Aeromonas hydrophila RIT668 and Citrobacter portucalensis RIT669—Potential Zoonotic Pathogens Isolated from Spotted Turtles
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Background
2.2. Characterization and Identification: Biochemical Assay and 16S rDNA Amplification
2.3. Supplemental Bioinformatics Methods
2.4. Antimicrobial Susceptibility/Multidrug Resistance Based on Disc Diffusion Assays
2.4.1. Disc Diffusion with Pre-Loaded Discs
2.4.2. Disc Diffusion with Stock Antibiotic Concentrations
2.5. Biofilm Formation and Biofilm Growth Check
2.6. MIC and MBEC Assay Device
2.7. Scanning Electron Microscopy (SEM)
2.8. Identification of Quorum Sensing Signals of the Acyl-Homoserine Lactone (AHL) Class
2.9. Bioinformatics Analysis of AHL-Related Genes
2.10. Computational Methods
3. Results
3.1. A. hydrophila
3.1.1. Growth
3.1.2. Taxonomy
3.1.3. Antimicrobial susceptibility screening with preloaded antibiotic discs
3.1.4. Antimicrobial Susceptibility Screening Using the Stock Concentration Method
3.1.5. MIC and MBEC
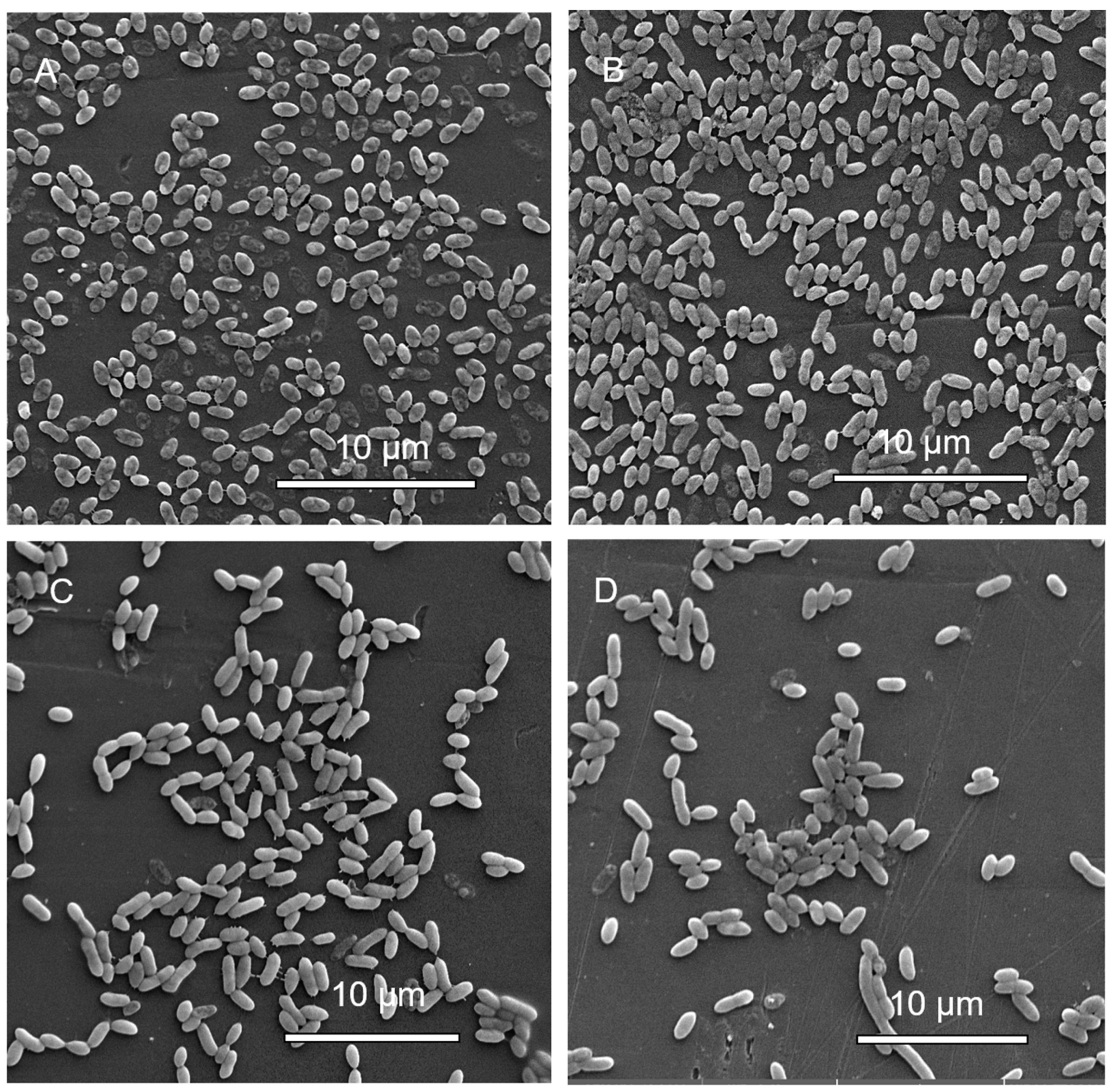
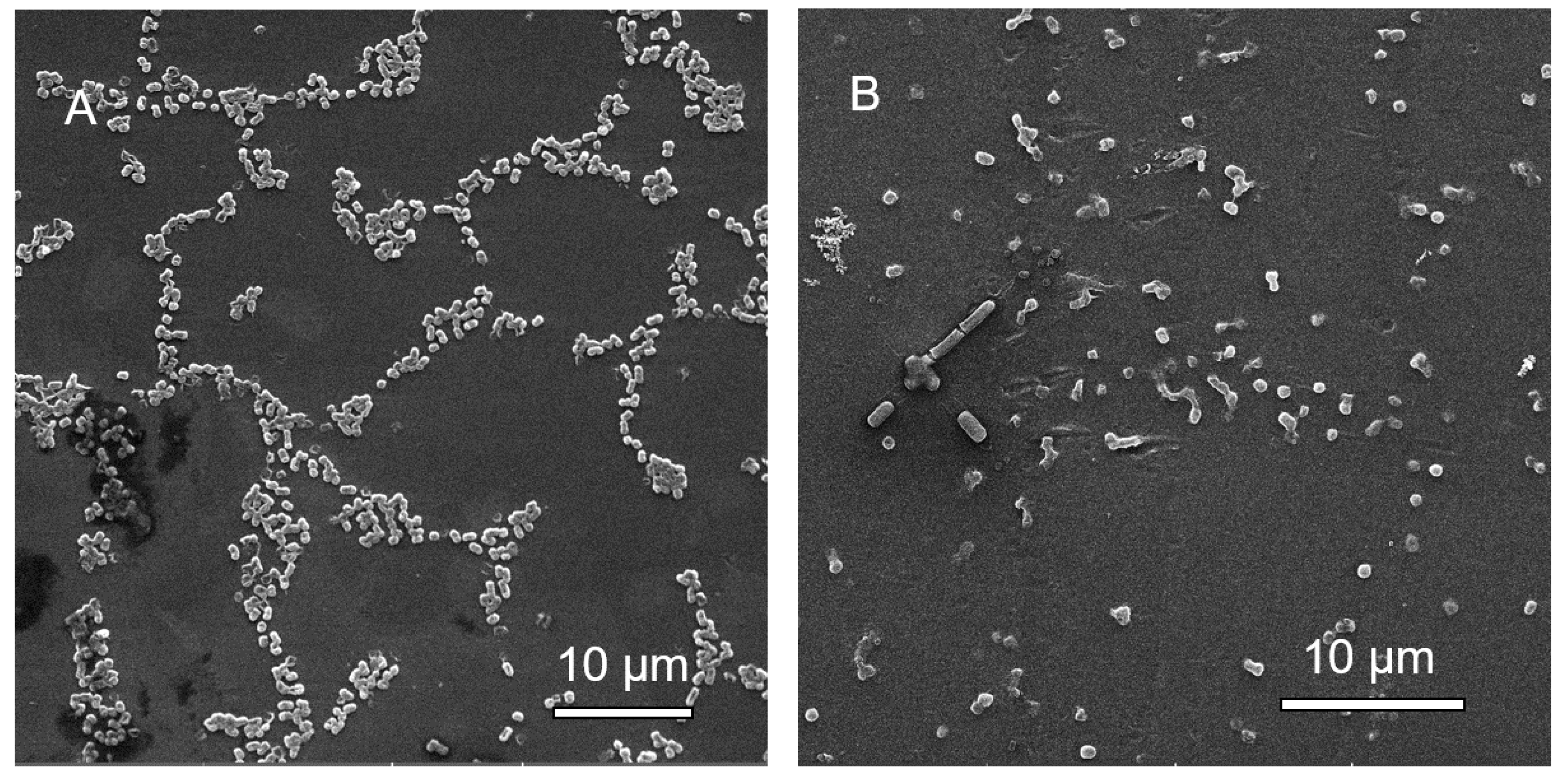
3.1.6. Scanning Electron Microscopy (SEM) Analysis of MBEC Samples
3.1.7. Detection of Quorum Sensing
3.2. C. portucalensis
3.2.1. Growth
3.2.2. Taxonomy
3.2.3. Antimicrobial Susceptibility Screening with Preloaded Antibiotic Discs
3.2.4. Antimicrobial Susceptibility Screening Using the Stock Concentration Method
3.2.5. MIC and MBEC
3.2.6. Scanning Electron Microscopy (SEM) Analysis of MBEC Samples
3.2.7. Detection of Quorum Sensing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cooper, M.A.; Shlaes, D. Fix the antibiotics pipeline. Nature 2011, 472, 32. [Google Scholar] [CrossRef]
- Wright, G.D. The antibiotic resistome. Expert Opin. Drug Discov. 2010, 5, 779–788. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.G.; Zazula, G.D.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nat. Cell Biol. 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Igbinosa, I.H.; Igumbor, E.U.; Aghdasi, F.; Tom, M.; Okoh, A.I. EmergingAeromonasSpecies Infections and Their Significance in Public Health. Sci. World J. 2012, 2012, 625023. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, K.P.; Ranjan, N. Citrobacter: An emerging health care associated urinary pathogen. Urol. Ann. 2013, 5, 313–314. [Google Scholar] [PubMed]
- Kregiel, D.; Rygala, A.; Kolesinska, B.; Nowacka, M.; Herc, A.S.; Kowalewska, A. Antimicrobial and Antibiofilm N-acetyl-L-cysteine Grafted Siloxane Polymers with Potential for Use in Water Systems. Int. J. Mol. Sci. 2019, 20, 2011. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Chen, L.; Zhou, S.; Li, H.; Long, J.; Yao, F.; Zhuang, Y.; Zhang, Z.; Huang, Y.; Duan, K. Co-existence of Citrobacter freundii exacerbated Pseudomonas aeruginosa infection in vivo. Int. J. Med. Microbiol. 2020, 310, 151379. [Google Scholar] [CrossRef]
- Pereira, A.L.; Silva, T.N.; Gomes, A.C.; Araujo, A.C.G.; Giugliano, L.G. Diarrhea-associated biofilm formed by enteroaggregative Escherichia coli and aggregative Citrobacter freundii: A consortium mediated by putative F pili. BMC Microbiol. 2010, 10, 57. [Google Scholar] [CrossRef]
- McCormick, A.; Hoellein, T.J.; Mason, S.A.; Schluep, J.; Kelly, J.J. Microplastic is an Abundant and Distinct Microbial Habitat in an Urban River. Environ. Sci. Technol. 2014, 48, 11863–11871. [Google Scholar] [CrossRef]
- Heo, G.-J.; Hossain, S.; Wimalasena, S. Virulence Factors and Antimicrobial Resistance Pattern of Citrobacter freundii Isolated from Healthy Pet Turtles and their Environment. Asian J. Anim. Veter- Adv. 2017, 12, 10–16. [Google Scholar] [CrossRef]
