Glutamate Decarboxylase from Lactic Acid Bacteria—A Key Enzyme in GABA Synthesis
Abstract
1. Introduction
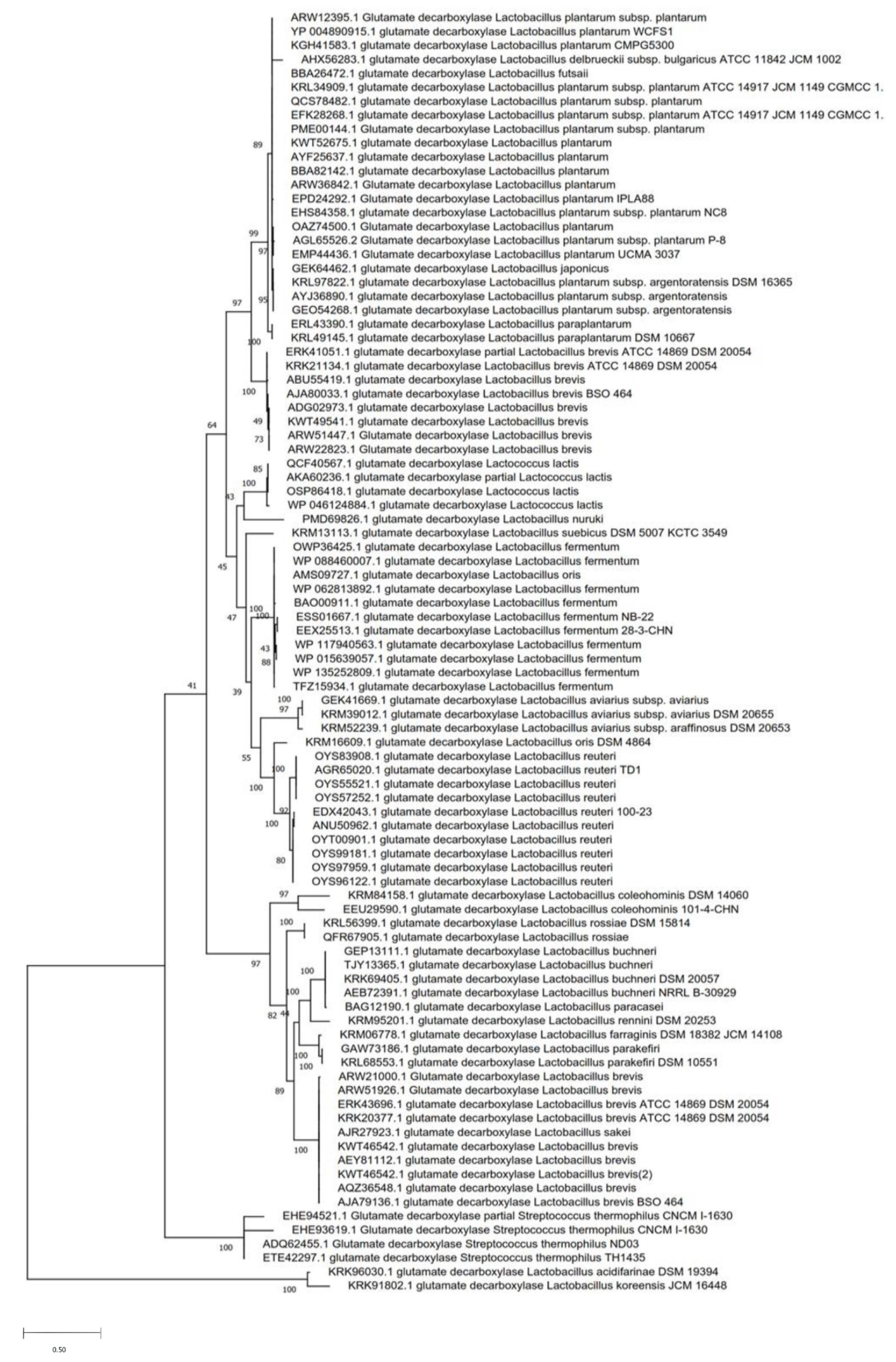
2. Biodiversity of Glutamate to γ-Aminobutyric Acid (GABA)-Producing Lactic Acid Bacteria
3. Occurrence and Organization of Glutamic Acid Decarboxylase (GAD) Genes
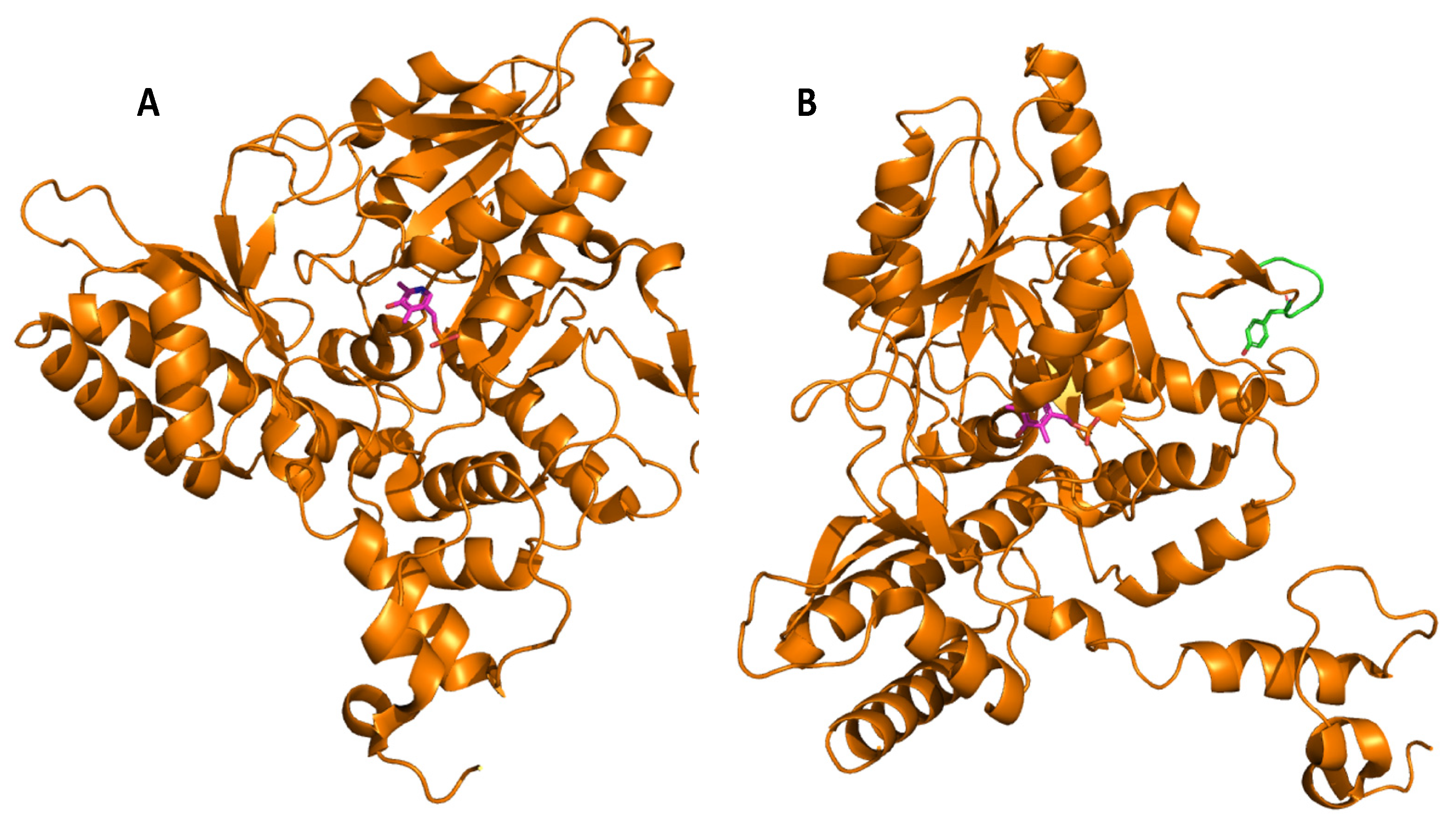
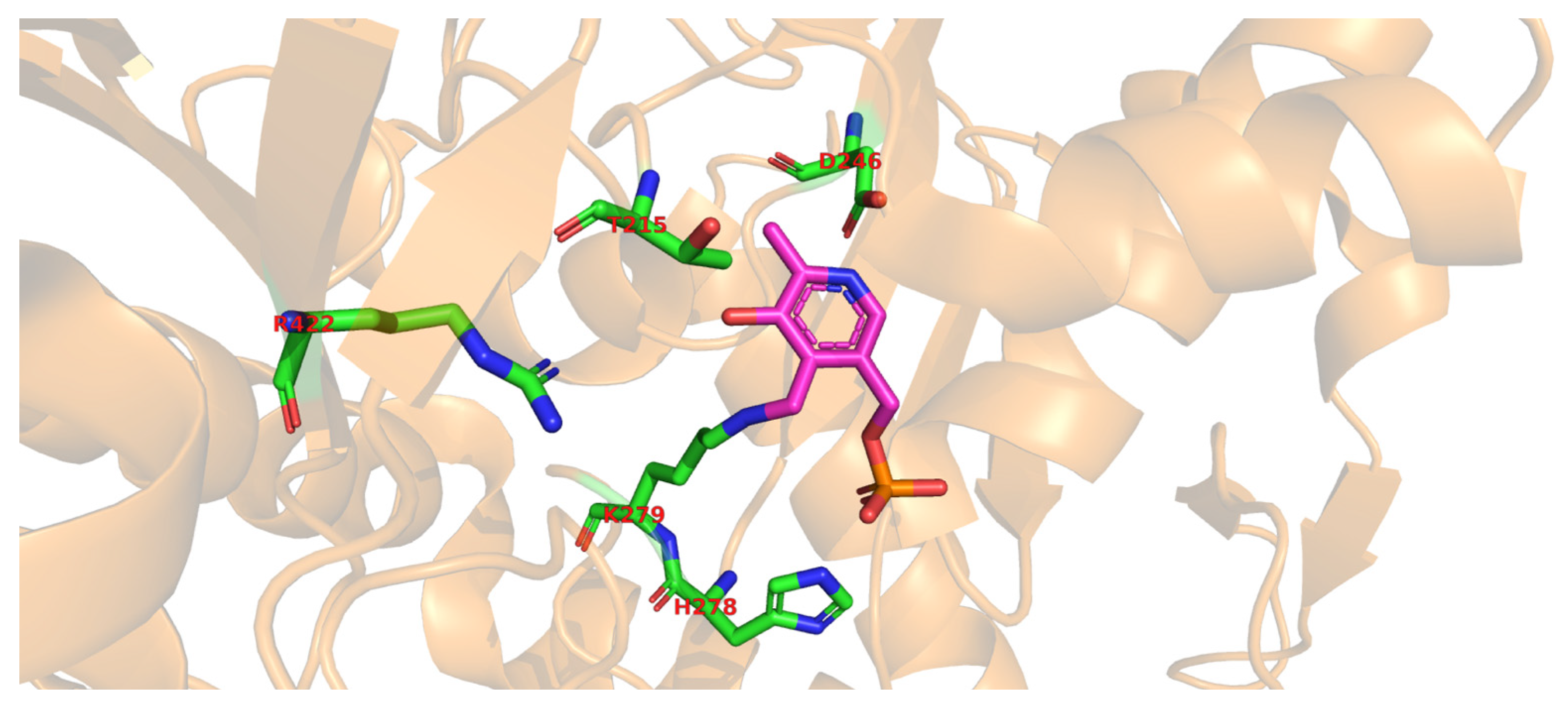
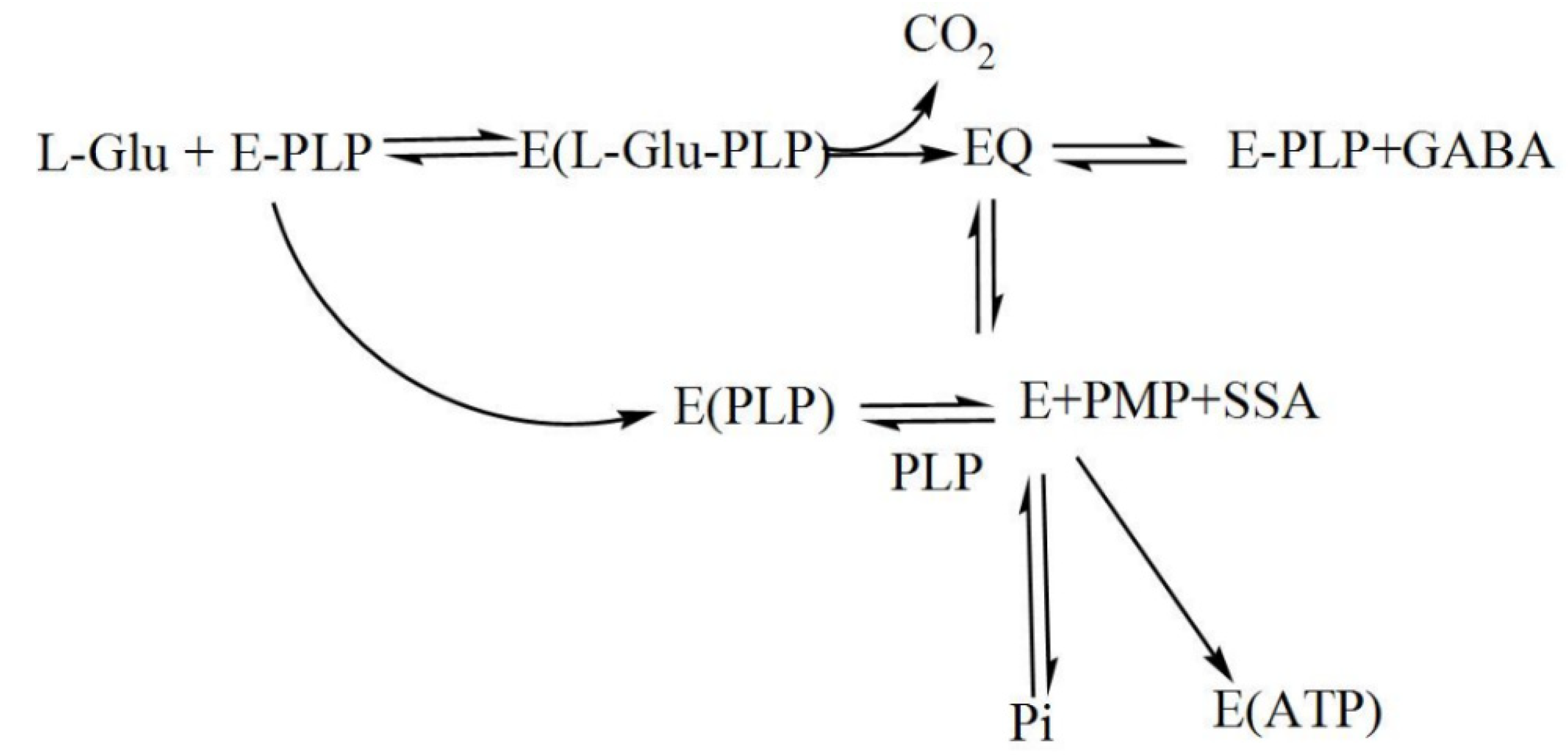
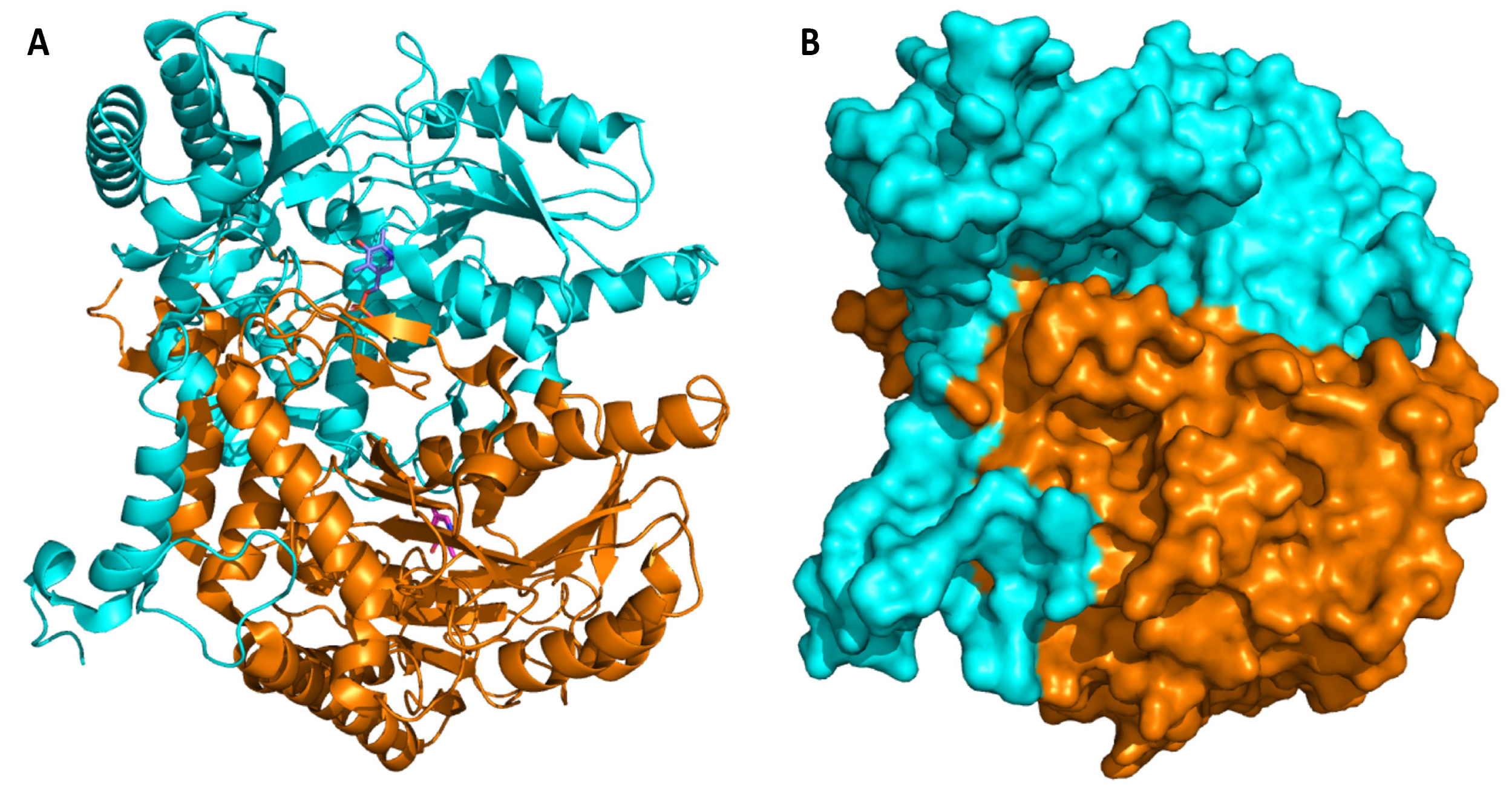
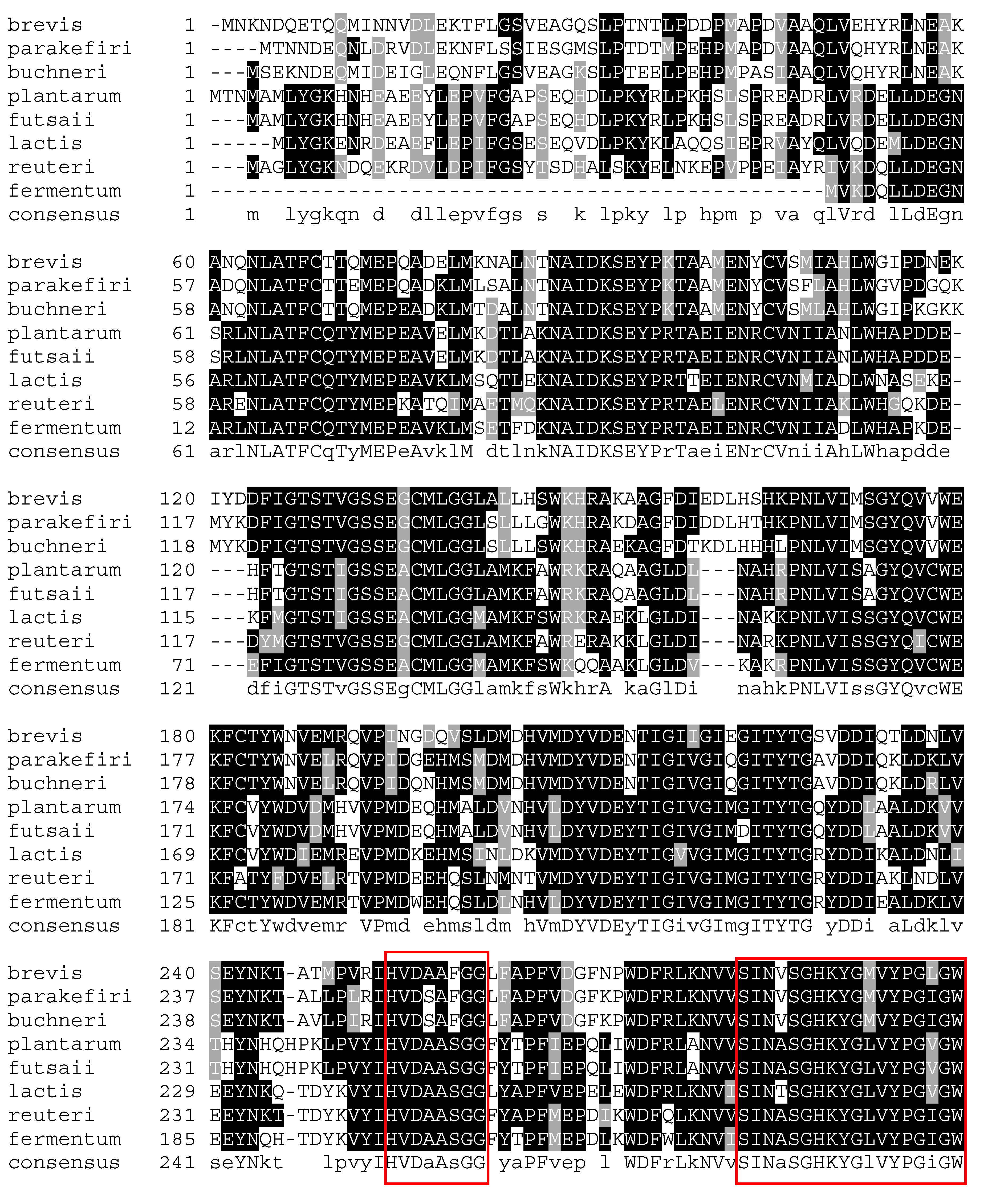
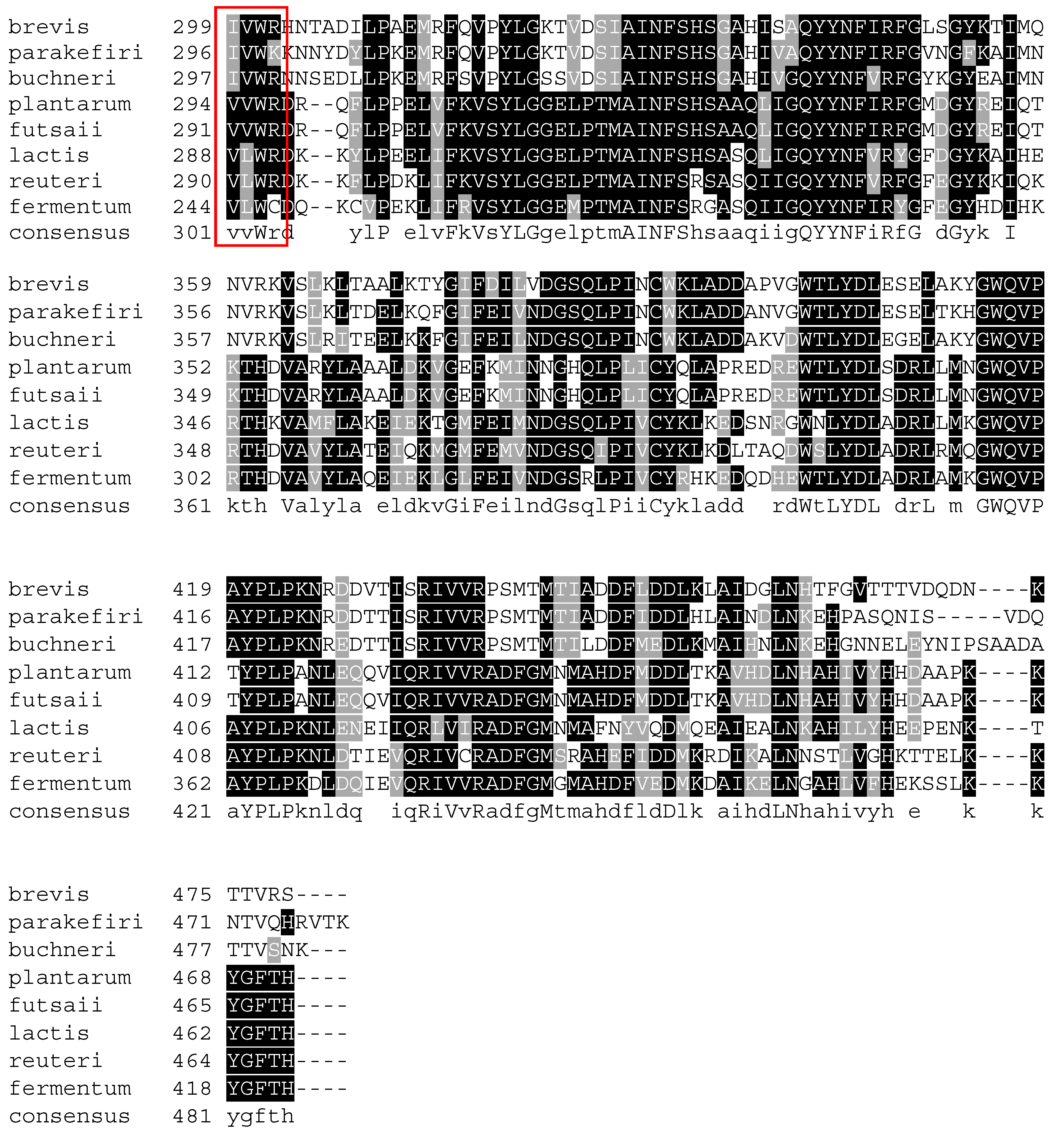
4. Glutamate Decarboxylase
5. Biochemical Insights into Glutamate Decarboxylase from Lactic Acid Bacteria
6. Improvement of GAD Activities and GABA Production
7. The Role of Glutamate Decarboxylase in the Manufacturing of Bio-Based Industrial Chemicals
8. Future Trends and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ly, D.; Mayrhofer, S.; Yogeswara, I.B.A.; Nguyen, T.-H.; Domig, K.J. Identification, classification and screening for γ-amino-butyric acid production in lactic acid bacteria from Cambodian fermented foods. Biomolecules 2019, 9, 768. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C. Surviving the acid test: Responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Nakajima, I.; Fujita, Y.; Kobayashi, M.; Kimoto, H.; Suzuki, I.; Aso, H. Lactococcus lactis contains only one glutamate decarboxylase gene. Microbiology 1999, 145, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Lu, P.; Shi, Y. Substrate selectivity of the acid-activated glutamate/γ-aminobutyric acid (GABA) antiporter GadC from Escherichia coli. J. Biol. Chem. 2013, 288, 15148–15153. [Google Scholar] [CrossRef] [PubMed]
- Somkuti, G.A.; Renye, J.A.; Steinberg, D.H. Molecular analysis of the glutamate decarboxylase locus in Streptococcus thermophilus ST110. J. Ind. Microbiol. Biotechnol. 2012, 39, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Ren, C.; Xu, Y. Deciphering the crucial roles of transcriptional regulator GadR on γ-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb. Cell Factories 2019, 18, 1–12. [Google Scholar] [CrossRef]
- Wu, Q.; Tun, H.M.; Law, Y.-S.; Khafipour, E.; Shah, N.P. Common distribution of gad operon in Lactobacillus brevis and its GadA contributes to efficient GABA synthesis toward cytosolic near-neutral pH. Front. Microbiol. 2017, 8, 206. [Google Scholar] [CrossRef]
- Brambilla, P.; Perez, J.; Barale, F.; Schettini, G.; Soares, J.C. GABAergic dysfunction in mood disorders. Mol. Psychiatry 2003, 8, 721–737. [Google Scholar] [CrossRef]
- Ko, C.Y.; Lin, H.-T.V.; Tsai, G.-J. γ-aminobutyric acid production in black soybean milk by Lactobacillus brevis FPA 3709 and the antidepressant effect of the fermented product on a forced swimming rat model. Process Biochem. 2013, 48, 559–568. [Google Scholar] [CrossRef]
- Inoue, K.; Shirai, T.; Ochiai, H.; Kasao, M.; Hayakawa, K.; Kimura, M.; Sansawa, H. Blood-pressure-lowering effect of a novel fermented milk containing γ-aminobutyric acid (GABA) in mild hypertensives. Eur. J. Clin. Nutr. 2003, 57, 490–495. [Google Scholar] [CrossRef]
- Tsai, J.S.; Lin, Y.S.; Pan, B.S.; Chen, T.J. Antihypertensive peptides and γ-aminobutyric acid from prozyme 6 facilitated lactic acid bacteria fermentation of soymilk. Process. Biochem. 2006, 41, 1282–1288. [Google Scholar] [CrossRef]
- Watanabe, S.; Katsube, T.; Sonomoto, K. Cholesterol-lowering effects of Lactobacillus brevis isolated from turnip “tsuda kabu”. Food Sci. Technol. Res. 2012, 18, 825–834. [Google Scholar] [CrossRef]
- Pouliot-Mathieu, K.; Gardner-Fortier, C.; Lemieux, S.; St-Gelais, D.; Champagne, C.P.; Vuillemard, J.-C. Effect of cheese containing γ-aminobutyric acid-producing lactic acid bacteria on blood pressure in men. PharmaNutrition 2013, 1, 141–148. [Google Scholar] [CrossRef]
- Xie, Z.; Xia, S.; Le, G.-W. γ-aminobutyric acid improves oxidative stress and function of the thyroid in high-fat diet fed mice. J. Funct. Foods 2014, 8, 76–86. [Google Scholar] [CrossRef]
- Xu, N.; Wei, L.; Liu, J. Biotechnological advances and perspectives of γ-aminobutyric acid production. World J. Microbiol. Biotechnol. 2017, 33, 64. [Google Scholar] [CrossRef]
- Diana, M.; Tres, A.; Quílez, J.; Llombart, M.; Rafecas, M. Spanish cheese screening and selection of lactic acid bacteria with high gamma-aminobutyric acid production. LWT Food Sci. Technol. 2014, 56, 351–355. [Google Scholar] [CrossRef]
