Phosphorus Reduces Negative Effects of Nitrogen Addition on Soil Microbial Communities and Functions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Soil Sampling
2.2. Soil Chemical Analyses
2.3. Soil DNA Extraction, Sequencing and Data Processing
2.4. Statistics
3. Results
3.1. Soil Characteristics under N and P Addition
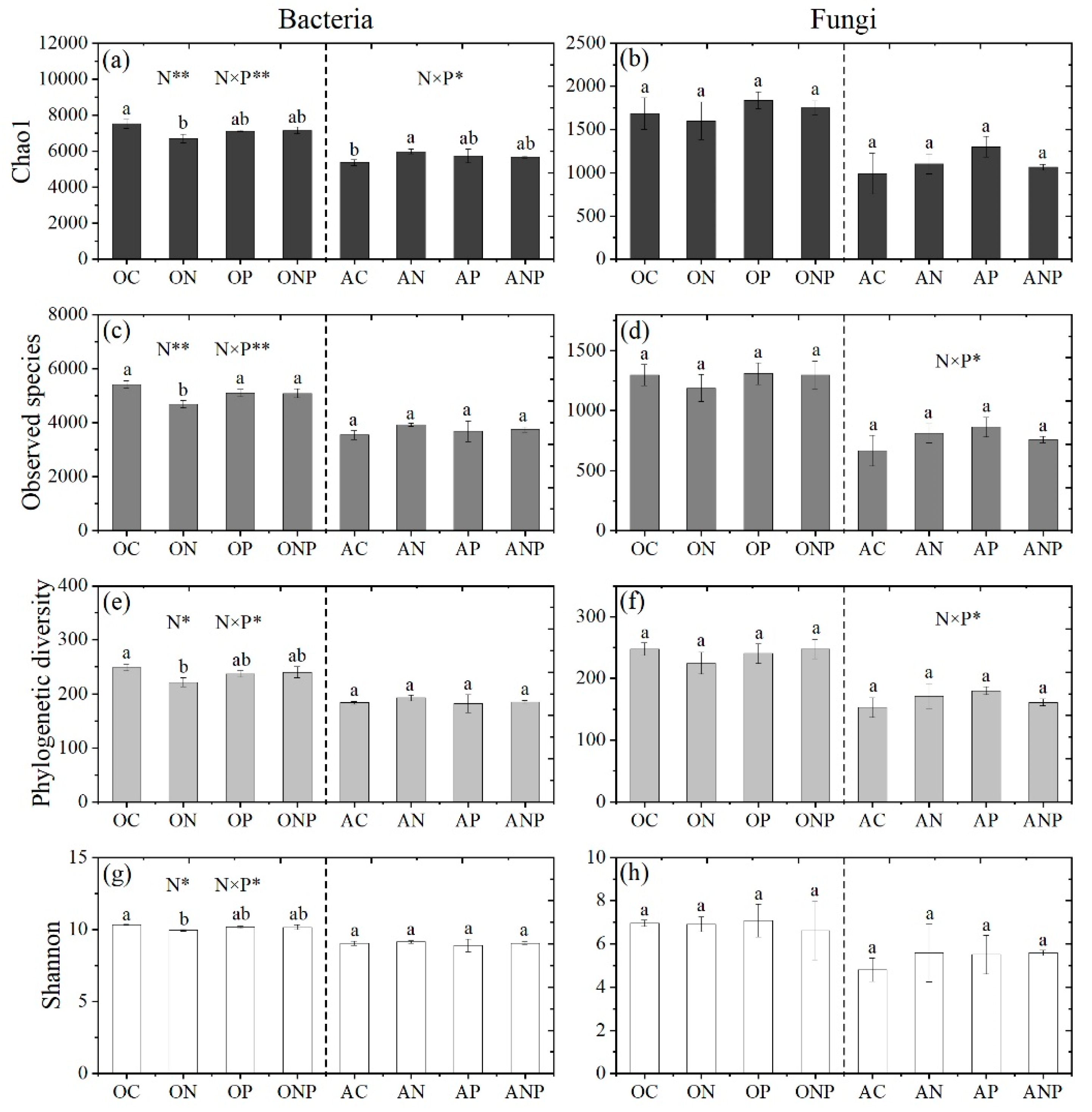
3.2. Microbial Diversity and Community Composition under N and P Addition
3.3. Dominant Microbial Taxa under N and P Addition
3.4. Microbial Functional Potentials under N and P Addition
4. Discussion
4.1. Microbial Diversity and Biomass under N and P Addition
4.2. Microbial Composition and Structure under N and P Addition
4.3. Microbial Functional Potentials under N and P Addition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, E.W.; Howarth, R.W.; Seitzinger, S.P.; Asner, G.P.; Cleveland, C.C.; Green, P.A.; Holland, E.A.; et al. Nitrogen Cycles: Past, Present, and Future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Galloway, J.N.; Winiwarter, W.; Leip, A.; Leach, A.M.; Bleeker, A.; Erisman, J.W. Nitrogen footprints: Past, present and future. Environ. Res. Lett. 2014, 9, 115003. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Aber, J.D.; Howarth, R.W.; Likens, G.E.; Matson, P.A.; Schindler, D.W.; Schlesinger, W.H.; Tilman, D.G. Human alteration of the global nitrogen cycle: Sources and consequences. Ecol. Appl. 1997, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Magill, A.H.; Aber, J.D.; Currie, W.S.; Nadelhoffer, K.J.; Martin, M.E.; McDowell, W.H.; Melillo, J.M.; Steudler, P. Ecosystem response to 15 years of chronic nitrogen additions at the Harvard Forest LTER, Massachusetts, USA. For. Ecol. Manag. 2004, 196, 7–28. [Google Scholar] [CrossRef]
- Hogberg, P.; Fan, H.; Quist, M.; Binkley, D.; Tamm, C.O. Tree growth and soil acidification in response to 30 years of experimental nitrogen loading on boreal forest. Glob. Chang. Biol. 2006, 12, 489–499. [Google Scholar] [CrossRef]
- Bobbink, R.; Hicks, K.; Galloway, J.; Spranger, T.; Alkemade, R.; Ashmore, M.; Bustamante, M.; Cinderby, S.; Davidson, E.; Dentener, F.; et al. Global assessment of nitrogen deposition effects on terrestrial plant diversity: A synthesis. Ecol. Appl. 2010, 20, 30–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Mao, Q.; Gilliam, F.S.; Luo, Y.; Mo, J. Nitrogen deposition contributes to soil acidification in tropical ecosystems. Glob. Chang. Biol. 2014, 20, 3790–3801. [Google Scholar] [CrossRef]
- Philippot, L.; Spor, A.; Hénault, C.; Bru, D.; Bizouard, F.; Jones, C.M.; Sarr, A.; Maron, P.-A. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 2013, 7, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Wagg, C.; Bender, S.F.; Widmer, F.; van der Heijden, M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Singh, B.K.; Maestre, F.T. Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol. Lett. 2017, 20, 1295–1305. [Google Scholar] [CrossRef] [Green Version]
