Chemical Ecology of Streptomyces albidoflavus Strain A10 Associated with Carpenter Ant Camponotus vagus
Abstract
:1. Introduction
2. Materials and Methods
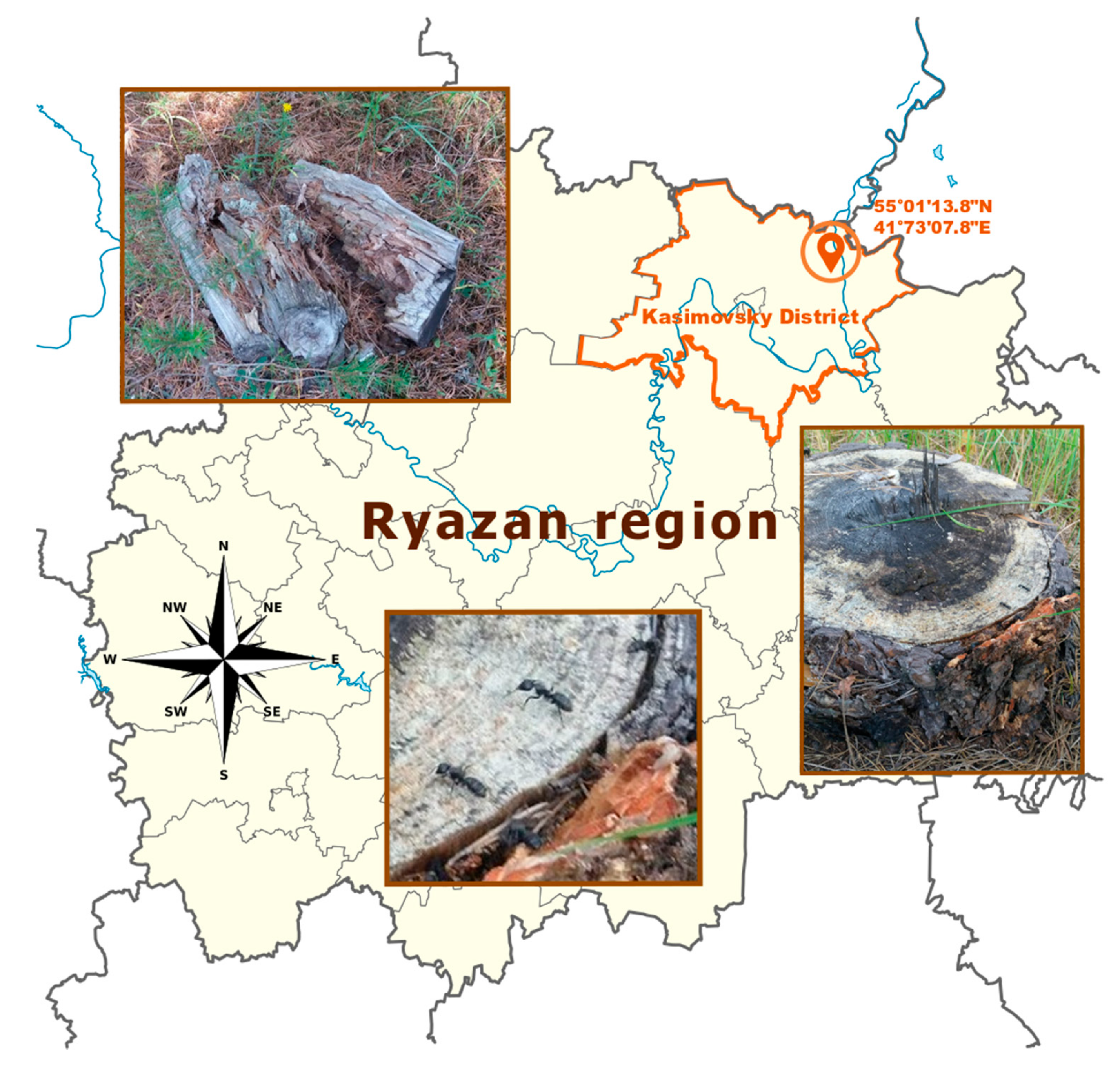
2.1. Collection and Microbial Isolation
2.2. Phenotypic Characterization
2.3. Gene Amplification and Phylogenetic Analysis
2.4. Fermentation and Antimicrobial Assay
2.5. Isolation and Identification of Antimycins
3. Results
3.1. Isolation and Characterization of Streptomyces Strain A10 Associated with Carpenter Ants
3.2. Antimicrobial Properties of Streptomyces sp. A10
3.3. Identification of Antifungal Compounds Produced by Strain A10
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Wu, C.; Shang, Z.; Lemetre, C.; Ternei, M.A.; Brady, S.F. Cadasides, calcium-dependent acidic lipopeptides from the soil metagenome that are active against multidrug-resistant bacteria. J. Am. Chem. Soc. 2019, 141, 3910–3919. [Google Scholar] [CrossRef] [PubMed]
- Hover, B.M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S.; et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Vikeli, E.; Widdick, D.A.; Batey, S.F.D.; Heine, D.; Holmes, N.A.; Bibb, M.J.; Martins, D.J.; Pierce, N.E.; Hutchings, M.I.; Wilkinson, B. In situ activation and heterologous production of a cryptic lantibiotic from an African plant ant-derived Saccharopolyspora species. Appl. Environ. Microbiol. 2019, 86, e01876-19. [Google Scholar] [CrossRef] [Green Version]
- Van Der Meij, A.; Worsley, S.F.; Hutchings, M.I.; Van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R.; Traxler, M.F.; Clardy, J.; Kolter, R. Old Meets New: Using interspecies interactions to detect secondary metabolite production in actinomycetes. Meth. Enzymol. 2012, 517, 89–109. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; Van Wezel, G. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Sandiford, S.K.; Van Wezel, G.P. Triggers and cues that activate antibiotic production by actinomycetes. J. Ind. Microbiol. Biotechnol. 2014, 41, 371–386. [Google Scholar] [CrossRef]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [CrossRef] [Green Version]
- Davies, J.; Spiegelman, G.B.; Yim, G. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 2006, 9, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomycesas symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seipke, R.F.; Barke, J.; Brearley, C.A.; Hill, L.; Yu, D.W.; Goss, R.J.M.; Hutchings, M.I. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS ONE 2011, 6, e22028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaltenpoth, M.; Engl, T. Defensive microbial symbionts in Hymenoptera. Funct. Ecol. 2014, 28, 315–327. [Google Scholar] [CrossRef]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4742–4746. [Google Scholar] [CrossRef] [Green Version]