- Le Vo, T.D.; Kim, T.W.; Hong, S.H. Effects of glutamate decarboxylase and γ-aminobutyric acid (GABA) transporter on the bioconversion of GABA in engineered Escherichia coli. Bioprocess Biosyst. Eng. 2012, 35, 645–650. [Google Scholar] [CrossRef]
- Komatsuzaki, N.; Nakamura, T.; Kimura, T.; Shima, J. Characterization of glutamate decarboxylase from a high γ-aminobutyric acid (GABA)-producer, Lactobacillus paracasei. Biosci. Biotechnol. Biochem. 2008, 72, 278–285. [Google Scholar] [CrossRef]
- Ratanaburee, A.; Kantachote, D.; Charernjiratrakul, W.; Sukhoom, A. Selection of γ-aminobutyric acid-producing lactic acid bacteria and their potential as probiotics for use as starter cultures in Thai fermented sausages (Nham). Int. J. Food Sci. Technol. 2013, 48, 1371–1382. [Google Scholar] [CrossRef]
- Komatsuzaki, N.; Shima, J.; Kawamoto, S.; Momose, H.; Kimura, T. Production of γ-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol. 2005, 22, 497–504. [Google Scholar] [CrossRef]
- Siragusa, S.; De Angelis, M.; Di Cagno, R.; Rizzello, C.G.; Coda, R.; Gobbetti, M. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 2007, 73, 7283–7290. [Google Scholar] [CrossRef] [PubMed]
- Fan, E.; Huang, J.; Hu, S.; Mei, L.; Yu, K. Cloning, sequencing and expression of a glutamate decarboxylase gene from the GABA-producing strain Lactobacillus brevis CGMCC 1306. Ann. Microbiol. 2012, 62, 689–698. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, M.Y.; Ji, G.E.; Lee, Y.S.; Hwang, K.T. Production of γ-aminobutyric acid in black raspberry juice during fermentation by Lactobacillus brevis GABA100. Int. J. Food Microbiol. 2009, 130, 12–16. [Google Scholar] [CrossRef]
- Kim, J.A.; Park, M.S.; Kang, S.A.; Ji, G.E. Production of γ-aminobutyric acid during fermentation of Gastrodia elata Bl. by co-culture of Lactobacillus brevis GABA 100 with Bifidobacterium bifidum BGN4. Food Sci. Biotechnol. 2014, 23, 459–466. [Google Scholar] [CrossRef]
- Lee, B.-J.; Kim, J.-S.; Kang, Y.M.; Lim, J.-H.; Kim, Y.-M.; Lee, M.-S.; Jeong, M.-H.; Ahn, C.-B.; Je, J.-Y. Antioxidant activity and γ-aminobutyric acid (GABA) content in sea tangle fermented by Lactobacillus brevis BJ20 isolated from traditional fermented foods. Food Chem. 2010, 122, 271–276. [Google Scholar] [CrossRef]
- Sanchart, C.; Rattanaporn, O.; Haltrich, D.; Phukpattaranont, P.; Maneerat, S. Lactobacillus futsaii CS3, a new GABA-producing strain isolated from Thai fermented shrimp (Kung-som). Indian J. Microbiol. 2016, 57, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.S.; Cha, I.T.; Roh, S.W.; Shin, H.H.; Seo, M.J. Enhanced production of γ-aminobutyric acid by optimizing culture conditions of Lactobacillus brevis HYE1 isolated from kimchi, a Korean fermented food. J. Microbiol. Biotechnol. 2017, 27, 450–459. [Google Scholar] [CrossRef]
- Li, H.; Gao, D.; Cao, Y. A high γ-aminobutyric acid-producing Lactobacillus brevis isolated from Chinese traditional paocai. Ann. Microbiol. 2008, 58, 649–653. [Google Scholar] [CrossRef]
- Sun, T.; Zhao, S.; Wang, H.; Cai, C.; Chen, Y.; Zhang, H. ACE-inhibitory activity and γ-aminobutyric acid content of fermented skim milk by Lactobacillus helveticus isolated from Xinjiang koumiss in China. Eur. Food Res. Technol. 2009, 228, 607–612. [Google Scholar] [CrossRef]
- Franciosi, E.; Carafa, I.; Nardin, T.; Schiavon, S.; Poznanski, E.; Cavazza, A.; Larcher, R.; Tuohy, K.M. Biodiversity and γ-aminobutyric acid production by lactic acid bacteria isolated from traditional alpine raw cow’s milk cheeses. Biomed. Res. Int. 2015, 2015, 625740. [Google Scholar] [CrossRef]
- Das, D.; Goyal, A. Antioxidant activity and γ-aminobutyric acid (GABA) producing ability of probiotic Lactobacillus plantarum DM5 isolated from Marcha of Sikkim. LWT Food Sci. Technol. 2015, 61, 263–268. [Google Scholar] [CrossRef]
- Han, S.H.; Hong, K.B.; Suh, H.J. Biotransformation of monosodium glutamate to γ-aminobutyric acid by isolated strain Lactobacillus brevis L-32 for potentiation of pentobarbital-induced sleep in mice. Food Biotechnol. 2017, 31, 80–93. [Google Scholar] [CrossRef]
