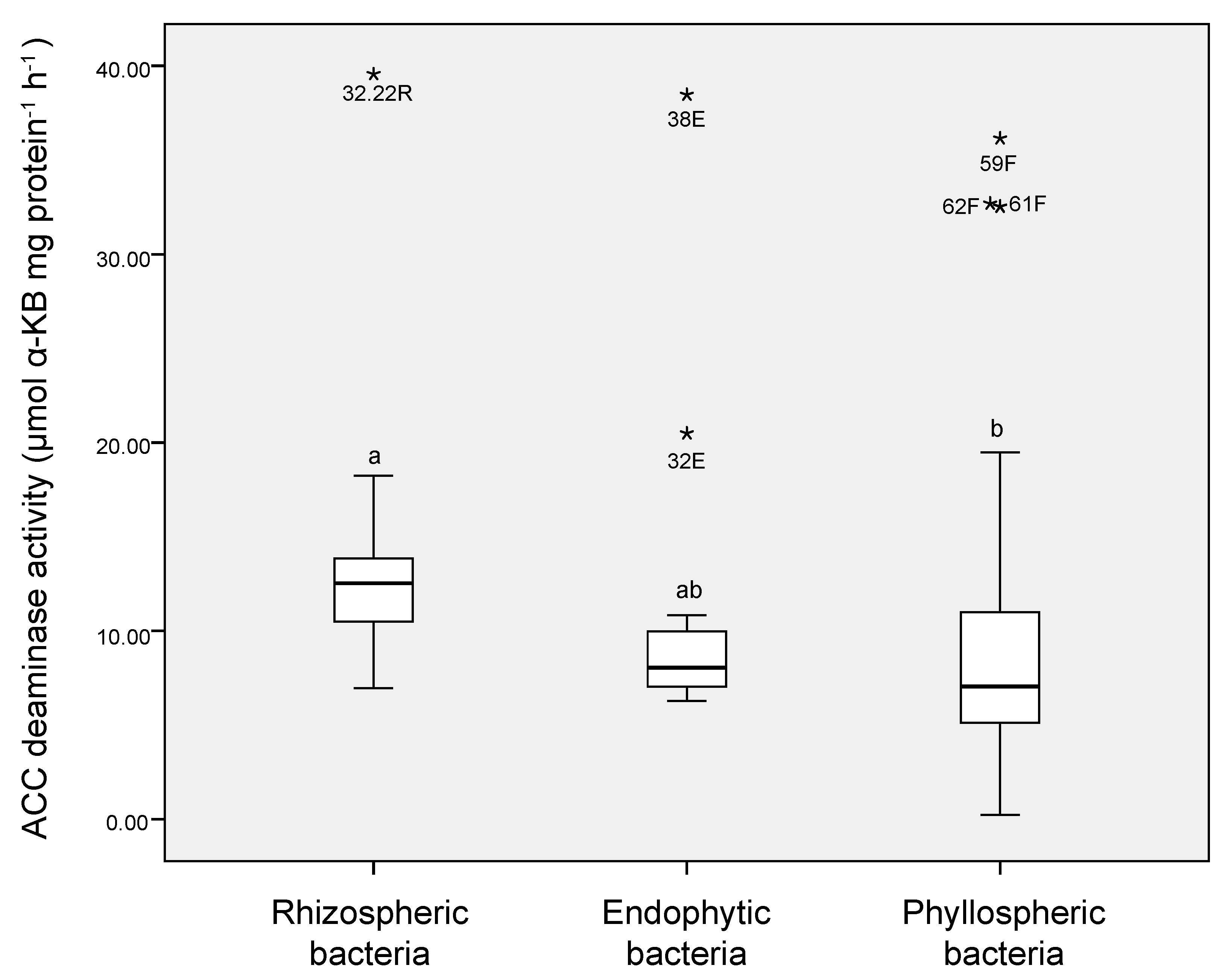
| Niche | No. of Total Isolates | Growth in DF + ACC | ERIC-PCR Genotyping | ||
|---|---|---|---|---|---|
| No. of Isolates | % | No. of Isolates | % | ||
| Endosphere | 86 | 45 | 52.32 | 23 | 51.11 |
| Phyllosphere | 89 | 52 | 58.42 | 37 | 71.15 |
| Rhizosphere | 403 | 83 | 20.59 | 41 | 49.40 |
| Total | 578 | 180 | 31.14 | 101 | 56.11 |
| Niche/Isolate | Plant | Closest Relatives or Cloned Sequences (Accession No.; % of Identity) * | Accession No. | ACC-Deaminase Ɏ | IRI § |
|---|---|---|---|---|---|
| Endosphere | |||||
| 38E | D. antarctica | Pseudomonas sp. PCH176 from soil of Himalayan region (MF774162; 100%) | MT786310 | 38.46 ¥ a | 0.80 a |
| 32E | D. antarctica | Ewingella sp. strain 3–24 from leaves collected from cold water pools of Huanglong park (KX378962; 99%) | MT786318 | 20.49 b | 0.57 b |
| Phyllosphere | |||||
| 59F | D. antarctica | Pseudomonas sp. L3_E02 from phyllosphere of Carpinus betulus L (MK216837; 100%) | MT786317 | 36.15 a | 0.76 a |
| 62F | D. antarctica | Pseudomonas gessardii strain P_B71/5 from Baltic sea (MT626814; 98%) | MT786316 | 32.53 a | 0.49 b |
| 61F | C. quitensis | Serratia marcescens strain BRM 046341 from corn stigma (MK461848; 100%) | MT786319 | 32.51 a | 0.55 b |
| M15-3A | D. antarctica | Rahnella sp. strain UASWS1864 from oak in Geneva (MK513744; 99%) | MT786312 | 14.08 b | 0.86 a |
| Rhizosphere | |||||
| 32.22R | D. antarctica | Serratia sp. JSC-N623-1 from soil (JF958141; 99%) | MT786315 | 39.56 a | 0.81 a |
| 32.17R | C. quitensis | Pseudomonas sp. strain C14-10 from sediment (MT255246; 99%) | MT786314 | 18.24 b | 0.59 b |
| 10.2R | D. antarctica | Staphylococcus sp. CSA7 from soil (HQ437165.1; 99%) | MT786311 | 16. 34 b | 0.57 b |