- Chung, T.; Yi, S.; Kim, B.; Kim, W.; Shin, G. Identification and antibiotic resistance profiling of bacterial isolates from septicaemic soft-shelled turtles (Pelodiscus sinensis). Vet. Med. 2017, 62, 169–177. [Google Scholar] [CrossRef]
- Wimalasena, S.H.M.P.; Shin, G.-W.; Hossain, S.; Heo, G.-J. Potential enterotoxicity and antimicrobial resistance pattern of Aeromonas species isolated from pet turtles and their environment. J. Vet. Med. Sci. 2017, 79, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Pace, A.; Dipineto, L.; Fioretti, A.; Hochscheid, S. Loggerhead sea turtles as sentinels in the western Mediterranean: Antibiotic resistance and environment-related modifications of Gram-negative bacteria. Mar. Pollut. Bull. 2019, 149, 110575. [Google Scholar] [CrossRef] [PubMed]
- Al-Bahry, S.; Mahmoud, I.; Al-Zadjali, M.; Elshafie, A.; Al-Harthy, A.; Al-Alawi, W. Antibiotic resistant bacteria as bio-indicator of polluted effluent in the green turtles, Chelonia mydas in Oman. Mar. Environ. Res. 2011, 71, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, P.M.; Conti, L. Links among Human Health, Animal Health, and Ecosystem Health. Annu. Rev. Public Health 2013, 34, 189–204. [Google Scholar] [CrossRef]
- Rosen, G.E.; Smith, K.F. Summarizing the Evidence on the International Trade in Illegal Wildlife. EcoHealth 2010, 7, 24–32. [Google Scholar] [CrossRef]
- Thomas, S.G.; Glover, M.A.; Parthasarathy, A.; Wong, N.H.; Shipman, P.A.; Hudson, A.O. Expression of a Shiga-Like Toxin during Plastic Colonization by Two Multidrug-Resistant Bacteria, Aeromonas hydrophila RIT668 and Citrobacter freundii RIT669, Isolated from Endangered Turtles (Clemmys guttata). Microorganisms 2020, 8, 1172. [Google Scholar] [CrossRef]
- McCoy, R.H.; Seidler, R.J. Potential Pathogens in the Environment: Isolation, Enumeration, and Identification of Seven Genera of Intestinal Bacteria Associated with Small Green Pet Turtles1. Appl. Microbiol. 1973, 25, 534–538. [Google Scholar] [CrossRef]
- Esteve, C.; Alcaide, E.; Giménez, M.J. Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): Diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 137–145. [Google Scholar] [CrossRef]
- Marchandin, H.; Godreuil, S.; Darbas, H.; Jean-Pierre, H.; Jumas-Bilak, E.; Chanal, C.; Bonnet, R. Extended-spectrum beta-lactamase TEM-24 in an Aeromonas clinical strain: Acquisition from the prevalent Enterobacter aerogenes clone in France. Antimicrob. Agents Chemother. 2003, 47, 3994–3995. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed]
- Epiotrowska, M.; Popowska, M. Insight into the mobilome of Aeromonas strains. Front. Microbiol. 2015, 6, 494. [Google Scholar] [CrossRef]
- Ribeiro, T.G.; Ribeiro-Gonçalves, B.; Da Silva, M.S.; Novais, Â.; Machado, E.; Carriço, J.A.; Peixe, L. Citrobacter portucalensis sp. nov., isolated from an aquatic sample. Int. J. Syst. Evol. Microbiol. 2017, 67, 3513–3517. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; Sultana, M.; Hossain, M.A. Complete genome arrangement revealed the emergence of a poultry origin superbug Citrobacter portucalensis strain NR-12. J. Glob. Antimicrob. Resist. 2019, 18, 126–129. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Rathje, J.; Habermann, D.; Brinks, E.; Cho, G.-S.; Franz, C.M. Draft Genome Sequence of Multidrug-Resistant Strain Citrobacter portucalensis MBTC-1222, Isolated from Uziza (Piper guineense ) Leaves in Nigeria. Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Bai, L.; Xia, S.; Lan, R.; Liu, L.; Ye, C.; Wang, Y.; Jin, D.; Cui, Z.; Jing, H.; Xiong, Y.; et al. Isolation and Characterization of Cytotoxic, Aggregative Citrobacter freundii. PLoS ONE 2012, 7, e33054. [Google Scholar] [CrossRef]
- Anderson, M.T.; Mitchell, L.A.; Zhao, L.; Mobley, H.L.T. Citrobacter freundii fitness during bloodstream infection. Sci. Rep. 2018, 8, 11792. [Google Scholar] [CrossRef]