- Zhao, A.; Hu, X.; Pan, L.; Wang, X. Isolation and characterization of a γ-aminobutyric acid producing strain Lactobacillus buchneri WPZ001 that could efficiently utilize xylose and corncob hydrolysate. Appl. Microbiol. Biotechnol. 2015, 99, 3191–3200. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xie, C.; Gu, Z. Optimisation of fermentative parameters for GABA enrichment by Lactococcus lactis. Czech J. Food Sci. 2009, 27, 433–442. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; Domingos-Lopes, M.F.P.; Stanton, C.; Ross, R.P.; Silva, C.C.G. Production of γ-aminobutryric acid (GABA) by Lactobacillus otakiensis and other Lactobacillus sp. isolated from traditional Pico cheese. Int. J. Dairy Technol. 2018, 70, 1–6. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Lü, F.-X.; Lu, Z.-X.; Bie, X.-M.; Jiao, Y.; Sun, L.-J.; Yu, B. Production of γ-aminobutyric acid by Streptococcus salivarius subsp. thermophilus Y2 under submerged fermentation. Amino Acids 2008, 34, 473–478. [Google Scholar] [CrossRef]
- Cho, Y.R.; Chang, J.Y.; Chang, H.C. Production of γ-aminobutyric acid (GABA) by Lactobacillus buchneri isolated from Kimchi and its neuroprotective effect on neuronal cells. J. Microbiol. Biotechnol. 2007, 17, 104–109. [Google Scholar]
- Lim, H.S.; Cha, I.T.; Lee, H.; Seo, M.J. Optimization of γ-aminobutyric acid production by Enterococcus faecium JK29 isolated from a traditional fermented foods. Korean J. Microbiol. Biotechnol. 2016, 44, 26–33. [Google Scholar] [CrossRef]
- Seo, M.-J.; Nam, Y.-D.; Park, S.-L.; Lee, S.-Y.; Yi, S.-H.; Lim, S.-I. γ-aminobutyric acid production in skim milk co-fermented with Lactobacillus brevis 877G and Lactobacillus sakei 795. Food Sci. Biotechnol. 2013, 22, 751–755. [Google Scholar] [CrossRef]
- Agung Yogeswara, I.B.; Kusumawati, I.G.A.W.; Sumadewi, N.L.U.; Rahayu, E.S.; Indrati, R. Isolation and identification of lactic acid bacteria from Indonesian fermented foods as γ-aminobutyric acid-producing bacteria. Int. Food Res. J. 2018, 25, 1753–1757. [Google Scholar]
- Barla, F.; Koyanagi, T.; Tokuda, N.; Matsui, H.; Katayama, T.; Kumagai, H.; Michihata, T.; Sasaki, T.; Tsuji, A.; Enomoto, T. The γ-aminobutyric acid-producing ability under low pH conditions of lactic acid bacteria isolated from traditional fermented foods of Ishikawa Prefecture, Japan, with a strong ability to produce ACE-inhibitory peptides. Biotechnol. Rep. 2016, 10, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Binh, T.T.T.; Ju, W.-T.; Jung, W.-J.; Park, R.D. Optimization of γ-amino butyric acid production in a newly isolated Lactobacillus brevis. Biotechnol. Lett. 2013, 36, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Shah, N.P. High γ-aminobutyric acid production from lactic acid bacteria: Emphasis on Lactobacillus brevis as a functional dairy starter. Crit. Rev. Food Sci. Nutr. 2017, 57, 3661–3672. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kim, K.-S. Isolation and identification of γ-aminobutyric acid (GABA)-producing lactic acid bacteria from Kimchi. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 777–785. [Google Scholar] [CrossRef]
- Tanamool, V.; Hongsachart, P.; Soemphol, W. Screening and characterisation of γ-aminobutyric acid (GABA) producing lactic acid bacteria isolated from Thai fermented fish (Plaa-som) in Nong Khai and its application in Thai fermented vegetables (Som-pak). Food Sci. Technol. 2019, 2061, 1–8. [Google Scholar] [CrossRef]
- Park, J.Y.; Jeong, S.-J.; Kim, J.H. Characterization of a glutamate decarboxylase (GAD) gene from Lactobacillus zymae. Biotechnol. Lett. 2014, 36, 1791–1799. [Google Scholar] [CrossRef]
- Sanchart, C.; Rattanaporn, O.; Haltrich, D.; Phukpattaranont, P.; Maneerat, S. Technological and safety properties of newly isolated GABA-producing Lactobacillus futsaii strains. J. Appl. Microbiol. 2016, 121, 734–745. [Google Scholar] [CrossRef]
- Li, H.; Cao, Y. Lactic acid bacterial cell factories for γ-aminobutyric acid. Amino Acids 2010, 39, 1107–1116. [Google Scholar] [CrossRef]
- Kono, I.; Himeno, K. Changes in γ-aminobutyric acid content during beni-koji making. Biosci. Biotechnol. Biochem. 2000, 64, 617–619. [Google Scholar] [CrossRef]
- Masuda, K.; Guo, X.-F.; Uryu, N.; Hagiwara, T.; Watabe, S. Isolation of marine yeasts collected from the Pacific Ocean showing a high production of γ-aminobutyric acid. Biosci. Biotechnol. Biochem. 2008, 72, 3265–3272. [Google Scholar] [CrossRef]