- Compton, J.E.; Watrud, L.S.; Arlene Porteous, L.; DeGrood, S. Response of soil microbial biomass and community composition to chronic nitrogen additions at Harvard forest. For. Ecol. Manag. 2004, 196, 143–158. [Google Scholar] [CrossRef]
- Janssens, I.A.; Dieleman, W.; Luyssaert, S.; Subke, J.-A.; Reichstein, M.; Ceulemans, R.; Ciais, P.; Dolman, A.J.; Grace, J.; Matteucci, G.; et al. Reduction of forest soil respiration in response to nitrogen deposition. Nat. Geosci. 2010, 3, 315–322. [Google Scholar] [CrossRef]
- Treseder, K.K. Nitrogen additions and microbial biomass: A meta-analysis of ecosystem studies. Ecol. Lett. 2008, 11, 1111–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, D.; Jiang, L.; Ma, S.; Fang, W.; Schmid, B.; Xu, L.; Zhu, J.; Li, P.; Losapio, G.; Jing, X.; et al. Effects of nitrogen deposition on soil microbial communities in temperate and subtropical forests in China. Sci. Total Environ. 2017, 607–608, 1367–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, D.; Niu, S. A global analysis of soil acidification caused by nitrogen addition. Environ. Res. Lett. 2015, 10, 024019. [Google Scholar] [CrossRef]
- Wang, C.; Liu, D.; Bai, E. Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 2018, 120, 126–133. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, H.; Gao, H.; An, S. Response and driving factors of soil microbial diversity related to global nitrogen addition. Land Degrad. Dev. 2020, 31, 190–204. [Google Scholar] [CrossRef]
- Eisenlord, S.D.; Zak, D.R. Simulated Atmospheric Nitrogen Deposition Alters Actinobacterial Community Composition in Forest Soils. Soil Sci. Soc. Am. J. 2010, 74, 1157–1166. [Google Scholar] [CrossRef] [Green Version]
- Entwistle, E.M.; Zak, D.R.; Edwards, I.P. Long-Term Experimental Nitrogen Deposition Alters the Composition of the Active Fungal Community in the Forest Floor. Soil Sci. Soc. Am. J. 2013, 77, 1648–1658. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bödeker, I.T.M.; Lindahl, B.D.; Olson, Å.; Clemmensen, K.E. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 2016, 30, 1967–1978. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Lan, Z.; Hu, S.; Bai, Y. Effects of nitrogen enrichment on belowground communities in grassland: Relative role of soil nitrogen availability vs. soil acidification. Soil Biol. Biochem. 2015, 89, 99–108. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Zheng, M.; Jiang, L.; Luo, Y. Patterns and mechanisms of responses by soil microbial communities to nitrogen addition. Soil Biol. Biochem. 2017, 115, 433–441. [Google Scholar] [CrossRef]
- Wardle, D.A.; Peltzer, D.A. Aboveground-Belowground Linkages, Ecosystem Development, and Ecosystem Restoration. In Linking Restoration and Ecological Succession; Walker, L.R., Walker, J., Hobbs, R.J., Eds.; Springer Series on Environmental Management; Springer: New York, NY, USA, 2007; pp. 45–68. ISBN 978-0-387-35302-9. [Google Scholar]