- Liu, L.; Chen, D.; Liu, L.; Lan, R.; Hao, S.; Jin, W.; Sun, H.; Wang, Y.; Liang, Y.; Xu, J. Genetic Diversity, Multidrug Resistance, and Virulence of Citrobacter freundii From Diarrheal Patients and Healthy Individuals. Front. Cell. Infect. Microbiol. 2018, 8, 233. [Google Scholar] [CrossRef]
- Mohanty, S.; Singhal, R.; Sood, S.; Dhawan, B.; Kapil, A.; Das, B.K. Citrobacter infections in a tertiary care hospital in Northern India. J. Infect. 2007, 54, 58–64. [Google Scholar] [CrossRef]
- Samonis, G.; Karageorgopoulos, D.E.; Kofteridis, D.P.; Matthaiou, D.K.; Sidiropoulou, V.; Maraki, S.; Falagas, M.E. Citrobacter infections in a general hospital: Characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 61–68. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Wong, W.-W.; Fung, C.-P.; Yu, K.-W.; Liu, C.-Y. Clinical features and antimicrobial susceptibility trends in Citrobacter freundii bacteremia. J. Microbiol. Immunol. Infect. 2002, 35, 109–114. [Google Scholar] [PubMed]
- Liu, L.-H.; Wang, N.-Y.; Wu, A.Y.-J.; Lin, C.-C.; Lee, C.-M.; Liu, C.-P. Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 2018, 51, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Joaquin, A.; Khan, S.; Russell, N.; Al Fayez, N. Neonatal Meningitis and Bilateral Cerebellar Abscesses due to Citrobacter freundii. Pediatr. Neurosurg. 1991, 17, 23–24. [Google Scholar] [CrossRef] [PubMed]
- Samonis, G.; Anaissie, E.; Elting, L.; Bodey, G.P. Review ofCitrobacter bacteremia in cancer patients over a sixteen-year period. Eur. J. Clin. Microbiol. Infect. Dis. 1991, 10, 479–485. [Google Scholar] [CrossRef]
- Aminharati, F.; Ehrampoush, M.H.; Dallal, M.M.S.; Yaseri, M.; Tafti, A.A.D.; Rajabi, Z. Citrobacter freundii Foodborne Disease Outbreaks Related to Environmental Conditions in Yazd Province, Iran. Iran J. Public Health 2019, 48, 1099–1105. [Google Scholar]
- Tschape, H.; Prager, R.; Streckel, W.; Fruth, A.; Tietze, E.; Böhme, G. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: Green butter as the infection source. Epidemiol. Infect. 1995, 114, 441–450. [Google Scholar] [CrossRef]
- Pardia, S.N.; Verma, I.C.; Deb, M.; Bhujwala, R.A. An outbreak of diarrhea due to citrobacter freundii in a neonatal special care nursery. Indian J. Pediatr. 1980, 47, 81–84. [Google Scholar] [CrossRef]
- Chen, Y.; Brook, T.C.; Alcon-Giner, C.; Clarke, P.; Hall, L.J.; Hoyles, L. Draft Genome Sequences of Citrobacter freundii and Citrobacter murliniae Strains Isolated from the Feces of Preterm Infants. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef]
- Shao, Y.; Xiong, Z.; Li, X.; Hu, L.; Shen, J.; Li, T.; Hu, F.; Chen, S. Prevalence of plasmid-mediated quinolone resistance determinants in Citrobacter freundii isolates from Anhui province, PR China. J. Med. Microbiol. 2011, 60, 1801–1805. [Google Scholar] [CrossRef]
- Park, Y.-J.; Yu, J.K.; Lee, S.; Oh, E.J.; Woo, G.-J. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 2007, 60, 868–871. [Google Scholar] [CrossRef]
- Park, Y.-J.; Park, S.Y.; Oh, E.-J.; Park, J.-J.; Lee, K.-Y.; Woo, G.-J.; Lee, K.-W. Occurrence of extended-spectrum β-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn. Microbiol. Infect. Dis. 2005, 51, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Moland, E.S.; Hanson, N.D.; Black, J.A.; Hossain, A.; Song, W.; Thomson, K.S. Prevalence of Newer -Lactamases in Gram-Negative Clinical Isolates Collected in the United States from 2001 to 2002. J. Clin. Microbiol. 2006, 44, 3318–3324. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Lee, J.E.; Park, S.J.; Kim, M.N.; Choo, E.J.; Kwak, Y.G.; Jeong, J.Y.; Woo, J.H.; Kim, N.J.; Kim, Y.S. Prevalence, microbiology, and clinical characteristics of extended-spectrum beta-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- ASTM International. ASTM E2799-17, Standard Method for Testing Disinfectant Efficacy against Pseudomonas Aeruginosa Biofilm Using the MBEC Assay; ASTM International: West Conshohocken, PA, USA, 2017. [Google Scholar] [CrossRef]