- Kinnersley, A.M.; Turano, F.J. γ-aminobutyric acid (GABA) and plant responses to stress. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Ueno, H. Enzymatic and structural aspects on glutamate decarboxylase. J. Mol. Catal. B Enzym. 2000, 10, 67–79. [Google Scholar] [CrossRef]
- Cotter, P.D.; O’Reilly, K.; Hill, C. Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J. Food Prot. 2001, 64, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- De Biase, D.; Pennacchietti, E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: Function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 2012, 86, 770–786. [Google Scholar] [CrossRef]
- Lim, H.S.; Seo, D.-H.; Cha, I.-T.; Lee, H.; Nam, Y.-D.; Seo, M.-J. Expression and characterization of glutamate decarboxylase from Lactobacillus brevis HYE1 isolated from kimchi. World J. Microbiol. Biotechnol. 2018, 34, 44. [Google Scholar] [CrossRef] [PubMed]
- Capitani, G.; De Biase, D.; Aurizi, C.; Gut, H.; Bossa, F.; Grütter, M.G. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 2003, 22, 4027–4037. [Google Scholar] [CrossRef]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of γ-aminobutyric acid from lactic acid bacteria: A systematic review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef]
- Yu, T.; Li, L.; Zhao, Q.; Wang, P.; Zuo, X. Complete genome sequence of bile-isolated Enterococcus avium strain 352. Gut Pathog. 2019, 11, 1–5. [Google Scholar] [CrossRef]
- Gao, D.; Chang, K.; Ding, G.; Wu, H.; Chen, Y.; Jia, M.; Liu, X.; Wang, S.; Jin, Y.; Pan, H.; et al. Genomic insights into a robust γ-aminobutyric acid-producer Lactobacillus brevis CD0817. AMB Express 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Liu, X.; Cao, Y. gadA gene locus in Lactobacillus brevis NCL912 and its expression during fed-batch fermentation. FEMS Microbiol. Lett. 2013, 349, 108–116. [Google Scholar] [CrossRef][Green Version]
- Lyu, C.; Zhao, W.; Peng, C.; Hu, S.; Fang, H.; Hua, Y.; Yao, S.; Huang, J.; Mei, L. Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production. Microb. Cell Factories 2018, 17, 180. [Google Scholar] [CrossRef] [PubMed]
- Yunes, R.A.; Poluektova, E.U.; Dyachkova, M.S.; Klimina, K.M.; Kovtun, A.S.; Averina, O.V.; Orlova, V.S.; Danilenko, V.N. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 2016, 42, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Yang, S.; Lü, F.; Lu, Z.; Bie, X.; Jiao, Y.; Zou, X. Cloning and expression of glutamate decarboxylase gene from Streptococcus thermophilus Y2. J. Gen. Appl. Microbiol. 2009, 55, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.W.; Leenhouts, K.; Burghoorn, J.; Brands, J.R.; Venema, G.; Kok, J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 1998, 27, 299–310. [Google Scholar] [CrossRef]
- Su, M.S.; Schlicht, S.; Gänzle, M.G. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Factories 2011, 10, S8. [Google Scholar] [CrossRef]
- Park, K.-B.; Oh, S.-H. Cloning, sequencing and expression of a novel glutamate decarboxylase gene from a newly isolated lactic acid bacterium, Lactobacillus brevis OPK-3. Bioresour. Technol. 2007, 98, 312–319. [Google Scholar] [CrossRef]
- Steffen-Munsberg, F.; Vickers, C.; Kohls, H.; Land, H.; Mallin, H.; Nobili, A.; Skalden, L.; Bergh, T.V.D.; Joosten, H.-J.; Berglund, P.; et al. Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol. Adv. 2015, 33, 566–604. [Google Scholar] [CrossRef]
- Huang, J.; Fang, H.; Gai, Z.-C.; Mei, J.-Q.; Li, J.-N.; Hu, S.; Lv, C.-J.; Zhao, W.-R.; Mei, L.-H. Lactobacillus brevis CGMCC 1306 glutamate decarboxylase: Crystal structure and functional analysis. Biochem. Biophys. Res. Commun. 2018, 503, 1703–1709. [Google Scholar] [CrossRef]
- Rocha, J.F.; Pina, A.F.; Sousa, S.F.; Cerqueira, N.M.F.S.A. PLP-dependent enzymes as important biocatalysts for the pharmaceutical, chemical and food industries: A structural and mechanistic perspective. Catal. Sci. Technol. 2019, 9, 4864–4876. [Google Scholar] [CrossRef]
- Sandmeier, E.; Hale, T.I.; Christen, P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 1994, 221, 997–1002. [Google Scholar] [CrossRef]