- LeBauer, D.S.; Treseder, K.K. Nitrogen limitation of net primary productivity in terrestrial ecosystems is globally distributed. Ecology 2008, 89, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Eisenhauer, N.; Cesarz, S.; Koller, R.; Worm, K.; Reich, P.B. Global change belowground: Impacts of elevated CO2, nitrogen, and summer drought on soil food webs and biodiversity. Glob. Chang. Biol. 2012, 18, 435–447. [Google Scholar] [CrossRef]
- Camenzind, T.; Hättenschwiler, S.; Treseder, K.K.; Lehmann, A.; Rillig, M.C. Nutrient limitation of soil microbial processes in tropical forests. Ecol. Monogr. 2018, 88, 4–21. [Google Scholar] [CrossRef]
- Allison, S.D.; Czimczik, C.I.; Treseder, K.K. Microbial activity and soil respiration under nitrogen addition in Alaskan boreal forest. Glob. Chang. Biol. 2008, 14, 1156–1168. [Google Scholar] [CrossRef] [Green Version]
- Crowley, K.F.; McNeil, B.E.; Lovett, G.M.; Canham, C.D.; Driscoll, C.T.; Rustad, L.E.; Denny, E.; Hallett, R.A.; Arthur, M.A.; Boggs, J.L.; et al. Do Nutrient Limitation Patterns Shift from Nitrogen Toward Phosphorus with Increasing Nitrogen Deposition Across the Northeastern United States? Ecosystems 2012, 15, 940–957. [Google Scholar] [CrossRef]
- Yang, K.; Zhu, J.; Gu, J.; Yu, L.; Wang, Z. Changes in soil phosphorus fractions after 9 years of continuous nitrogen addition in a Larix gmelinii plantation. Ann. For. Sci. 2015, 72, 435–442. [Google Scholar] [CrossRef]
- Zhou, W.; Guo, Y.; Zhu, B.; Wang, X.; Zhou, L.; Yu, D.; Dai, L. Seasonal variations of nitrogen flux and composition in a wet deposition forest ecosystem on Changbai Mountain. Acta Ecol. Sin. 2015, 35. [Google Scholar] [CrossRef] [Green Version]
- Aber, J.; McDowell, W.; Nadelhoffer, K.; Magill, A.; Berntson, G.; Kamakea, M.; McNulty, S.; Currie, W.; Rustad, L.; Fernandez, I. Nitrogen Saturation in Temperate Forest Ecosystems. BioScience 1998, 48, 921–934. [Google Scholar] [CrossRef]
- Bai, E.; Li, W.; Li, S.; Sun, J.; Peng, B.; Dai, W.; Jiang, P.; Han, S. Pulse Increase of Soil N2O Emission in Response to N Addition in a Temperate Forest on Mt Changbai, Northeast China. PLoS ONE 2014, 9, e102765. [Google Scholar] [CrossRef] [PubMed]
- Kuo, S. Phosphorus. In Methods of Soil Analysis: Chemical Methods. Part 3. Chemical Methods; Sparks, D.L., Ed.; SSSA Book Series: Madison, WI, USA, 1996; pp. 869–919. [Google Scholar]
- Brookes, P.C.; Powlson, D.S.; Jenkinson, D.S. Measurement of microbial biomass phosphorus in soil. Soil Biol. Biochem. 1982, 14, 319–329. [Google Scholar] [CrossRef]
- Brookes, P.C.; Landman, A.; Pruden, G.; Jenkinson, D.S. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Jenkinson, D.S. Determination of Microbial Biomass Carbon and Nitrogen in Soil. In Advances in Nitrogen Cycling in Agricultural Ecosystems; Wilson, J.B., Ed.; CAB International: Wallingford, UK, 1988; pp. 368–386. [Google Scholar]
