Innovation and Application of the Type III Secretion System Inhibitors in Plant Pathogenic Bacteria
Abstract
:1. Introduction
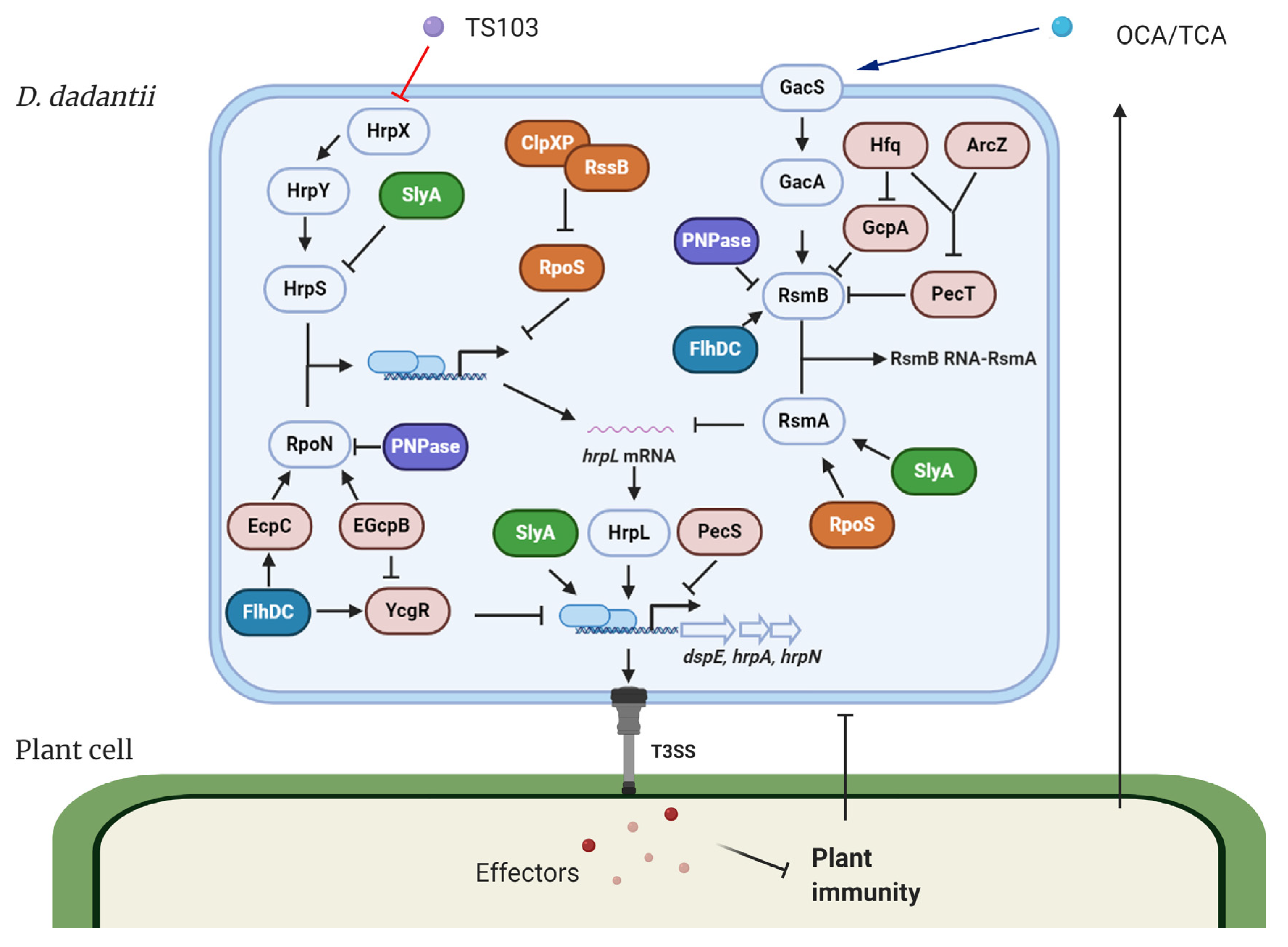
2. Regulation of T3SS in D. dadantii
2.1. Bacterial Second Messengers Regulate T3SS
2.2. Transcriptional Regulators Control T3SS
2.3. Post-Transcriptional Regulators Control T3SS
3. Discovery of T3SS Inhibitors in Plant Pathogens and Their Regulatory Mechanisms
3.1. Plant Phenolic Compounds as T3SS Inhibitors
3.2. Salicylidene Acylhydrazides and Their Derivatives as T3SS Inhibitors
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323. [Google Scholar] [CrossRef] [PubMed]
- Hueck, C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 1998, 62, 379–433. [Google Scholar] [CrossRef] [Green Version]
- Coburn, B.; Sekirov, I.; Finlay, B.B. Type III secretion systems and disease. Clin. Microbiol. Rev. 2007, 20, 535–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Büttner, D.; He, S.Y. Type III protein secretion in plant pathogenic bacteria. Plant. Physiol. 2009, 150, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef] [Green Version]
- Oh, C.S.; Kim, J.F.; Beer, S.V. The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol. Plant. Pathol. 2005, 6, 125–138. [Google Scholar] [CrossRef]
- Oh, C.-S.; Beer, S.V. Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 2005, 253, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Cunnac, S.; Lindeberg, M.; Collmer, A. Pseudomonassyringae type III secretion system effectors: Repertoires in search of functions. Curr. Opin. Microbiol. 2009, 12, 53–60. [Google Scholar] [CrossRef]
- Jin, L.; Ham, J.H.; Hage, R.; Zhao, W.; Soto-Hernandez, J.; Lee, S.Y.; Paek, S.-M.; Kim, M.G.; Boone, C.; Coplin, D.L. Direct and indirect targeting of PP2A by conserved bacterial type-III effector proteins. PLOS Pathog. 2016, 12, e1005609. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.-F.; Nomura, K.; Ding, X.; Chen, X.; Wang, K.; Aung, K.; Uribe, F.; Rosa, B.; Yao, J.; Chen, J.; et al. Pseudomonassyringae effector avirulence protein E localizes to the host plasma membrane and down-regulates the expression of the nonrace-specific disease resistance1/harpin-induced1-like13 gene required for antibacterial immunity in Arabidopsis. Plant. Physiol. 2015, 169, 793–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, M.C.; Linington, R.G.; Auerbuch, V. Chemical inhibitors of the type three secretion system: Disarming bacterial pathogens. Antimicrob. Agents Chemother. 2012, 56, 5433–5441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pendergrass, H.A.; May, A.E. Natural product type III secretion system inhibitors. Antibiotics 2019, 8, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, L.; Zhou, S.; Zhu, L.; Liang, C.; Chen, X. Small-molecule inhibitors of the type III secretion system. Mol. Cells 2015, 20, 17659–17674. [Google Scholar] [CrossRef] [PubMed]