- Yu, K.; Lin, L.; Hu, S.; Huang, J.; Mei, L. C-terminal truncation of glutamate decarboxylase from Lactobacillus brevis CGMCC 1306 extends its activity toward near-neutral pH. Enzym. Microb. Technol. 2012, 50, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Tramonti, A.; John, R.A.; Bossa, F.; De Biase, D. Contribution of Lys276 to the conformational flexibility of the active site of glutamate decarboxylase from Escherichia coli. Eur. J. Biochem. 2002, 269, 4913–4920. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Shin, B.-H.; Kim, Y.-H.; Nam, S.-W.; Jeon, S.-J. Cloning and expression of a full-length glutamate decarboxylase gene from Lactobacillus brevis BH2. Biotechnol. Bioprocess Eng. 2007, 12, 707–712. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, R.; Sun, H.; Li, B. Quantum chemistry studies of the catalysis mechanism differences between the two isoforms of glutamic acid decarboxylase. J. Mol. Model. 2013, 19, 705–714. [Google Scholar] [CrossRef]
- Chang, C.; Zhang, J.; Ma, S.; Wang, L.; Wang, D.; Zhang, J.; Gao, Q. Purification and characterization of glutamate decarboxylase from Enterococcus raffinosus TCCC11660. J. Ind. Microbiol. Biotechnol. 2017, 44, 817–824. [Google Scholar] [CrossRef]
- Battaglioli, G.; Liu, H.; Martin, D.L. Kinetic differences between the isoforms of glutamate decarboxylase: Implications for the regulation of GABA synthesis. Neurochemistry 2003, 86, 879–887. [Google Scholar] [CrossRef]
- Jansonius, J.N. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 1998, 8, 759–769. [Google Scholar] [CrossRef]
- Hiraga, K.; Ueno, Y.; Oda, K. Glutamate decarboxylase from Lactobacillus brevis: Activation by ammonium sulfate. Biosci. Biotechnol. Biochem. 2008, 72, 1299–1306. [Google Scholar] [CrossRef]
- Shin, S.; Kim, H.; Joo, Y.; Lee, S.; Lee, Y.; Lee, S.J.; Lee, D.W. Charactrization of glutamate decarboxylase from Lactobacillus plantarum and its C-terminal function for the pH dependence of activity. J. Agric. Food Chem. 2014, 62, 12186–12193. [Google Scholar] [CrossRef]
- Sa, H.D.; Park, J.Y.; Jeong, S.J.; Lee, K.W.; Kim, J.H. Characterization of glutamate decarboxylase (GAD) from Lactobacillus sakei A156 isolated from jeot-gal. J. Microbiol. Biotechnol. 2015, 25, 696–703. [Google Scholar] [CrossRef]
- Seo, M.-J.; Nam, Y.-D.; Lee, S.-Y.; Park, S.-L.; Yi, S.-H.; Lim, S.-I. Expression and characterization of a glutamate decarboxylase from Lactobacillus brevis 877G producing γ-aminobutyric acid. Biosci. Biotechnol. Biochem. 2013, 77, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Shim, J.M.; Yao, Z.; Kim, J.A.; Kim, H.J.; Kim, J.H. Characterization of a glutamate decarboxylase (GAD) from Enterococcus avium M5 isolated from Jeotgal, a Korean fermented seafood. J. Microbiol. Biotechnol. 2017, 27, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Lin, Q.; Lu, Z.X.; Lu, F.X.; Bie, X.M.; Zou, X.K.; Sun, L.J. Characterization of a novel glutamate decarboxyase from Streptococcus salivarius ssp. thermophilus Y2. J. Chem. Technol. Biotechnol. 2008, 83, 855–861. [Google Scholar] [CrossRef]
- Shi, F.; Xie, Y.; Jiang, J.; Wang, N.; Li, Y.; Wang, X. Directed evolution and mutagenesis of glutamate decarboxylase from Lactobacillus brevis Lb85 to broaden the range of its activity toward a near-neutral pH. Enzym. Microb. Technol. 2014, 61–62, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Zhao, W.; Hu, S.; Huang, J.; Lu, T.; Jin, Z.; Mei, L.; Yao, S. Physiology-oriented engineering strategy to improve gamma-aminobutyrate production in Lactobacillus brevis. J. Agric. Food Chem. 2017, 65, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xin, Y.; Zhang, F.; Feng, Z.; Fu, J.; Luo, L.; Yin, Z. Enhanced γ-aminobutyric acid-forming activity of recombinant glutamate decarboxylase (gadA) from Escherichia coli. World J. Microbiol. Biotechnol. 2011, 27, 693–700. [Google Scholar] [CrossRef]
- Kook, M.C.; Seo, M.J.; Cheigh, C.I.; Lee, S.J.; Pyun, Y.R.; Park, H. Enhancement of γ-aminobutyric acid production by Lactobacillus sakei B2-16 expressing glutamate decarboxylase from Lactobacillus plantarum ATCC 14917. J. Appl. Biol. Chem. 2010, 53, 816–820. [Google Scholar]
- Tajabadi, N.; Baradaran, A.; Ebrahimpour, A.; Rahim, R.A.; Bakar, F.A.; Manap, M.Y.A.; Mohammed, A.S.; Saari, N. Overexpression and optimization of glutamate decarboxylase in Lactobacillus plantarum Taj-Apis362 for high γ-aminobutyric acid production. Microb. Biotechnol. 2015, 8, 623–632. [Google Scholar] [CrossRef]