- Sparling, G.P.; West, A.W. A direct extraction method to estimate soil microbial C: Calibration in situ using microbial respiration and 14C labelled cells. Soil Biol. Biochem. 1988, 20, 337–343. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. AEM 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Keller, K.; Brodie, E.L.; Larsen, N.; Piceno, Y.M.; Phan, R.; Andersen, G.L. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006, 34, W394–W399. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R 21. Foundation for Statistical Computing; R Development Core Team: Vienna, Austria, 2006. [Google Scholar]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. AEM 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Bai, E.; Wang, Q.; Gao, D.; Zhou, J.; Jiang, P.; Wu, J. Biogeographic Distribution Patterns of Bacteria in Typical Chinese Forest Soils. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Lucas, R.W.; Klaminder, J.; Futter, M.N.; Bishop, K.H.; Egnell, G.; Laudon, H.; Högberg, P. A meta-analysis of the effects of nitrogen additions on base cations: Implications for plants, soils, and streams. For. Ecol. Manag. 2011, 262, 95–104. [Google Scholar] [CrossRef]
- Kaspari, M.; Bujan, J.; Weiser, M.D.; Ning, D.; Michaletz, S.T.; Zhili, H.; Enquist, B.J.; Waide, R.B.; Zhou, J.; Turner, B.L.; et al. Biogeochemistry drives diversity in the prokaryotes, fungi, and invertebrates of a Panama forest. Ecology 2017, 98, 2019–2028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nottingham, A.T.; Turner, B.L.; Stott, A.W.; Tanner, E.V.J. Nitrogen and phosphorus constrain labile and stable carbon turnover in lowland tropical forest soils. Soil Biol. Biochem. 2015, 80, 26–33. [Google Scholar] [CrossRef]
- Schimel, J.; Balser, T.C.; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Kuperman, R.G.; Edwards, C.A. Effects of Acidic Deposition on Soil Invertebrates and Microorganisms. In Reviews of Environmental Contamination and Toxicology; Ware, G.W., Nigg, H.N., Bevenue, A., Eds.; Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 1997; Volume 148, pp. 35–138. ISBN 978-1-4612-7478-0. [Google Scholar]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, T.; Gilliam, F.S.; Gundersen, P.; Zhang, W.; Chen, H.; Mo, J. Interactive Effects of Nitrogen and Phosphorus on Soil Microbial Communities in a Tropical Forest. PLoS ONE 2013, 8, e61188. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, Z.; Wang, F.; Zou, B.; Chen, Y.; Zhao, J.; Mo, Q.; Li, Y.; Li, X.; Xia, H. Effects of nitrogen and phosphorus addition on soil microbial community in a secondary tropical forest of China. Biol. Fertil. Soils 2015, 51, 207–215. [Google Scholar] [CrossRef]