| 9.9R | D. antarctica | Staphylococcus condimenti strain HBUAS56223 from vegetables (MT229638; 100%) | MT786321 | 16.12 b | 0.59 b |
| 29.13R | C. quitensis | Enterobacter sp. CV87 from soil (KJ482866; 99%) | MT786320 | 13.89 b | 0.65 b |
| 21.24R | C. quitensis | Pseudomonas sp. strain PAMC 27327 from Antarctic soil (MT555366; 99%) | MT786313 | 13.21 b | 0.66 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araya, M.A.; Valenzuela, T.; Inostroza, N.G.; Maruyama, F.; Jorquera, M.A.; Acuña, J.J. Isolation and Characterization of Cold-Tolerant Hyper-ACC-Degrading Bacteria from the Rhizosphere, Endosphere, and Phyllosphere of Antarctic Vascular Plants. Microorganisms 2020, 8, 1788. https://doi.org/10.3390/microorganisms8111788
Araya MA, Valenzuela T, Inostroza NG, Maruyama F, Jorquera MA, Acuña JJ. Isolation and Characterization of Cold-Tolerant Hyper-ACC-Degrading Bacteria from the Rhizosphere, Endosphere, and Phyllosphere of Antarctic Vascular Plants. Microorganisms. 2020; 8(11):1788. https://doi.org/10.3390/microorganisms8111788
Chicago/Turabian StyleAraya, Macarena A., Tamara Valenzuela, Nitza G. Inostroza, Fumito Maruyama, Milko A. Jorquera, and Jacquelinne J. Acuña. 2020. "Isolation and Characterization of Cold-Tolerant Hyper-ACC-Degrading Bacteria from the Rhizosphere, Endosphere, and Phyllosphere of Antarctic Vascular Plants" Microorganisms 8, no. 11: 1788. https://doi.org/10.3390/microorganisms8111788
APA StyleAraya, M. A., Valenzuela, T., Inostroza, N. G., Maruyama, F., Jorquera, M. A., & Acuña, J. J. (2020). Isolation and Characterization of Cold-Tolerant Hyper-ACC-Degrading Bacteria from the Rhizosphere, Endosphere, and Phyllosphere of Antarctic Vascular Plants. Microorganisms, 8(11), 1788. https://doi.org/10.3390/microorganisms8111788