- Choi, J.W.; Yim, S.S.; Lee, S.H.; Kang, T.J.; Park, S.J.; Jeong, K.J. Enhanced production of γ-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb. Cell Fact. 2015, 14, 21. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, J.; Chen, L.; Lu, F.; Lu, Z. Biosynthesis of γ-aminobutyric acid by a recombinant Bacillus subtilis strain expressing the glutamate decarboxylase gene derived from Streptococcus salivarius ssp. thermophilus Y2. Process. Biochem. 2014, 49, 1851–1857. [Google Scholar] [CrossRef]
- Li, H.; Qiu, T.; Liu, X.; Cao, Y. Continuous cultivation of Lactobacillus brevis NCL912 for production of γ-aminobutyric acid. Ann. Microbiol. 2013, 63, 1649–1652. [Google Scholar] [CrossRef]
- Li, H.; Qiu, T.; Huang, G.; Cao, Y. Production of γ-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb. Cell Factories 2010, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y.-H.; Liaw, W.-C.; Kuo, J.-M.; Deng, C.-S.; Wu, C.-H. Hydrogel film-immobilized Lactobacillus brevis RK03 for γ-aminobutyric acid production. Int. J. Mol. Sci. 2017, 18, 2324. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.M.; Lee, J.W.; Park, S.M.; Lee, M.Y.; Ji, G.E.; Park, M.S.; Heo, T.R. Improvement of γ-aminobutyric acid (GABA) production using cell entrapment of Lactobacillus brevis GABA 057. J. Microbiol. Biotechnol. 2006, 16, 562–568. [Google Scholar]
- Thangrongthong, S.; Puttarat, N.; Ladda, B.; Itthisoponkul, T.; Pinket, W.; Kasemwong, K.; Taweechotipatr, M. Microencapsulation of probiotic Lactobacillus brevis ST-69 producing GABA using alginate supplemented with nanocrystalline starch. Food Sci. Biotechnol. 2020, 29, 1475–1478. [Google Scholar] [CrossRef]
- Villegas, J.M.; Brown, L.; de Giori, G.S.; Hebert, E.M. Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. LWT Food Sci. Technol. 2016, 67, 22–26. [Google Scholar] [CrossRef]
- Yang, X.; Ke, C.; Zhu, J.; Wang, Y.; Zeng, W.; Huang, J. Enhanced productivity of γ-amino butyric acid by cascade modifications of a whole-cell biocatalyst. Appl. Microbiol. Biotechnol. 2018, 102, 3623–3633. [Google Scholar] [CrossRef]
- Fan, L.-Q.; Li, M.-W.; Qiu, Y.-J.; Chen, Q.-M.; Jiang, S.-J.; Shang, Y.-J.; Zhao, L.-M. Increasing thermal stability of glutamate decarboxylase from Escherichia coli by site-directed saturation mutagenesis and its application in GABA production. J. Biotechnol. 2018, 278, 1–9. [Google Scholar] [CrossRef]
- Ke, C.; Wei, J.; Ren, Y.; Yang, X.; Chen, J.; Huang, J. Efficient γ-aminobutyric acid bioconversion by engineered Escherichia coli. Biotechnol. Biotechnol. Equip. 2018, 32, 566–573. [Google Scholar] [CrossRef]
- Ho, N.A.T.; Hou, C.Y.; Kim, W.H.; Kang, T.J. Expanding the active pH range of Escherichia coli glutamate decarboxylase by breaking the cooperativeness. J. Biosci. Bioeng. 2013, 115, 154–158. [Google Scholar] [CrossRef]
- Lin, L.; Hu, S.; Yu, K.; Huang, J.; Yao, S.; Lei, Y.; Hu, G.; Mei, L. Enhancing the activity of glutamate decarboxylase from Lactobacillus brevis by directed evolution. Chin. J. Chem. Eng. 2014, 22, 1322–1327. [Google Scholar] [CrossRef]
- Zhang, Q.-F.; Hu, S.; Zhao, W.-R.; Huang, J.; Mei, J.-Q.; Mei, L.-H. Parallel strategy increases the thermostability and activity of glutamate decarboxylase. Molecules 2020, 25, 690. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Pasamontes, L.; Lassen, S.F.; Wyss, M. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta Protein Struct. Mol. Enzym. 2000, 1543, 408–415. [Google Scholar] [CrossRef]
- Lammens, T.M.; De Biase, D.; Franssen, M.C.R.; Scott, E.L.; Sanders, J.P.M. The application of glutamic acid α-decarboxylase for the valorization of glutamic acid. Green Chem. 2009, 11, 1562–1567. [Google Scholar] [CrossRef]
- Park, H.; Ahn, J.; Lee, J.; Lee, H.; Kim, C.; Jung, J.-K.; Lee, H.; Lee, E.G. Expression, immobilization and enzymatic properties of glutamate decarboxylase fused to a cellulose-binding domain. Int. J. Mol. Sci. 2012, 13, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, H.S.; Lee, D.W. Production of γ-aminobutyric acid using immobilized glutamate decarboxylase from Lactobacillus plantarum. Microbiol. Biotechnol. Lett. 2015, 43, 300–305. [Google Scholar] [CrossRef]
- Lammens, T.M.; Franssen, M.C.R.; Scott, E.L.; Sanders, J.P.M. Availability of protein-derived amino acids as feedstock for the production of bio-based chemicals. Biomass Bioenergy 2012, 44, 168–181. [Google Scholar] [CrossRef]