- Kroiss, J.; Kaltenpoth, M.; Schneider, B.; Schwinger, M.-G.; Hertweck, C.; Maddula, R.K.; Strohm, E.; Svatoš, A. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 2010, 6, 261–263. [Google Scholar] [CrossRef]
- Schoenian, I.; Spiteller, M.; Ghaste, M.; Wirth, R.; Herz, H.; Spiteller, D. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc. Natl. Acad. Sci. USA 2011, 108, 1955–1960. [Google Scholar] [CrossRef] [Green Version]
- Schultz, T.R. In search of ant ancestors. Proc. Natl. Acad. Sci. USA 2000, 97, 14028–14029. [Google Scholar] [CrossRef] [Green Version]
- Caldera, E.J.; Poulsen, M.; Suen, G.; Currie, C.R. Insect symbioses: A case study of past, present, and future Fungus-growing ant research. Environ. Entomol. 2009, 38, 78–92. [Google Scholar] [CrossRef]
- Currie, C.R. A Community of ants, fungi, and bacteria: A multilateral approach to studying symbiosis. Annu. Rev. Microbiol. 2001, 55, 357–380. [Google Scholar] [CrossRef] [Green Version]
- Ronque, M.U.V.; Lyra, M.L.; Migliorini, G.H.; Bacci, M.; Oliveira, P.S. Symbiotic bacterial communities in rainforest fungus-farming ants: Evidence for species and colony specificity. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zakalyukina, Y.V.; Biryukov, M.V.; Lukianov, D.A.; Shiriaev, D.I.; Komarova, E.S.; Skvortsov, D.A.; Kostyukevich, Y.; Tashlitsky, V.N.; Polshakov, V.I.; Nikolaev, E.; et al. Nybomycin-producing Streptomyces isolated from carpenter ant Camponotus vagus. Biochimie 2019, 160, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Osterman, I.A.; Wieland, M.; Maviza, T.P.; Lashkevich, K.A.; Lukianov, D.A.; Komarova, E.S.; Zakalyukina, Y.V.; Buschauer, R.; Shiriaev, D.I.; Leyn, S.A.; et al. Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel. Nat. Chem. Biol. 2020, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef] [Green Version]
- Prauser, H.; Falta, R. Phagensensibilität, Zellwand-Zusammensetzung und Taxonomie von Actinomyceten. J. Basic Microbiol. 1968, 8, 39–46. [Google Scholar] [CrossRef]
- Zakalyukina, Y.V.; Biryukov, M.V.; Golichenkov, M.V.; Netrusov, A.I. Phenotypic and phylogenetic characterization of actinomycetes isolated from Lasius niger and Formica cunicularia ants. Mosc. Univ. Biol. Sci. Bull. 2017, 72, 13–19. [Google Scholar] [CrossRef]