- Arnold, D.L.; Pitman, A.; Jackson, R.W. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 2003, 4, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Troisfontaines, P.; Cornelis, G.R. Type III secretion: More systems than you think. Physiology 2005, 20, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Collmer, A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: Trafficking harpins, Avr proteins, and death. J. Bacteriol. 1997, 179, 5655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.-M.; Beer, S.V. hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J. Bacteriol. 1995, 177, 6201–6210. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Hutcheson, S.W. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 1994, 176, 3089–3091. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Heu, S.; Yi, J.; Lu, Y.; Hutcheson, S. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 1994, 176, 1025–1036. [Google Scholar]
- Brito, B.; Aldon, D.; Barberis, P.; Boucher, C.; Genin, S. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol. Plant-Microbe Interact. 2002, 15, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Wengelnik, K.; Bonas, U. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 1996, 178, 3462–3469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNally, R.R.; Zhao, Y.; Sundin, G.W. Towards understanding fire blight: Virulence mechanisms and their regulation in Erwinia amylovora. In Bacteria-Plant Interactions: Advanced Research and Future Trends; Caister Academy Press: Norfolk, UK, 2015; pp. 61–82. [Google Scholar]
- Zhao, Y.; Sundin, G. Exploring linear and cyclic (di)-nucleotides as messengers for regulation of T3SS and biofilm formation in Erwinia amylovora. J. Plant. Pathol. 2017, 99, 25–35. [Google Scholar]
- Zhao, Y. Genomics of Erwinia amylovora and related Erwinia species associated with pome fruit trees. In Genomics of Plant-Associated Bacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–36. [Google Scholar]
- Malnoy, M.; Martens, S.; Norelli, J.L.; Barny, M.-A.; Sundin, G.W.; Smits, T.H.; Duffy, B. Fire blight: Applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. 2012, 50, 475–494. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Shao, X.; Deng, X. Regulation of type III secretion system in Pseudomonas syringae. Environ. Microbiol. 2019, 21, 4465–4477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, H.N.; Chakravarthy, S.; Wei, H.-L.; BuiNguyen, H.; Stodghill, P.V.; Collmer, A.; Swingle, B.M.; Cartinhour, S.W. Global analysis of the HrpL regulon in the plant pathogen Pseudomonas syringae pv. tomato DC3000 reveals new regulon members with diverse functions. PLoS ONE 2014, 9, e106115. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Peng, Q.; Zhang, Q.; Zou, L.; Li, Y.; Robert, C.; Pritchard, L.; Liu, H.; Hovey, R.; Wang, Q.; et al. Genome-Wide Identification of HrpL-Regulated Genes in the Necrotrophic Phytopathogen Dickeya dadantii 3937. PLoS ONE 2010, 5, e13472. [Google Scholar] [CrossRef]