- Sanchart, C.; Watthanasakphuban, N.; Boonseng, O.; Nguyen, T.-H.; Haltrich, D.; Maneerat, S. Tuna condensate as a promising low-cost substrate for glutamic acid and GABA formation using Candida rugosa and Lactobacillus futsaii. Process. Biochem. 2018, 70, 29–35. [Google Scholar] [CrossRef]
- Laroute, V.; Yasaro, C.; Narin, W.; Mazzoli, R.; Pessione, E.; Cocaign-Bousquet, M.; Loubière, P. GABA production in Lactococcus lactis is enhanced by arginine and co-addition of malate. Front. Microbiol. 2016, 7, 1050. [Google Scholar] [CrossRef]
- Lammens, T.M.; Franssen, M.C.R.; Scott, E.L.; Sanders, J.P.M. Synthesis of biobased N-methylpyrrolidone by one-pot cyclization and methylation of γ-aminobutyric acid. Green Chem. 2010, 12, 1430–1436. [Google Scholar] [CrossRef]
- Park, S.J.; Kim, E.Y.; Noh, W.; Oh, Y.H.; Kim, H.Y.; Song, B.K.; Cho, K.M.; Hong, S.H.; Lee, S.H.; Jegal, J. Synthesis of nylon 4 from γ-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess Biosyst. Eng. 2013, 36, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Tokiwa, Y.; Calabia, B.P.; Ugwu, C.U.; Aiba, S. Biodegradability of plastics. Int. J. Mol. Sci. 2009, 10, 3722–3742. [Google Scholar] [CrossRef] [PubMed]


| LAB Species and Strain | Sources | Fermentation Conditions | GABA Production | References |
|---|---|---|---|---|
| L. brevis HY1 | Kimchi | 30 °C, 48 h | 18.76 mM | [27] |
| L. brevis NCL912 | Paocai | pH 5.0, 32 °C, 36 h Fed-batch fermentation | 149.05 mM | [28] |
| L. helveticus NDO1 | Koumiss | pH 3.5, 30 °C, 30 h | 0.16 g/L | [29] |
| L. brevis BJ20 | Fermented jotgal | 30 °C, 24 h | 2.465 mg/L | [25] |
| L. paracasei 15C | Raw milk cheese | pH 5.5, 30 °C, 48 h, anaerobe | 14.8 mg/kg | [30] |
| L. rhamnosus 21D-B | Raw milk cheese | pH 5.5, 30 °C, 48 h, anaerobe | 11.3 mg/kg | [30] |
| S. thermophilus 84C | Raw milk cheese | pH 5.5, 30 °C, 48 h, anaerobe | 80 mg/kg | [30] |
| L. plantarum DM5 | Marcha Sikkim | pH 6.4, 30 °C, 30 h | NR | [31] |
| L. brevis L-32 | Kimchi | 30 °C, 24 h | 38 g/L | [32] |
| L. buchneri WPZ001 | Chinese fermented sausage | 30 °C, 72 h | 129 g/L | [33] |
| L. lactis | Kimchi | pH 5.5, 30 °C, 20 h | 6.41 g/L | [34] |
| L. otakiensis | Pico cheese | 30 °C, 48 h | 659 mg/L | [35] |
| S. thermophilus Y2 | Yoghurt | pH 4.5, 40 °C, 100 h | 7.98 g/L | [36] |
| L. buchneri MS | Kimchi | pH 5.0, 30 °C, 36 h | 251 mM | [37] |
| E. faecium JK29 | Kimchi | 30 °C, 72 h | 14.86 mM | [38] |
| L. brevis 877G | Kimchi | 30 °C, 24 h | 18.94 mM | [39] |
| L. plantarum IFK 10 | fermented soybean | pH 6.5, 37 °C, 48 h | 2.68 g/L | [40] |
| Weissella hellenica | ika-kurozukuri | 30 °C, 96 h | 7.69 g/L | [41] |
| L. brevis K203 | Kimchi | pH 5.25, 37 °C, 48 h | 44.4 g/L | [42] |
| L. futsaii CS3 | Kung-som | 37 °C, 108 h | 25 g/L | [26] |
| L. paracasei NFR7415 | Fermented fish | 30 °C, 144 h | 302 mM | [20] |
| L. plantarum C48 | Cheese | 30 °C, 48 h | 16 mg/kg | [21] |
| L. paracasei PF6 | Cheese | 30 °C, 48 h | 99.9 mg/kg | [21] |
| L. brevis PM17 | Cheese | 30 °C, 48 h | 15 mg/kg | [21] |
| L. lactis PU1 | Cheese | 30 °C, 72 h | 36 mg/kg | [21] |
| L. delbrueckii subsp. bulgaricus PR1 | Cheese | 42 °C, 48 h | 63 mg/kg | [21] |
| L. lactis subsp. lactis | Cheese starter | 30 °C, 48 h | 27.1 mg/L | [3] |
| L. brevis CECT 8183 | Goat cheese | pH 4.7, 30 °C, 48 h | 0.96 mM | [16] |
| L. brevis CECT 8182 | Sheep cheese | pH 4.7, 30 °C, 48 h | 0.94 mM | [16] |
| L. brevis CECT 8182 | Goat cheese | pH 4.7, 30 °C, 48 h | 0.99 mM | [16] |
| L. lactis CECT 8184 | Goat cheese | pH 4.7, 30 °C, 48 h | 0.93 mM | [16] |
| L. namurensis NH2 | Nham | 30 °C, 24 h | 9.06 g/L | [17] |
| P. pentosaceus HN8 | Nham | 30 °C, 24 h | 7.34 g/L | [17] |
| L. plantarum | paork kampeus | pH 6.5, 37 °C, 72 h | 20 mM | [1] |
| Source | Molecular Mass of Subunit (kDa) | Optimal pH | Optimal Temperature | Effect of Metal Ions (Increased Activity) | Effect of Metal Ions (Decreased Activity) | Km (Mm) | Vmax | References |
|---|---|---|---|---|---|---|---|---|
| L. zymae | 53 | 4.5 | 41 | NH4+, Ca2+, Mg2+, Mn2+, Na+ | Co2+, Cu2+, Ag+ | 1.7 | 0.01 mM/min | [46] |
| L. paracasei NFRI 7415 | 57 | 5 | 50 | NH4+, Ca2+ | EDTA, Na+ | 5 | NR | [18] |
| L. sakei A156 | 54.4 | 5 | 55 | Mn2+, Co2+, Ca2+, Zn2+ | NH4+, Mg2+, Ag+ | 0.045 | 0.011 mM/min | [80] |
| L. brevis CGMCC 1306 | 53 | 4.8 | 48 | NR | NR | 10.26 | 8.86 U/mg | [22] |