- Li, Y.; Tian, D.; Wang, J.; Niu, S.; Tian, J.; Ha, D.; Qu, Y.; Jing, G.; Kang, X.; Song, B. Differential mechanisms underlying responses of soil bacterial and fungal communities to nitrogen and phosphorus inputs in a subtropical forest. PeerJ 2019, 7, e7631. [Google Scholar] [CrossRef]
- Leff, J.W.; Jones, S.E.; Prober, S.M.; Barberán, A.; Borer, E.T.; Firn, J.L.; Harpole, W.S.; Hobbie, S.E.; Hofmockel, K.S.; Knops, J.M.H.; et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. USA 2015, 112, 10967–10972. [Google Scholar] [CrossRef] [Green Version]
- Navarrete, A.A.; Soares, T.; Rossetto, R.; van Veen, J.A.; Tsai, S.M.; Kuramae, E.E. Verrucomicrobial community structure and abundance as indicators for changes in chemical factors linked to soil fertility. Antonie Leeuwenhoek 2015, 108, 741–752. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Guan, D.; Zhou, B.; Zhao, B.; Ma, M.; Qin, J.; Jiang, X.; Chen, S.; Cao, F.; Shen, D.; et al. Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in northeast China. Soil Biol. Biochem. 2015, 90, 42–51. [Google Scholar] [CrossRef]
- Wu, J.; Liu, W.; Zhang, W.; Shao, Y.; Duan, H.; Chen, B.; Wei, X.; Fan, H. Long-term nitrogen addition changes soil microbial community and litter decomposition rate in a subtropical forest. Appl. Soil Ecol. 2019, 142, 43–51. [Google Scholar] [CrossRef]
- Li, H.; Xu, Z.; Yang, S.; Li, X.; Top, E.M.; Wang, R.; Zhang, Y.; Cai, J.; Yao, F.; Han, X.; et al. Responses of Soil Bacterial Communities to Nitrogen Deposition and Precipitation Increment Are Closely Linked with Aboveground Community Variation. Microb. Ecol. 2016, 71, 974–989. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.L.; Sa, P.T.; Beattie, G.A.C.; Milham, P.J.; Riegler, M.; Spooner-Hart, R.N.; Holford, P. Additions of sugar and nitrogenous fertiliser affect plant nitrogen status and soil microbial communities. Appl. Soil Ecol. 2019, 139, 47–55. [Google Scholar] [CrossRef]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi; CAB International: Wallingford, UK, 2008. [Google Scholar]
- van Diepen, L.T.A.; Lilleskov, E.A.; Pregitzer, K.S.; Miller, R.M. Simulated Nitrogen Deposition Causes a Decline of Intra- and Extraradical Abundance of Arbuscular Mycorrhizal Fungi and Changes in Microbial Community Structure in Northern Hardwood Forests. Ecosystems 2010, 13, 683–695. [Google Scholar] [CrossRef]
- Camenzind, T.; Hempel, S.; Homeier, J.; Horn, S.; Velescu, A.; Wilcke, W.; Rillig, M.C. Nitrogen and phosphorus additions impact arbuscular mycorrhizal abundance and molecular diversity in a tropical montane forest. Glob. Chang. Biol. 2014, 20, 3646–3659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treseder, K.K. A meta-analysis of mycorrhizal responses to nitrogen, phosphorus, and atmospheric CO2 in field studies. New Phytol. 2004, 164, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Jach-Smith, L.C.; Jackson, R.D. N addition undermines N supplied by arbuscular mycorrhizal fungi to native perennial grasses. Soil Biol. Biochem. 2018, 116, 148–157. [Google Scholar] [CrossRef]
