A Role for the Microbiota in the Immune Phenotype Alteration Associated with the Induction of Disease Tolerance and Persistent Asymptomatic Infection of Salmonella in the Chicken
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals, Housing, and Treatments
2.2. Bacteria Preparation
2.3. Bacterial Detection
2.4. Sample Collection and Processing
2.5. Microbiota Sequencing
2.6. Microbiota Bioinformatic Analysis
2.7. Real-Time Quantitative RT-PCR
2.8. Statistical Analysis for qRT-PCR and Microbiota Analysis
3. Results
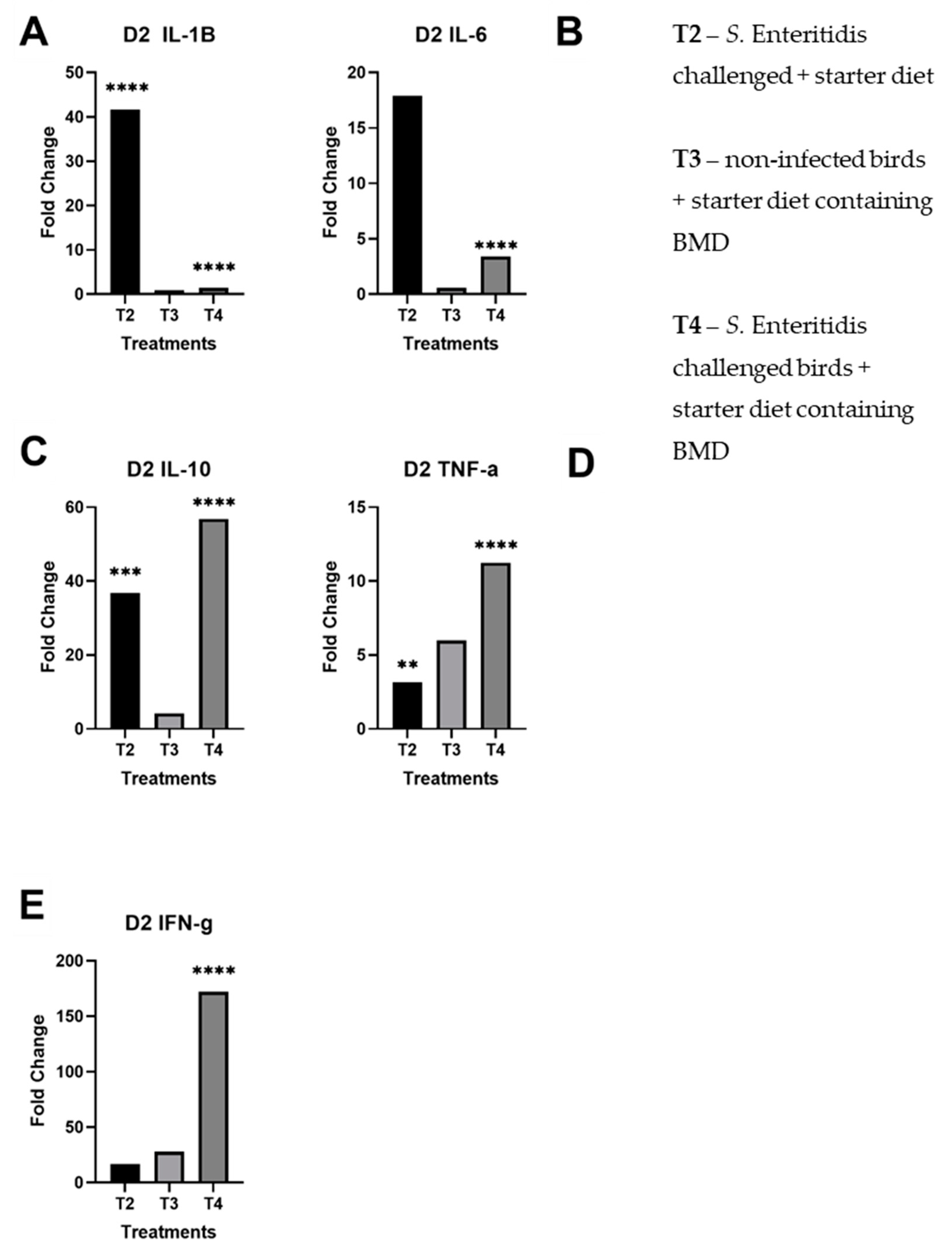
3.1. Real-Time Quantitative RT-PCR Results
3.2. Microbiota Composition
3.3. Beta Diversity Index of Cecal Composition
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boore, A.L.; Hoekstra, R.M.; Iwamoto, M.; Fields, P.I.; Bishop, R.D.; Swerdlow, D.L.; Chaturvedi, V. Salmonella enterica infections in the United States and assessment of coefficients of variation: A novel approach to identify epidemiologic characteristics of individual serotypes, 1996–2011. PLoS ONE 2015, 10, e0145416. [Google Scholar] [CrossRef] [Green Version]
- Crhanova, M.; Hradecka, H.; Faldynova, M.; Matulova, M.; Havlickova, H.; Sisak, F.; Rychlik, I. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar Enteritidis infection. Infect. Immun. 2011, 79, 2755–2763. [Google Scholar] [CrossRef] [Green Version]
- Kogut, M.H.; Arsenault, R.J. Immunometabolic phenotype alterations associated with the induction of disease tolerance and persistent asymptomatic infection of Salmonella in the chicken intestine. Front. Immunol. 2017, 8, 372. [Google Scholar] [CrossRef] [Green Version]