- Olson, M.E.; Ceri, H.; Morck, D.W.; Buret, A.G.; Read, R.R. Biofilm bacteria: Formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 2002, 66, 86–92. [Google Scholar]
- Donlan, R. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Hancock, V.; Ferriã¨res, L.; Klemm, P. Biofilm formation by asymptomatic and virulent urinary tract infectiousEscherichia colistrains. FEMS Microbiol. Lett. 2007, 267, 30–37. [Google Scholar] [CrossRef]
- Shunmugaperumal, T. Biofilm Eradication and Prevention: A Pharmaceutical Approach to Medical Device Infections; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010. [Google Scholar]
- Jaff, M.R. Advances in the management of patients with vascular disease. Expert Rev. Cardiovasc. Ther. 2012, 10, 151–153. [Google Scholar] [CrossRef]
- Mani, G.; Feldman, M.D.; Patel, D.; Agrawal, C.M. Coronary stents: A materials perspective. Biomaterials 2007, 28, 1689–1710. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- De Beer, D.; Stoodley, P.; Roe, F.; Lewandowski, Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 1994, 43, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Hudson, I. The efficacy of intranasal mupirocin in the prevention of staphylococcal infections: A review of recent experience. J. Hosp. Infect. 1994, 27, 81–98. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R.; Chandler, J.R.; Blodgett, J.A.V.; Lima, P.S.; Duerkop, B.A.; Oinuma, K.-I.; Greenberg, E.P.; Clardy, J. Quorum-Sensing-Regulated Bactobolin Production byBurkholderia thailandensisE264. Org. Lett. 2010, 12, 716–719. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Song, L.; Sass, A.; White, J.; Wilmot, C.; Marchbank, A.; Boaisha, O.; Paine, J.; Knight, D.; Challis, G.L. Enacyloxins Are Products of an Unusual Hybrid Modular Polyketide Synthase Encoded by a Cryptic Burkholderia ambifaria Genomic Island. Chem. Biol. 2011, 18, 665–677. [Google Scholar] [CrossRef]
- Lee, R.S.; Choi, S.-M.; Jo, S.J.; Lee, J.; Cho, S.-Y.; Kim, S.-H.; Lee, D.-G.; Jeong, H. Clinical Characteristics and Antimicrobial Susceptibility Trends in Citrobacter Bacteremia: An 11-Year Single-Center Experience. Infect. Chemother. 2019, 51, 1–9. [Google Scholar] [CrossRef]
- Huttner, B.; Haustein, T.; Uçkay, I.; Renzi, G.; Stewardson, A.J.; Schaerrer, D.; Agostinho, A.; Andremont, A.; Schrenzel, J.; Pittet, D.; et al. Decolonization of intestinal carriage of extended-spectrum -lactamase-producing Enterobacteriaceae with oral colistin and neomycin: A randomized, double-blind, placebo-controlled trial. J. Antimicrob. Chemother. 2013, 68, 2375–2382. [Google Scholar] [CrossRef]
- Igbinosa, I.H. Antibiogram profiling and pathogenic status of Aeromonas species recovered from Chicken. Saudi J. Biol. Sci. 2014, 21, 481–485. [Google Scholar] [CrossRef]
- Stratev, D.; Odeyemi, O.A. Antimicrobial resistance of Aeromonas hydrophila isolated from different food sources: A mini-review. J. Infect. Public Health 2016, 9, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.L.; Wilcox, M.H. Aeromonas infections and their treatment. J. Antimicrob. Chemother. 1995, 35, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, S.; Tsurumoto, T.; Yonekura, A.; Adachi, K.; Shindo, H. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus epidermidis biofilms isolated from infected total hip arthroplasty cases. J. Orthop. Sci. 2006, 11, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. Biofilms and Antimicrobial Resistance. Clin. Orthop. Relat. Res. 2005, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Ceri, H.; Gibb, A.P.; Olson, M.E.; Sepandj, F. MIC versus MBEC to Determine the Antibiotic Sensitivity ofStaphylococcus aureusin Peritoneal Dialysis Peritonitis. Perit. Dial. Int. 2010, 30, 652–656. [Google Scholar] [CrossRef]