- Guo, Y.; Zheng, W.; Rong, X.; Huang, B. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: Use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 2008, 58, 149–159. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gao, Y.; Zeng, X.D.; Ren, B.; Zeng, J.R.; Xu, T.; Yang, Y.Z.; Hu, X.C.; Zhu, Z.Y.; Shi, L.M.; Zhou, G.Y.; et al. Antagonistic activity against rice blast disease and elicitation of host-defence response capability of an endophytic Streptomyces albidoflavus OsiLf-2. Plant Pathol. 2019, 69, 259–271. [Google Scholar] [CrossRef]
- Muthukrishnan, P.; Chithra Devi, D.; Mostafa, A.A.; Alsamhary, K.I.; Abdel-Raouf, N.; Nageh Sholkamy, E. Antimicrobial efficacy of Nocardiopsis sp. MK_MSt033 against selected multidrug resistant clinical microbial pathogens. J. Infect. Public Health 2020, 13, 1522–1532. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fifth Informational Supplement; CLSI Document M100-S25; CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; Approved Standard; CLSI Document M07-A10; CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; Approved Standard; CLSI Document M27–A3; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Vanner, S.A.; Li, X.; Zvanych, R.; Torchia, J.; Sang, J.; Andrews, D.W.; Magarvey, N.A. Chemical and biosynthetic evolution of the antimycin-type depsipeptides. Mol. BioSyst. 2013, 9, 2712–2719. [Google Scholar] [CrossRef] [PubMed]
- Becerril, A.; Álvarez, S.; Braña, A.F.; Rico, S.; Díaz, M.; Santamaría, R.I.; Salas, J.A.; Méndez, C. Uncovering production of specialized metabolites by Streptomyces argillaceus: Activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS ONE 2018, 13, e0198145. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Labeda, D.P.; Dunlap, C.A.; Rong, X.; Huang, Y.; Doroghazi, J.R.; Ju, K.-S.; Metcalf, W.W. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Leeuwenhoek 2016, 110, 563–583. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Guo, Y.; Huang, B. Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA–DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp solvifaciens. Syst. Appl. Microbiol. 2009, 32, 314–322. [Google Scholar] [CrossRef]
- Labeda, D.P.; Goodfellow, M.; Brown, R.; Ward, A.C.; Lanoot, B.; Vanncanneyt, M.; Swings, J.; Kim, S.-B.; Liu, Z.; Chun, J.; et al. Phylogenetic study of the species within the family Streptomycetaceae. Antonie Leeuwenhoek 2012, 101, 73–104. [Google Scholar] [CrossRef]