- Carrasco, J.M.D.; Redondo, E.A.; Viso, N.D.P.; Redondo, L.M.; Farber, M.D.; Miyakawa, M.E.F. Tannins and bacitracin differentially modulate gut microbiota of broiler chickens. BioMed Res. Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Niewold, T.A. The nonantibiotic anti-inflammatory effect of antimicrobial growth promoters, the real mode of action? A hypothesis. Poult. Sci. 2007, 86, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.B.; Kogut, M.H. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front. Vet. Sci. 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Wan, M.L.Y.; Ling, K.H.; El-Nezami, H.; Wang, M.F. Influence of functional food components on gut health. Crit. Rev. Food Sci. Nutr. 2019, 59, 1927–1936. [Google Scholar] [CrossRef]
- Broom, L.J. The sub-inhibitory theory for antibiotic growth promoters. Poult. Sci. 2017, 96, 3104–3108. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Butz, N.; Cadenas, M.B.; Koci, M.; Ballou, A.; Mendoza, M.; Ali, R.; Hassan, H. An attenuated Salmonella enterica serovar Typhimurium strain and galacto-oligosaccharides accelerate clearance of Salmonella infections in poultry through modifications to the gut microbiome. Appl. Environ. Microbiol. 2017, 84, e02526-17. [Google Scholar] [CrossRef] [Green Version]
- Lillehoj, H.; Liu, Y.; Calsamiglia, S.; Fernandez-Miyakawa, M.E.; Chi, F.; Cravens, R.L.; Oh, S.; Gay, C.G. Phytochemicals as antibiotic alternatives to promote growth and enhance host health. Vet. Res. 2018, 49. [Google Scholar] [CrossRef] [Green Version]
- Center for Veterinary Medicine. Veterinary Feed Directive (VFD) [Online]. FDA 2020. Available online: https://www.fda.gov/animal-veterinary/development-approval-process/veterinary-feed-directive-vfd (accessed on 18 October 2020).