- Fux, C.A.; Wilson, S.; Stoodley, P. Detachment Characteristics and Oxacillin Resistance of Staphyloccocus aureus Biofilm Emboli in an In Vitro Catheter Infection Model. J. Bacteriol. 2004, 186, 4486–4491. [Google Scholar] [CrossRef]
- Evans, R.C.; Holmes, C.J. Effect of vancomycin hydrochloride on Staphylococcus epidermidis biofilm associated with silicone elastomer. Antimicrob. Agents Chemother. 1987, 31, 889–894. [Google Scholar] [CrossRef]
- Enneson, J.J.; Litzgus, J.D. Using long-term data and a stage-classified matrix to assess conservation strategies for an endangered turtle (Clemmys guttata). Biol. Conserv. 2008, 141, 1560–1568. [Google Scholar] [CrossRef]
- Howell, J.; McKnight, D.; Seigel, R. A novel method of collecting spotted turtles (Clemmys guttata). Herpetol. Rev. 2016, 47, 28–31. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program: Table 1. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Holland, B.R.; Huber, K.T.; Dress, A.; Moulton, V. Delta plots: A tool for analyzing phylogenetic distance data. Mol. Biol. Evol. 2002, 19, 2051–2059. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement; CLSI: Wayne, PA, USA, 2013. [Google Scholar]
- EUCAST; The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 25 June 2020).
- Parthasarathy, A. Scanning Electron Microscopy (SEM) for Microbes—A Simple and Inexpensive Method for Sample Preparation; ResearchGate: Berlin, Germany, 2019. [Google Scholar] [CrossRef]
- Steindler, L.; Venturi, V. Detection of quorum-sensingN-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 2007, 266, 1–9. [Google Scholar] [CrossRef]
- Gan, H.M.; Dailey, L.K.; Halliday, N.; Williams, P.; Hudson, A.O.; Savka, M.A. Genome sequencing-assisted identification and the first functional validation of N-acyl-homoserine-lactone synthases from the Sphingomonadaceae family. PeerJ 2016, 4, e2332. [Google Scholar] [CrossRef]
- Scott, R.A.; Weil, J.; Le, P.T.; Williams, P.; Fray, R.G.; Von Bodman, S.B.; Savka, M.A. Long- and Short-Chain Plant-Produced Bacterial N-Acyl-Homoserine Lactones Become Components of Phyllosphere, Rhizosphere, and Soil. Mol. Plant-Microbe Interact. 2006, 19, 227–239. [Google Scholar] [CrossRef]
- Gan, H.M.; Buckley, L.; Szegedi, E.; Hudson, A.O.; Savka, M.A. Identification of an rsh Gene from a Novosphingobium sp. Necessary for Quorum-Sensing Signal Accumulation. J. Bacteriol. 2009, 191, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.P.; Silo-Suh, L.; Woods, D.E.; Ohman, D.E.; Sokol, P.A. Comparative Analysis of Plant and Animal Models for Characterization of Burkholderia cepacia Virulence. Infect. Immun. 2003, 71, 5306–5313. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.; Locascio, P.F.; Land, M.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Multiple alignment using hidden Markov models. Proc. Int. Conf. Intell. Syst. Mol. Boil. 1995, 3, 114–120. [Google Scholar]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Egan, H.M.; Gan, H.Y.; Ahmad, N.H.; Aziz, N.A.; Hudson, A.O.; Savka, M.A. Whole genome sequencing and analysis reveal insights into the genetic structure, diversity and evolutionary relatedness of luxI and luxR homologs in bacteria belonging to the Sphingomonadaceae family. Front. Cell. Infect. Microbiol. 2014, 4, 188. [Google Scholar] [CrossRef]
- Roth, V. Doubling Time Computing. 2006. Available online: http://www.doubling-time.com/compute.php (accessed on 25 June 2020).