- Joynt, R.; Seipke, R.F. A phylogenetic and evolutionary analysis of antimycin biosynthesis. Microbiology 2018, 164, 28–39. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, J.Y.; Jung, H.W.; Hwang, B.K. Streptomyces koyangensis sp. nov., a novel actinomycete that produces 4-phenyl-3-butenoic acid. Int. J. Syst. Evol. Microbiol. 2005, 55, 257–262. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhu, X.; Kim, S.J.; Zhang, W. Antimycin-type depsipeptides: Discovery, biosynthesis, chemical synthesis, and bioactivities. Nat. Prod. Rep. 2016, 33, 1146–1165. [Google Scholar] [CrossRef]
- Abidi, S.; Ha, S.; Rosen, R. Liquid chromatography—thermospray mass spectrometric study of N-acylamino dilactones and 4-butyrolactones derived from antimycin A. J. Chromatogr. A 1990, 522, 179–194. [Google Scholar] [CrossRef]
- Radchenko, A.G. Review of ants of the genus Camponotus (Hymenoptera, Formicidae) of the Palearctic. Subgenus Camponotus Zool. Zhurnal 1997, 76, 554–564. [Google Scholar]
- Khan, S.; Guo, L.; Maimaiti, Y.; Mijit, M.; Qiu, D. Entomopathogenic fungi as microbial biocontrol agent. Mol. Plant Breed. 2012, 3, 63–79. [Google Scholar] [CrossRef]
- Oh, D.-C.; Scott, J.J.; Currie, C.R.; Clardy, J. Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp. Org. Lett. 2009, 11, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Kermarrec, A.; Mauleon, H. Quelques aspects de la pathogénie d’ Entomophthora coronata Cost. Kervork. pour la Fourmi-Manioc de la Guadeloupe: Acromyrmex octospinosus (Formicidae, Attini). Ann. Parasitol. 1975, 50, 351–360. [Google Scholar] [CrossRef] [Green Version]
- Sung, G.-H.; Hywel-Jones, N.L.; Sung, J.-M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [Green Version]
- Labeda, D.P.; Doroghazi, J.R.; Ju, K.-S.; Metcalf, W.W. Taxonomic evaluation of Streptomyces albus and related species using multilocus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 894–900. [Google Scholar] [CrossRef] [Green Version]
- Skinner, F.A. Inhibition of Fusarium culmorum by Streptomyces albidoflavus. Nat. Cell Biol. 1953, 172, 1191. [Google Scholar] [CrossRef]
- Yan, L.-L.; Han, N.-N.; Zhang, Y.-Q.; Yu, L.-Y.; Chen, J.; Wei, Y.-Z.; Li, Q.-P.; Tao, L.; Zheng, G.-H.; Yang, S.-E.; et al. Antimycin A18 produced by an endophytic Streptomyces albidoflavus isolated from a mangrove plant. J. Antibiot. 2010, 63, 259–261. [Google Scholar] [CrossRef]