- Barrow, P.A.; Methner, U. Salmonella in Domestic Animals, 2nd ed.; CABI: Wallingsford, UK, 2013. [Google Scholar]
- Juricova, H.; Videnska, P.; Lukac, M.; Faldynova, M.; Babak, V.; Havlickova, H.; Sisak, F.; Rychlik, I. Influence of Salmonella enterica serovar Enteritidis infection on the development of the cecum microbiota in newly hatched chicks. Appl. Environ. Microbiol. 2013, 79, 745–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mon, K.K.Z.; Saelao, P.; Halstead, M.M.; Chanthavixay, G.; Chang, H.; Garas, L.; Maga, E.A.; Zhou, H. Salmonella enterica serovars Enteritidis infection alters the indigenous microbiota diversity in young layer chicks. Front. Vet. Sci. 2015, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogut, M.H. The effect of microbiome modulation on the intestinal health of poultry. Anim. Feed Sci. Technol. 2019, 250, 32–40. [Google Scholar] [CrossRef]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.H.; Arsenault, R.J. Gut health: The new paradigm in food animal production. Front. Vet. Sci. 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Kogut, M.H.; Yin, X.; Yuan, J.; Broom, L. Gut health in poultry. CAB Rev. 2017, 12, 1–7. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.; McMurdie, P.; Han, A.; Johnson, A.J.A.; Holmes, S. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2020, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B. The RDP and GreenGenes taxonomic training sets formatted for DADA2 [software]. Zenodo 2016. [Google Scholar] [CrossRef]
- Eldaghayes, I.; Rothwell, L.; Williams, A.; Withers, D.; Balu, S.; Davison, F.; Kaiser, P. Infectious bursal disease virus: Strains that differ in virulence differentially modulate the innate immune response to infection in the chicken bursa. Viral Immunol. 2003, 19, 83–91. [Google Scholar] [CrossRef] [Green Version]
- Kogut, M.H.; Rothwell, L.; Kaiser, P. Differential regulation of cytokine gene expression by avian heterophils during receptor-mediated phagocytosis of opsonized and nonopsonized Salmonella enteritidis. J. Interferon Cytokine Res. 2003, 23, 319–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser, P.; Wigley, P.; Burnside, J.; Barrow, P.A.; Galyov, E.E.; Rothwell, L. Differential 498 cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology 2000, 146, 3217–3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swaggerty, C.L.; Kogut, M.H.; He, H.; Genovese, K.J.; Johnson, C.; Arsenault, R.J. Differential levels of cecal colonization by Salmonella Enteritidis in chickens triggers distinct immune kinome profiles. Front. Vet. Sci. 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Moody, A.; Sellers, S.; Bumstead, N. Measuring infectious bursal disease virus RNA in blood by multiplex real-time quantitative RT-PCR. J. Virol. Methods 2000, 85, 55–64. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.C.; Bessegatto, J.A.; Alfieri, A.A.; Weese, J.S.; Filho, J.A.B.; Oba, A. Different antibiotic growth promoters induce specific changes in the cecal microbiota membership of broiler chicken. PLoS ONE 2017, 12, e0171642. [Google Scholar] [CrossRef]
- Gong, J.; Yu, H.; Liu, T.; Gill, J.J.; Chambers, J.R.; Wheatcroft, R.; Sabour, P.M. Effects of zinc bacitracin, bird age and access to range on bacterial microbiota in the ileum and caeca of broiler chickens. J. Appl. Microbiol. 2008, 104, 1372–1382. [Google Scholar] [CrossRef]