- Hudson, J.A. Effect of pre-incubation temperature on the lag time of Aeromonas hydrophila. Lett. Appl. Microbiol. 1993, 16, 274–276. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Del Castillo, C.S.; Hikima, J.-I.; Jang, H.-B.; Nho, S.-W.; Jung, T.S.; Wongtavatchai, J.; Kondo, H.; Hirono, I.; Takeyama, H.; Aoki, T. Comparative Sequence Analysis of a Multidrug-Resistant Plasmid from Aeromonas hydrophila. Antimicrob. Agents Chemother. 2013, 57, 120–129. [Google Scholar] [CrossRef]
- Majumdar, T.; Das, B.; Bhadra, R.K.; Dam, B.; Mazumder, S. Complete nucleotide sequence of a quinolone resistance gene (qnrS2) carrying plasmid of Aeromonas hydrophila isolated from fish. Plasmid 2011, 66, 79–84. [Google Scholar] [CrossRef]
- Hernould, M.; Gagné, S.; Fournier, M.; Quentin, C.; Arpin, C. Role of the AheABC Efflux Pump in Aeromonas hydrophila Intrinsic Multidrug Resistance. Antimicrob. Agents Chemother. 2008, 52, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Zervosen, A.; Valladares, M.H.; Devreese, B.; Prosperi-Meys, C.; Adolph, H.-W.; Mercuri, P.S.; Vanhove, M.; Amicosante, G.; Van Beeumen, J.; Frère, J.M.; et al. Inactivation of Aeromonas hydrophila metallo-beta-lactamase by cephamycins and moxalactam. JBIC J. Biol. Inorg. Chem. 2001, 268, 3840–3850. [Google Scholar]
- Huddleston, J.R.; Brokaw, J.M.; Zak, J.C.; Jeter, R.M. Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst. Appl. Microbiol. 2013, 36, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Silver, A.C.; Williams, D.; Faucher, J.; Horneman, A.J.; Gogarten, J.P.; Graf, J. Complex Evolutionary History of the Aeromonas veronii Group Revealed by Host Interaction and DNA Sequence Data. PLoS ONE 2011, 6, e16751. [Google Scholar] [CrossRef] [PubMed]
- Palma-Martínez, I.; Guerrero-Mandujano, A.; Ruiz-Ruiz, M.J.; Hernández-Cortez, C.; Molina-López, J.; Bocanegra-García, V.; Castro-Escarpulli, G. Active Shiga-Like Toxin Produced by Some Aeromonas spp., Isolated in Mexico City. Front. Microbiol. 2016, 7, 1522. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, J.-P.; Defez, C.; Bouziges, N.; Mahamat, A.; Sotto, A. Clinical and molecular epidemiology of multidrug-resistant Citrobacter spp. infections in a French university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.K.; Yadav, A.; Choudhary, U.; Aropa, D.R. Citrobacter Bacteremia in a Tertiary Care Hospital. Scand. J. Infect. Dis. 2003, 35, 765–768. [Google Scholar] [CrossRef]
- Barlow, M.; Hall, B.G. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 2002, 46, 1190–1198. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, C.; Sun, Q.; Schwarz, S.; Ou, Y.; Yang, L.; Huang, Z.; Eichhorn, I.; Walsh, T.R.; Wang, Y.; et al. Prevalence and Genetic Analysis of mcr-3-Positive Aeromonas Species from Humans, Retail Meat, and Environmental Water Samples. Antimicrob. Agents Chemother. 2018, 62, AAC.00404–18. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.J.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Alekshun, M.N.; Levy, S.B. Molecular Mechanisms of Antibacterial Multidrug Resistance. Cell 2007, 128, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Nikaido, H. Efflux-Mediated Drug Resistance in Bacteria. Drugs 2009, 69, 1555–1623. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [PubMed]
- Renier, S.; Hébraud, M.; Desvaux, M. Molecular biology of surface colonization by Listeria monocytogenes: An additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 2011, 13, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Wormser, G.P.; Keusch, G.T.; Heel, R.C. Co-trimoxazole (trimethoprim-sulfamethoxazole): An updated review of its antibacterial activity and clinical efficacy. Drugs 1982, 24, 459–518. [Google Scholar] [CrossRef]
- Drelichman, V. Bacteremias due to Citrobacter diversus and Citrobacter freundii. Arch. Intern. Med. 1985, 145, 1808–1810. [Google Scholar] [CrossRef]
- Cai, L.; Wang, H.; Liang, L.; Wang, G.; Xu, X.; Wang, H. Response of Formed-Biofilm of Enterobacter cloacae, Klebsiella oxytoca, and Citrobacter freundii to Chlorite-Based Disinfectants. J. Food Sci. 2018, 83, 1326–1332. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, J.; Wu, J.; Wang, J.; Luo, Y. Potential risks of microplastics combined with superbugs: Enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol. Environ. Saf. 2020, 187, 109852. [Google Scholar] [CrossRef]