- Yan, G.; Zhou, M.; Wang, M.; Han, S.; Liu, G.; Zhang, X.; Sun, W.; Huang, B.; Wang, H.; Xing, Y.; et al. Nitrogen deposition and decreased precipitation altered nutrient foraging strategies of three temperate trees by affecting root and mycorrhizal traits. Catena 2019, 181, 104094. [Google Scholar] [CrossRef]
- Jach-Smith, L.C.; Jackson, R.D. Inorganic N addition replaces N supplied to switchgrass (Panicum virgatum) by arbuscular mycorrhizal fungi. Ecol. Appl. 2020, 30. [Google Scholar] [CrossRef]
- Li, W.; Sheng, H.; Liu, Y.; Zhang, R.; Ekawati, D.; Qian, Y.; Lou, Y. Ecostoichiometry Reveals the Separation of Microbial Adaptation Strategies in a Bamboo Forest in an Urban Wetland under Simulated Nitrogen Deposition. Forests 2020, 11, 428. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; Knight, R.; Gilbert, J.A.; McCulley, R.L. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science 2013, 342, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Nostrand, J.D.; Yin, H.; Wu, L.; Yuan, T.; Zhou, J. Hybridization of Environmental Microbial Community Nucleic Acids by GeoChip. In Microbial Environmental Genomics (MEG); Martin, F., Uroz, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1399, pp. 183–196. ISBN 978-1-4939-3367-9. [Google Scholar]





| Soil Layer | Diversity Index | SOC | TN | TP | NO3− | NH4+ | pH | MBC | MBN | MBP | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | O horizon | Chao1 | 0.556 | 0.301 | −0.069 | −0.684 * | 0.784 ** | 0.770 ** | 0.281 | 0.601 * | 0.41 |
| Observed species | 0.526 | 0.316 | 0.000 | −0.716 ** | 0.808 ** | 0.867 *** | 0.261 | 0.634 * | 0.501 | ||
| Phylogenetic diversity | 0.521 | 0.232 | 0.153 | −0.641 * | 0.710 ** | 0.729 ** | 0.223 | 0.557 | 0.388 | ||
| Shannon | 0.546 | 0.371 | −0.008 | −0.678 * | 0.753 ** | 0.894 *** | 0.266 | 0.601 * | 0.538 | ||
| A horizon | Chao1 | 0.033 | −0.075 | 0.245 | 0.297 | −0.375 | −0.759 ** | 0.353 | 0.115 | 0.148 | |
| Observed species | 0.196 | 0.037 | 0.338 | 0.525 | −0.626 * | −0.626 * | 0.401 | 0.098 | 0.334 | ||
| Phylogenetic diversity | −0.01 | −0.196 | 0.209 | 0.375 | −0.541 | −0.552 | 0.243 | −0.072 | 0.197 | ||
| Shannon | 0.305 | 0.11 | 0.208 | 0.429 | −0.634 * | −0.127 | 0.326 | 0.165 | 0.416 | ||
| Fungi | O horizon | Chao1 | −0.061 | −0.276 | 0.584 * | −0.088 | 0.034 | 0.11 | −0.184 | −0.108 | 0.061 |
| Observed species | 0.22 | −0.081 | 0.477 | −0.289 | 0.283 | 0.415 | −0.045 | 0.107 | 0.097 | ||
| Phylogenetic diversity | 0.422 | 0.1 | 0.391 | −0.294 | 0.443 | 0.497 | 0.132 | 0.282 | 0.21 | ||
| Shannon | −0.021 | −0.29 | 0.249 | −0.28 | −0.045 | 0.229 | −0.166 | −0.072 | −0.31 | ||
| A horizon | Chao1 | 0.033 | −0.075 | 0.245 | 0.297 | −0.375 | −0.759 ** | 0.353 | 0.115 | 0.148 | |
| Observed species | 0.196 | 0.037 | 0.338 | 0.525 | −0.626 * | −0.626 * | 0.401 | 0.098 | 0.334 | ||