- Bratburd, J.R.; Keller, C.; Vivas, E.; Gemperline, E.; Li, L.; Rey, F.E.; Currie, C.R.; Blaser, M.J. Gut microbial and metabolic responses to Salmonella enterica serovar Typhimurium and Candida albicans. mBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Gadde, U.D.; Oh, S.; Lillehoj, H.S.; Lillehoj, E.P. Antibiotic growth promoters virginiamycin and bacitracin methylene disalicylate alter the chicken intestinal metabolome. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Arsenault, R.J.; Napper, S.; Kogut, M.H. Salmonella enterica Typhimurium infection causes metabolic changes in chicken muscle involving AMPK, fatty acid and insulin/mTOR signaling. Vet. Res. 2013, 44, 35. [Google Scholar] [CrossRef] [Green Version]
- Kogut, M.H.; Genovese, K.J.; He, H.; Arsenault, R.J. AMPK and mTOR: Sensors and regulators of immunometabolic changes during Salmonella infection in the chicken. Poult. Sci. 2016, 95, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, W.N.; Beal, R.K.; Powers, C.; Wu, X.; Humphrey, T.; Watson, M.; Bailey, M.; Friedman, A.; Smith, A.L. Regional and global changes in TCRαβ T cell repertoires in the gut are dependent upon the complexity of the enteric microflora. Dev. Comp. Immunol. 2010, 34, 406–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huyghebaert, G.; Ducatelle, R.; Immerseel, F.V. An update on alternatives to antimicrobial growth promoters for broilers. Vet. J. 2011, 187, 182–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorbara, M.T.; Pamer, E.G. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal. Immunol. 2019, 12, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rychlik, I.; Elsheimer-Matulova, M.; Kyrova, K. Gene expression in the chicken caecum in response to infections with non-typhoid Salmonella. Vet. Res. 2014, 45, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blacher, E.; Levy, M.; Tatirovsky, E.; Elinav, E. Microbiome-modulated metabolites at the interface of host immunity. J. Immunol. 2017, 198, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Buck, M.D.; O’Sullivan, D.; Pearce, E.L. T cell metabolism drives immunity. J. Exp. Med. 2015, 212, 1345–1360. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Chen, C.; Indugu, N.; Werlang, G.O.; Singh, M.; Kim, W.K.; Thippareddi, H. Effect of antibiotic withdrawal in feed on chicken gut microbial dynamics, immunity, growth performance and prevalence of foodborne pathogens. PLoS ONE 2018, 13, e0192450. [Google Scholar] [CrossRef] [Green Version]
- Ballou, A.L.; Ali, R.A.; Mendoza, M.A.; Ellis, J.C.; Hassan, H.M.; Croom, W.J.; Koci, M.D. Development of the chick microbiome: How early exposure influences future microbial diversity. Front. Vet. Sci. 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Stanley, D.; Hughes, R.J.; Geier, M.S.; Moore, R.J. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: Challenges presented for the identification of performance enhancing probiotic bacteria. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; de Angelis, M. The controversial role of human gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wilson, J.; Koenigsknecht, M.; Chou, W. The intracellular innate immune sensor NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol. 2017, 18, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Danzeisen, J.L.; Kim, H.B.; Isaacson, R.E.; Tu, Z.J.; Johnson, T.J. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS ONE 2011, 6, e27949. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Hunkapiller, A.A.; Layton, A.C.; Chang, Y.J.; Robbins, K.R. Response of intestinal microbiota to antibiotic growth promoters in chickens. Foodborne Pathog. Dis. 2013, 10, 331–337. [Google Scholar] [CrossRef] [PubMed]