- Duncan, E.M.; Broderick, A.C.; Fuller, W.J.; Galloway, T.S.; Godfrey, M.H.; Hamann, M.; Limpus, C.J.; Lindeque, P.K.; Mayes, A.G.; Omeyer, L.C.M.; et al. Microplastic ingestion ubiquitous in marine turtles. Glob. Chang. Biol. 2019, 25, 744–752. [Google Scholar] [CrossRef]
- Norton, T.; Thompson, R.; Pope, J.; Veltkamp, C.; Banks, B.; Howard, C.V.; Hawkins, S. Using confocal laser scanning microscopy, scanning electron microscopy and phase contrast light microscopy to examine marine biofilms. Aquat. Microb. Ecol. 1998, 16, 199–204. [Google Scholar] [CrossRef]
- Hannig, C.; Follo, M.; Hellwig, E.; Al-Ahmad, A. Visualization of adherent micro-organisms using different techniques. J. Med. Microbiol. 2010, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.R.; Korber, D.R.; Hoyle, B.D.; Costerton, J.W.; Caldwell, D.E. Optical sectioning of microbial biofilms. J. Bacteriol. 1991, 173, 6558–6567. [Google Scholar] [CrossRef] [PubMed]
- Atshan, S.S.; Shamsudin, M.N.; Karunanidhi, A.; Van Belkum, A.; Lung, L.T.T.; Sekawi, Z.; Nathan, J.J.; Ling, K.H.; Seng, J.S.C.; Ali, A.M.; et al. Quantitative PCR analysis of genes expressed during biofilm development of methicillin resistant Staphylococcus aureus (MRSA). Infect. Genet. Evol. 2013, 18, 106–112. [Google Scholar] [CrossRef]
- Li, Z.; Nair, S.K. Quorum sensing: How bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 2012, 21, 1403–1417. [Google Scholar] [CrossRef]
- Swift, S.; Karlyshev, A.V.; Fish, L.; Durant, E.L.; Winson, M.K.; Chhabra, S.R.; Williams, P.; MacIntyre, S.; Stewart, G.S. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: Identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 1997, 179, 5271–5281. [Google Scholar] [CrossRef]
- Gui, M.; Liu, L.; Wu, R.; Hu, J.; Wang, S.; Li, P. Detection of New Quorum Sensing N-Acyl Homoserine Lactones From Aeromonas veronii. Front. Microbiol. 2018, 9, 1712. [Google Scholar] [CrossRef]
- Holden, M.T.; Chhabra, S.R.; De Nys, R.; Stead, P.; Bainton, N.J.; Hill, P.J.; Manefield, M.; Kumar, N.; Labatte, M.; England, D.; et al. Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 1999, 33, 1254–1266. [Google Scholar] [CrossRef]
- Heimpel, H.; Raghavachar, A. Hematological side effects of co-trimoxazole. Infection 1987, 15 (Suppl. 5), S248–S253. [Google Scholar] [CrossRef]
- National Institute of Diabetes and Digestive and Kidney Diseases LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. 2012. Available online: https://pubmed.ncbi.nlm.nih.gov/31643176/ (accessed on 5 November 2020).
- Bjarnsholt, T.; Givskov, M.C. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1213–1222. [Google Scholar] [CrossRef]
- Ponnusamy, K.; Paul, D.; Kweon, J.H. Inhibition of Quorum Sensing Mechanism andAeromonas hydrophilaBiofilm Formation by Vanillin. Environ. Eng. Sci. 2009, 26, 1359–1363. [Google Scholar] [CrossRef]
- Truchado, P.; Gil-Izquierdo, A.; Tomás-Barberán, F.; Allende, A. Inhibition by Chestnut Honey ofN-Acyl-l-homoserine Lactones and Biofilm Formation in Erwinia carotovora, Yersinia enterocolitica, and Aeromonas hydrophila. J. Agric. Food Chem. 2009, 57, 11186–11193. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Salighehzadeh, R.; Sharifiyazdi, H. Antimicrobial Resistance and Incidence of Integrons in Aeromonas Species Isolated from Diseased Freshwater Animals and Water Samples in Iran. Antibiotics 2019, 8, 198. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, I.; Feudi, C.; Wang, Y.; Kaspar, H.; Feßler, A.T.; Lübke-Becker, A.; Michael, G.B.; Shen, J.; Schwarz, S. Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-Vet and from diagnostic submissions. J. Antimicrob. Chemother. 2018, 73, 1217–1221. [Google Scholar] [CrossRef]
- Yang, L.; Li, P.; Liang, B.; Hu, X.; Li, J.; Xie, J.; Yang, C.; Hao, R.; Wang, L.; Jia, L.; et al. Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci. Rep. 2018, 8, 10653. [Google Scholar] [CrossRef]





| Antibiotic | Amount of Antibiotic (μg) | Minimum Inhibition Zone Diameter (mm) for Sensitivity | A. hydrophila | C. portucalensis | ||
|---|---|---|---|---|---|---|
| Zone of Inhibition (mm) | Response | Zone of Inhibition (mm) | Response | |||
| Gentamicin | 10 | 15 | 0 | R | 0 | R |
| Tetracycline | 30 | 15 | 0 | R | 0 | R |
| Doxycycline | 30 | 14 | 0 | R | 0 | R |
| Kanamycin | 30 | 18 | 0 | R | 0 | R |
| Streptomycin | 10 | 15 | 0 | R | 0 | R |
| Tobramycin | 10 | 15 | 0 | R | 0 | R |