- Cheng, K.; Rong, X.; Pinto-Tomás, A.A.; Fernández-Villalobos, M.; Murillo-Cruz, C.; Huang, Y. Population genetic analysis of Streptomyces albidoflavus reveals habitat barriers to homologous recombination in the diversification of streptomycetes. Appl. Environ. Microbiol. 2015, 81, 966–975. [Google Scholar] [CrossRef] [Green Version]
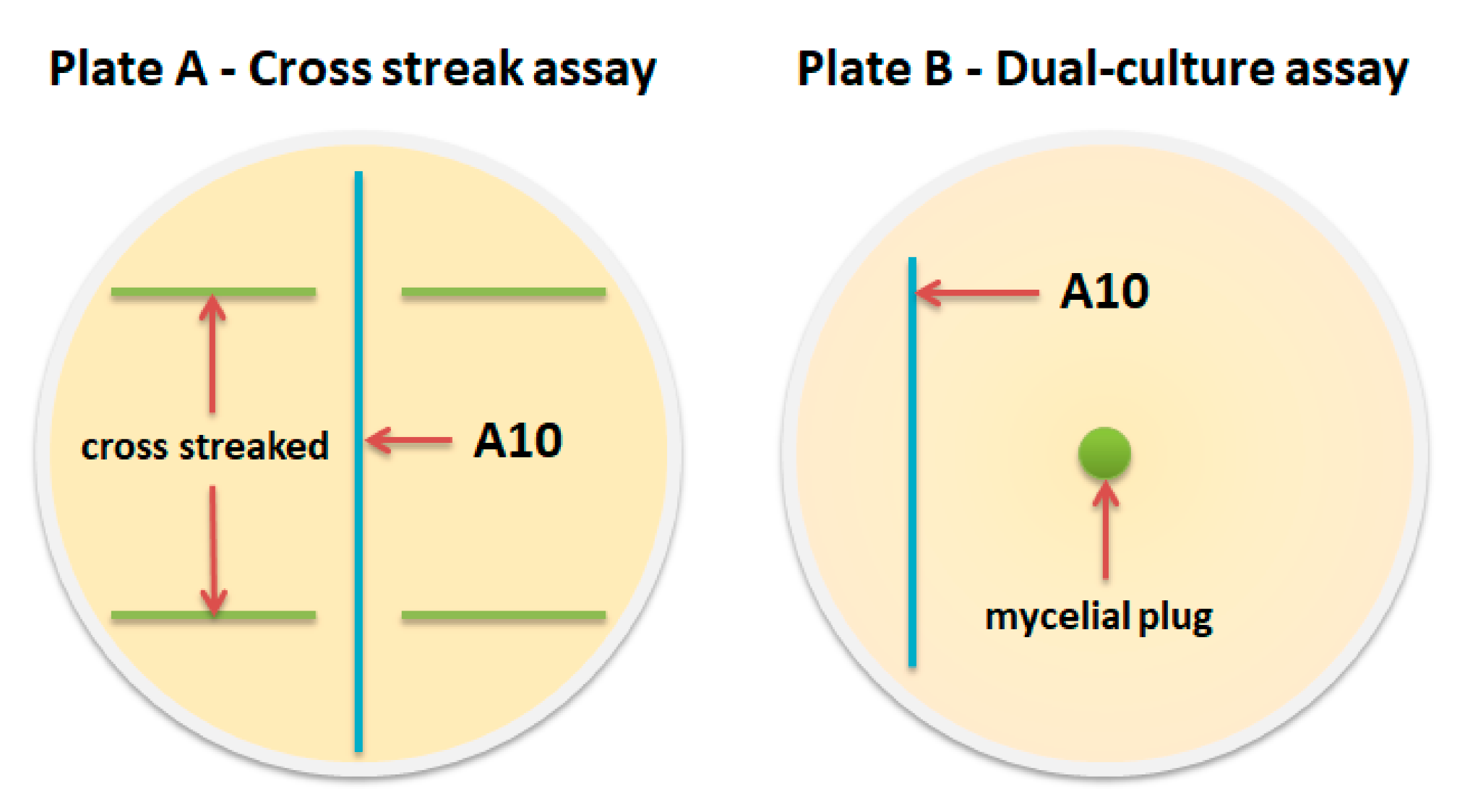





| Strains | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Yeast extract-malt extract agar (ISP-2) | ||||
| Growth | good | good | good | good |
| Aerial mycelium | greenish beige | sparse, white | white yellow | grey |
| Substrate mycelium | pale yellow | colorless | brown | ivory |
| Diffusible pigment | none | none | none | none |
| Oatmeal agar (ISP-3) | ||||
| Growth | good | good | good | good |
| Aerial mycelium | greenish beige | none | white yellow | grey |
| Substrate mycelium | light yellow | colorless | brown | ivory |
| Diffusible pigment | none/reddish | none/reddish | none | none |
| Inorganic salts-starch agar (ISP-4) | ||||
| Growth | moderate | good | good | good |
| Aerial mycelium | none | sparse, white | white yellow | grey |
| Substrate mycelium | pale yellow | colorless | brown | ivory |
| Diffusible pigment | none | none | none | none |
| Glycerol-asparagine agar (ISP-5) | ||||
| Growth | good | good | weak | good |
| Aerial mycelium | beige | white | white yellow | white/yellow |
| Substrate mycelium | yellowish white | colorless | creamy | yellow |
| Diffusible pigment | none | none | none | none |
| Peptone-yeast extract-iron agar (ISP-6) | ||||
| Growth | good | good | good | n/d |
| Aerial mycelium | slightly pink | white | white/grey | n/d |
| Substrate mycelium | brownish pink | brown | dark brown | n/d |
| Diffusible pigment | none | brown | dark brown | n/d |
| Tyrosine agar (ISP-7) | ||||
| Growth | good | good | good | n/d |
| Aerial mycelium | beige | sparse, white | white | n/d |
| Substrate mycelium | slightly pink | colorless | brown | n/d |
| Diffusible pigment | none | none | brown | n/d |
| Phenotypic Tests | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Morphology spore chains | RF | RF | RF | RF |
| Spore surface | smooth | smooth | smooth | smooth |
| Melanoid pigment | - | - | + | - |
| Growth: | ||||
| 1% NaCl | + | + | + | + |
| 5% NaCl | + | + | + | + |
| 8% NaCl | + | w | + | n/d |
| 10 °C | + | n/d | + | n/d |
| 45 °C | w | n/d | n/d | n/d |
| Acid from: | ||||
| Adonitol | - | n/d | - | n/d |
| Arabinose | - | -/+ | + | + |
| Dulcit | - | n/d | n/d | n/d |