| RNA Target | Probe/Primer Sequences | Accession No. | |
|---|---|---|---|
| 28S | Probe | 5′-(FAM)-AGGACCGCTACGGACCTCCACCA-(TAMRA)-3′ | X59733 |
| F | 5′-GGCGAAGCCAGAGGAAACT-3′ | ||
| R | 5′-GACGACCGATTGCACGTC-3′ | ||
| IL-1β | Probe | 5′-(FAM)-CCACACTGCAGCTGGAGGAAGCC-(TAMRA)-3′ | AJ245728 |
| F | 5′-GCTCTACATGTCGTGTGTGATGAG-3′ | ||
| R | 5′-TGTCGATGTCCCGCATGA-3′ | ||
| IL-6 | Probe | 5′-(FAM)-AGGAGAAATGCCTGACGAAGCTCTCCA-(TAMRA)-3′ | AJ250838 |
| F | 5′-GCTCGCCGGCTTCGA-3′ | ||
| R | 5′-GGTAGGTCTGAAAGGCGAACAG-3′ | ||
| TNF-α | Probe | 5′-(FAM)-TGCTGAGAAGGAACAAACTGGTGGT-(TAMRA)-3′ | MF000729 |
| F | 5′-CCCATCCCTGGTCCGTAA-3′ | ||
| R | 5′- GGCGGCGTATACGAAGTAAAG-3′ | ||
| IL-10 | Probe | 5′-(FAM)-CGACGATGCGGCGCTGTCA-(TAMRA)-3′ | AJ621614 |
| F | 5′-CATGCTGCTGGGCCTGAA-3′ | ||
| R | 5′-CGTCTCCTTGATCTGCTTGATG-3′ | ||
| IFN-γ | Probe | 5′-(FAM)-TGGCCAAGCTCCCGATGAACGA-(TAMRA)-3′ | Y07922 |
| F | 5′-GTGAAGAAGGTGAAAGATATATCATGGA-3′ | ||
| R | 5′-GCTTTGCGCTGGATTCTCA-3′ | ||
| Group | Paenibacillaceae | Lachnospiraceae | Clostridiaceae | Enterobacteriaceae | Bacillaceae | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Day 2 | Day 4 | Day 2 | Day 4 | Day 2 | Day 4 | Day 2 | Day 4 | Day 2 | Day 4 | |
| T1 | 0.11 a,b | 8.23 a | 0 | 1.32 b | 95.9 a | 69.1 a | 0.55 a | 0.94 a,c | 0.14 a,c | 1.70 a |
| T2 | 0 b,d | 3.02 a,b | 0 | 0.14 b | 23.5 b | 23.8 b | 73.6 b | 58.9 b | 0 b | 0.02 b |
| T3 | 0.18 a,c | 7.25 a,b | 0.04 | 12.33 a | 86.5 c | 40.3 b | 4.45 c | 0.19 c | 0.20 c | 0.07 b |
| T4 | 0 d | 1.98 b | 0 | 1.25 b | 25.0 b | 34.3 b | 68.1 b | 48.5 b | 0 b | 0 b |
| Age | Comparison | R 1 | Probability 2 |
|---|---|---|---|
| Day 2 | T1 vs. T2 | 0.96 | 0.001 |
| T1 vs. T3 | 0.1 | 0.002 | |
| T1 vs. T4 | 0.98 | 0.001 | |
| Day 4 | T1 vs. T2 | 0.62 | 0.001 |
| T1 vs. T3 | 0.15 | 0.002 | |
| T1 vs. T4 | 0.59 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, A.; Bortoluzzi, C.; Pilla, R.; Kogut, M.H. A Role for the Microbiota in the Immune Phenotype Alteration Associated with the Induction of Disease Tolerance and Persistent Asymptomatic Infection of Salmonella in the Chicken. Microorganisms 2020, 8, 1879. https://doi.org/10.3390/microorganisms8121879
Lee A, Bortoluzzi C, Pilla R, Kogut MH. A Role for the Microbiota in the Immune Phenotype Alteration Associated with the Induction of Disease Tolerance and Persistent Asymptomatic Infection of Salmonella in the Chicken. Microorganisms. 2020; 8(12):1879. https://doi.org/10.3390/microorganisms8121879
Chicago/Turabian StyleLee, Annah, Cristiano Bortoluzzi, Rachel Pilla, and Michael H. Kogut. 2020. "A Role for the Microbiota in the Immune Phenotype Alteration Associated with the Induction of Disease Tolerance and Persistent Asymptomatic Infection of Salmonella in the Chicken" Microorganisms 8, no. 12: 1879. https://doi.org/10.3390/microorganisms8121879
APA StyleLee, A., Bortoluzzi, C., Pilla, R., & Kogut, M. H. (2020). A Role for the Microbiota in the Immune Phenotype Alteration Associated with the Induction of Disease Tolerance and Persistent Asymptomatic Infection of Salmonella in the Chicken. Microorganisms, 8(12), 1879. https://doi.org/10.3390/microorganisms8121879