| Phylogenetic diversity | −0.01 | −0.196 | 0.209 | 0.375 | −0.541 | −0.552 | 0.243 | −0.072 | 0.197 | ||
| Shannon | 0.305 | 0.11 | 0.208 | 0.429 | −0.634 * | −0.127 | 0.326 | 0.165 | 0.416 |
| Soil Layer | Effect | R2 | F | P | |
|---|---|---|---|---|---|
| Bacteria | O horizon | N | 0.170 | 2.396 | 0.020 |
| P | 0.099 | 1.391 | 0.158 | ||
| N × P | 0.165 | 2.337 | 0.023 | ||
| A horizon | N | 0.162 | 3.112 | 0.028 | |
| P | 0.259 | 4.979 | 0.001 | ||
| N × P | 0.162 | 3.118 | 0.032 | ||
| Fungi | O horizon | N | 0.182 | 2.533 | 0.003 |
| P | 0.134 | 1.855 | 0.038 | ||
| N × P | 0.107 | 1.492 | 0.105 | ||
| A horizon | N | 0.157 | 2.338 | 0.004 | |
| P | 0.161 | 2.393 | 0.005 | ||
| N × P | 0.144 | 2.135 | 0.017 |
| Variable | Bacteria | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| O | A | O | A | |||||
| r | P | r | P | r | P | r | P | |
| SOC | 0.2234 | 0.066 | 0.1408 | 0.151 | 0.1420 | 0.115 | 0.3204 | 0.011 |
| TN | 0.3859 | 0.013 | 0.1138 | 0.204 | 0.1238 | 0.212 | 0.1628 | 0.138 |
| TP | −0.0627 | 0.637 | 0.3370 | 0.029 | −0.1213 | 0.762 | 0.1978 | 0.115 |
| NO3− | 0.3291 | 0.026 | 0.7606 | 0.001 | 0.3815 | 0.005 | 0.2734 | 0.042 |
| NH4+ | 0.4572 | 0.007 | 0.504 | 0.001 | 0.2456 | 0.058 | 0.1714 | 0.143 |
| AN | 0.3148 | 0.018 | 0.6085 | 0.002 | 0.386 | 0.005 | 0.2073 | 0.062 |
| pH | 0.7428 | 0.001 | 0.3829 | 0.019 | 0.3277 | 0.009 | 0.4481 | 0.001 |
| MBC | 0.0483 | 0.327 | 0.455 | 0.005 | 0.1533 | 0.16 | 0.1961 | 0.102 |
| MBN | 0.3642 | 0.014 | 0.0473 | 0.334 | 0.2502 | 0.037 | 0.0845 | 0.238 |
| MBP | 0.6281 | 0.001 | 0.5463 | 0.004 | 0.2686 | 0.032 | 0.2334 | 0.062 |
| NO3− + pH | 0.3291 | 0.016 | 0.7658 | 0.004 | 0.3815 | 0.003 | 0.2774 | 0.033 |
| All | 0.1872 | 0.097 | 0.5723 | 0.003 | 0.2383 | 0.045 | 0.2155 | 0.061 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, Z.; Yang, J.; Sang, C.; Wang, X.; Sun, L.; Jiang, P.; Wang, C.; Bai, E. Phosphorus Reduces Negative Effects of Nitrogen Addition on Soil Microbial Communities and Functions. Microorganisms 2020, 8, 1828. https://doi.org/10.3390/microorganisms8111828
Xia Z, Yang J, Sang C, Wang X, Sun L, Jiang P, Wang C, Bai E. Phosphorus Reduces Negative Effects of Nitrogen Addition on Soil Microbial Communities and Functions. Microorganisms. 2020; 8(11):1828. https://doi.org/10.3390/microorganisms8111828
Chicago/Turabian StyleXia, Zongwei, Jingyi Yang, Changpeng Sang, Xu Wang, Lifei Sun, Ping Jiang, Chao Wang, and Edith Bai. 2020. "Phosphorus Reduces Negative Effects of Nitrogen Addition on Soil Microbial Communities and Functions" Microorganisms 8, no. 11: 1828. https://doi.org/10.3390/microorganisms8111828
APA StyleXia, Z., Yang, J., Sang, C., Wang, X., Sun, L., Jiang, P., Wang, C., & Bai, E. (2020). Phosphorus Reduces Negative Effects of Nitrogen Addition on Soil Microbial Communities and Functions. Microorganisms, 8(11), 1828. https://doi.org/10.3390/microorganisms8111828