| Neomycin | 30 | 12 # | 13 | S | 16 | S |
| Novobiocin | 30 | 12 * | 5 | R | 0 | R |
| Erythromycin | 15 | 21 ^ | 10 | R | 7 | R |
| Sulfamethoxazole + Trimethoprim | 23.75 + 1.25 | 16 | 18 | S | 12 | I |
| Penicillin | 10 U | 15 | 6 | R | 0 | R |
| Sterile disc | 0 | N/A | 0 | N/A | 0 | N/A |
| Antibiotic (Concentration in mg/mL) | Amount on Disc (μg) | Minimum Inhibition Zone Diameter for Sensitivity (mm) | A. hydrophila | C. portucalensis | ||
|---|---|---|---|---|---|---|
| Zone of Inhibition (mm) | Response | Zone of Inhibition (mm) | Response | |||
| Tetracycline (30) | 600 | 15 | 31 | S | 30.7 | S |
| Rifamycin (30) | 600 | 20 * | 0 | R | 0 | R |
| Cefaclor (50) | 1000 | 18 | 0 | R | 0 | R |
| Fusidic acid (12.5) | 500 | 24 # | 0 | R | 0 | R |
| Kanamycin (50) | 1000 | 18 | 0 | R | 0 | R |
| Clindamycin (50) | 1000 | 17 ^ | 8 | R | 0 | R |
| Neomycin (30) | 600 | 12 § | 22.5 | S | 23.5 | S |
| Cotrimoxazole (30) | 600 | 16 | 30.5 | S | 35 | S |
| Control | N/A | N/A | 0 | N/A | 0 | N/A |
| Antibiotic | Antibiotic Class/Family | Therapeutic Target | A. hydrophila | C. portucalensis | ||
|---|---|---|---|---|---|---|
| Response | RGI Prediction | Response | RGI Prediction | |||
| Gentamycin | Aminoglycoside | 30S ribosomal subunit (protein synthesis) | R | No | R | Yes |
| Tetracycline | Tetracycline | Aminoacyl tRNA binding to RNA-ribosome complex (30S subunit; protein synthesis) | R, but S at 600 μg | Yes | R, but S at 600 μg | Yes |
| Doxycycline | Tetracycline | Aminoacyl tRNA binding to RNA-ribosome complex (30S subunit; protein synthesis) | R | Yes | R | Yes |
| Kanamycin | Aminoglycoside | 30S ribosomal subunit (protein synthesis) | R | No | R | Yes |
| Streptomycin | Aminoglycoside | 30S ribosomal subunit (protein synthesis) | R | No | R | Yes |
| Tobramycin | Aminoglycoside | 30S ribosomal subunit (protein synthesis) | R | No | R | Yes |
| Neomycin | Aminoglycoside | 30S ribosomal subunit (protein synthesis) | S | No | S | Yes (R predicted) |
| Novobiocin | Aminocoumarin | DNA synthesis | R | No | R | Yes |
| Erythromycin | Macrolide | Growth | R | No | R | Yes |
| Penicillin | Beta-lactams | Final stages of cell wall synthesis | R | No | R | Yes |
| Rifamycin | Ansamycin | DNA-dependent RNA polymerase | R | No | R | Yes |
| Cefaclor | Cephalosporin | Peptidoglycan synthesis | R | Yes | R | Yes |
| Fusidic acid | Fusidane | Translocation of elongation factor G (Protein synthesis) | R | No | R | No |
| Clindamycin | Lincomycin | 50S ribosomal subunit (protein synthesis) | R | No | R | No |
| Cotrimoxazole | Sulfonamides | Dihydropteroate synthase (DHPS) | R, but S at 600 μg | No | R, but S at 600 μg | No |
| Assay | A. hydrophila RIT668 | C. portucalensis RIT669 | ||
|---|---|---|---|---|
| Cotrimoxazole (μg/mL) | Neomycin (μg/mL) | Cotrimoxazole (μg/mL) | Neomycin (μg/mL) | |
| MIC | 500–1000 | 32.3–62.5 | 7.8–15.6 | 7.8–31.3 |
| MBEC | >1000 | >1000 | >1000 | >1000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, S.G.; Abajorga, M.; Glover, M.A.; Wengert, P.C.; Parthasarathy, A.; Savka, M.A.; Wadsworth, C.B.; Shipman, P.A.; Hudson, A.O. Aeromonas hydrophila RIT668 and Citrobacter portucalensis RIT669—Potential Zoonotic Pathogens Isolated from Spotted Turtles. Microorganisms 2020, 8, 1805. https://doi.org/10.3390/microorganisms8111805
Thomas SG, Abajorga M, Glover MA, Wengert PC, Parthasarathy A, Savka MA, Wadsworth CB, Shipman PA, Hudson AO. Aeromonas hydrophila RIT668 and Citrobacter portucalensis RIT669—Potential Zoonotic Pathogens Isolated from Spotted Turtles. Microorganisms. 2020; 8(11):1805. https://doi.org/10.3390/microorganisms8111805
Chicago/Turabian StyleThomas, Seema G., Milky Abajorga, Maryah A. Glover, Peter C. Wengert, Anutthaman Parthasarathy, Michael A. Savka, Crista B. Wadsworth, Paul A. Shipman, and André O. Hudson. 2020. "Aeromonas hydrophila RIT668 and Citrobacter portucalensis RIT669—Potential Zoonotic Pathogens Isolated from Spotted Turtles" Microorganisms 8, no. 11: 1805. https://doi.org/10.3390/microorganisms8111805
APA StyleThomas, S. G., Abajorga, M., Glover, M. A., Wengert, P. C., Parthasarathy, A., Savka, M. A., Wadsworth, C. B., Shipman, P. A., & Hudson, A. O. (2020). Aeromonas hydrophila RIT668 and Citrobacter portucalensis RIT669—Potential Zoonotic Pathogens Isolated from Spotted Turtles. Microorganisms, 8(11), 1805. https://doi.org/10.3390/microorganisms8111805