| S. salivarius subsp. thermophilus Y2 | 46.9 | 4 | 55 | Ba2+ | Fe2+, Zn2+, Cu2+, Mn 2+, Na+, Ag+, Co2+, Li+, K+ | 2.3 | NR | [83] |
| Enterococcus avium M5 | 53 | 4.5 | 55 | Ca2+, Mg2+, Mn2+, Zn2+ | Cu2+, Ag+ | 3.26 | 0.012 mM/min | [82] |
| E. raffinosus TCCC11660 | 55 | 4.6 | 45 | Mo6+, Mg2+ | Fe2+, Zn2+, Cu2+, Co2+ | 5.26 | 3.45 µM/min | [75] |
| Lactococcus lactis | NR | 4.7 | NR | NR | NR | 0.51 | NR | [3] |
| L. brevis 877G | 50 | 5.2 | 45 | Ca2+, Mg2+, Mn2+, Na+ | Ag+, Zn2+, Cu2+, K+ | 3.6 | 0.06 mM/min | [81] |
| Strain | GABA Enhancement Techniques | Reaction Conditions | GABA Production | References |
|---|---|---|---|---|
| L. plantarum Taj-apis 362 | GAD was expressed in pMG36e vector | Resting cells, reaction mixtures contain 1.32 mM glutamic acid and 200 mM sodium acetate, incubated at 37 °C for 60 min | 1.14 g/L | [88] |
| L. plantarum ATCC 14917 | L. sakei expression host | MRS supplemented with 1% MSG, incubated at 30 °C for 48 h, initial pH 6.0 | 27.36 g/L | [87] |
| S. salivarius ssp, thermophilus Y2 | B. subtilis expression host | Resting cells, reaction mixtures contain 0.4 M sodium glutamate and 0.4 M acetate buffer, incubated at 37 °C for 6 h | 5.26 g/L | [90] |
| L. brevis NCL 912 | Continuous cultivation method | Fermentation medium with glucose, yeast extract, soy peptone, MnSO4, Tween 80 and MSG, initial pH 5.0. incubated at 32 °C with 150 rpm agitation. | 5.11 g/L | [91] |
| L. brevis NCL 912 | Fed-batch fermentation | Seed medium containing glucose, soya peptone, MnSO4, 4H2O, l-glutamate. Incubated at 32 °C for 84 h with initial pH 5.0 | 103.72 g/L | [92] |
| L. brevis RK03 | Cell immobilization with hydrogels 2-hydroxyethyl methacrylate/polyethylene glycol diacrylate (HEMA/PEGDA) | MRS medium containing 450 mM MSG, incubated for 84 h at 30 °C. | 39.7 g/L | [93] |
| L. brevis GABA 057 | Cell immobilization with alginate beads + isomaltooligosaccharide | GYP medium (pH 4.5) containing MSG incubated for 48 h at 37 °C. | 23 g/L | [94] |
| L. lactis | optimizing fermentative condition (temperature 31.9 °C, pH 7.1, 15 g/L of MSG) | Growth on optimized MRS medium containing brown rice, germinated soy bean and skim milk. | 7.2 g/L | [34] |
| L. brevis CRL 1942 | optimizing culture conditions (30 °C, 48 h, 270 mM MSG) | Growth on optimized MRS medium | 26.30 g/L | [96] |
| E. faecium JK29 | optimizing MRS medium (0.5% sucrose, 2% yeast extract, 0.5% MSG, pH 7.5, 30 °C) | Growth on optimized MRS medium | 1.53 g/L | [38] |
| L. brevis HYE1 | optimizing MRS medium (2.14% maltose, 4.01% tryptone, 2.38% MSG, pH 4.74) | Growth on optimized MRS medium | 2.21 g/L | [27] |
| L. brevis | modifiying MRS medium containing 6% l-glutamic acid, 4% maltose, 2% yeast extract, 1% NaCl, 1% CaCl2, 2 g Tween 80, 0.02 mM PLP, pH 5.25, 37 °C, 72 h | Growth on optimized MRS medium | 44.4 g/L | [42] |
| L. brevis Lb85 | directed evolution and mutagenesis | Growth on LBG medium supplemented with glucose, kanamycin and l-glutamate, incubated at 30 °C with 200 rpm agitation. | 7.13 g/L | [84] |
| L. lactis FJNUGA01 | whole-cell bioconversion with pET28a | Resting cells in deionized water with 2 mol/L glutamate, incubated at 45 °C for 6 h | 34 g/L | [99] |
| L. plantarum WCFS1 | immobilized enzymes to porous silica beads | Enzymatic conversion of 0.5 M MSG, 0.2 mM PLP and 0.02 µg GAD/µL in sodium acetate buffer (pH 5.0), incubated at 37 °C for 20 min. | 41.7 g/L | [106] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yogeswara, I.B.A.; Maneerat, S.; Haltrich, D. Glutamate Decarboxylase from Lactic Acid Bacteria—A Key Enzyme in GABA Synthesis. Microorganisms 2020, 8, 1923. https://doi.org/10.3390/microorganisms8121923
Yogeswara IBA, Maneerat S, Haltrich D. Glutamate Decarboxylase from Lactic Acid Bacteria—A Key Enzyme in GABA Synthesis. Microorganisms. 2020; 8(12):1923. https://doi.org/10.3390/microorganisms8121923
Chicago/Turabian StyleYogeswara, Ida Bagus Agung, Suppasil Maneerat, and Dietmar Haltrich. 2020. "Glutamate Decarboxylase from Lactic Acid Bacteria—A Key Enzyme in GABA Synthesis" Microorganisms 8, no. 12: 1923. https://doi.org/10.3390/microorganisms8121923
APA StyleYogeswara, I. B. A., Maneerat, S., & Haltrich, D. (2020). Glutamate Decarboxylase from Lactic Acid Bacteria—A Key Enzyme in GABA Synthesis. Microorganisms, 8(12), 1923. https://doi.org/10.3390/microorganisms8121923