| Fructose | + | + | + | - |
| Galactose | + | + | n/d | n/d |
| Glucose | + | + | n/d | + |
| Inositol | - | - | - | - |
| Lactose | - | n/d | n/d | n/d |
| Maltose | + | n/d | n/d | n/d |
| Mannitol | + | - | + | - |
| Mannose | w | + | n/d | n/d |
| Raffinose | - | - | - | - |
| Rhamnose | - | - | - | - |
| Salicin | - | n/d | n/d | n/d |
| Sorbitol | - | n/d | n/d | n/d |
| Sucrose | w | w | - | - |
| Xylose | + | + | + | - |
| Enzymes: | ||||
| L-arginine | + | + | n/d | n/d |
| L-ornitin | + | + | n/d | n/d |
| L-lysin | + | + | n/d | n/d |
| Decomposition of: | ||||
| Cellulose | - | - | - | - |
| Gelatin | + | + | + | n/d |
| Starch | + | + | + | n/d |
| Urea | + | + | n/d | n/d |
| Production of: | ||||
| Soluble pigments | + | + | + | n/d |
| Citrate utilization | + | + | n/d | n/d |
| Malonate utilization | + | n/d | n/d | n/d |
| β-glucosidase | - | n/d | n/d | n/d |
| Test Strain | Growth Inhibition Rate (%) * from Dual-Culture Assay | Growth Inhibition from Cross Streak Assay |
|---|---|---|
| O. sinensis VKPM F-1479 | 5.0 ± 3.2 b | - |
| C. coronatus VKPM F-1359 | 15.7 ± 1.8 a | + |
| B. bassiana VKPM F-1357 | 18.2 ± 3.6 b | + |
| C. coronatus VKPM F-442 | 28.9 ± 3.4 a | ++ |
| M. rileyi VKPM F-1360 | n/a | +++ |
| L. lecanii VKPM F-837 | n/a | - |
| C. albicans CBS 8836 | n/a | +++ |
| A. niger INA 00760 | n/a | - |
| St. aureus ATCC 25923 | n/a | - |
| B. subtilis ATCC 6633 | n/a | - |
| B. thuringiensis VKPM B-6650 | n/a | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baranova, A.A.; Chistov, A.A.; Tyurin, A.P.; Prokhorenko, I.A.; Korshun, V.A.; Biryukov, M.V.; Alferova, V.A.; Zakalyukina, Y.V. Chemical Ecology of Streptomyces albidoflavus Strain A10 Associated with Carpenter Ant Camponotus vagus. Microorganisms 2020, 8, 1948. https://doi.org/10.3390/microorganisms8121948
Baranova AA, Chistov AA, Tyurin AP, Prokhorenko IA, Korshun VA, Biryukov MV, Alferova VA, Zakalyukina YV. Chemical Ecology of Streptomyces albidoflavus Strain A10 Associated with Carpenter Ant Camponotus vagus. Microorganisms. 2020; 8(12):1948. https://doi.org/10.3390/microorganisms8121948
Chicago/Turabian StyleBaranova, Anna A., Alexey A. Chistov, Anton P. Tyurin, Igor A. Prokhorenko, Vladimir A. Korshun, Mikhail V. Biryukov, Vera A. Alferova, and Yuliya V. Zakalyukina. 2020. "Chemical Ecology of Streptomyces albidoflavus Strain A10 Associated with Carpenter Ant Camponotus vagus" Microorganisms 8, no. 12: 1948. https://doi.org/10.3390/microorganisms8121948