- Yap, M.-N.; Yang, C.-H.; Barak, J.D.; Jahn, C.E.; Charkowski, A.O. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 2005, 187, 639–648. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Xiao, Y.; Zhou, J.-M. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant.-Microbe Interact. 2006, 19, 1159–1166. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Peng, Q.; Zhang, Q.; Yi, X.; Choi, C.J.; Reedy, R.M.; Charkowski, A.O.; Yang, C.-H. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant.-Microbe Interact. 2008, 21, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Peng, Q.; San Francisco, M.; Wang, Y.; Zeng, Q.; Yang, C.-H. Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS ONE 2008, 3, e2973. [Google Scholar] [CrossRef]
- Chatterjee, A.; Cui, Y.; Chatterjee, A.K. Regulation of Erwinia carotovora hrpLEcc (sigma-LEcc), Which Encodes an Extracytoplasmic Function Subfamily of Sigma Factor Required for Expression of the HRP Regulon. Mol. Plant.-Microbe Interact. 2002, 15, 971–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, A.; Cui, Y.; Liu, Y.; Dumenyo, C.K.; Chatterjee, A.K. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl. Environ. Microbiol. 1995, 61, 1959–1967. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.Y.; Gui, G.; Wei, B.; Preston, J.F.; Oakford, L.; Yüksel, Ü.; Giedroc, D.P.; Romeo, T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997, 272, 17502–17510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czajkowski, R.; Perombelon, M.C.; van Veen, J.A.; van der Wolf, J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant. Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Reverchon, S.; Nasser, W. Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 2013, 5, 622–636. [Google Scholar] [PubMed]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [Green Version]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef]
- Paul, R.; Weiser, S.; Amiot, N.C.; Chan, C.; Schirmer, T.; Giese, B.; Jenal, U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004, 18, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, C.G.; Lee, D.-J. Bacterial diguanylate cyclases: Structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol. Adv. 2015, 33, 124–141. [Google Scholar] [CrossRef]
- Ryan, R.P.; Fouhy, Y.; Lucey, J.F.; Crossman, L.C.; Spiro, S.; He, Y.-W.; Zhang, L.-H.; Heeb, S.; Cámara, M.; Williams, P.; et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 2006, 103, 6712–6717. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, A.J.; Ryjenkov, D.A.; Gomelsky, M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J. Bacteriol. 2005, 187, 4774–4781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamayo, R.; Tischler, A.D.; Camilli, A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 2005, 280, 33324–33330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Povolotsky, T.L.; Hengge, R. ‘Life-style’control networks in Escherichia coli: Signaling by the second messenger c-di-GMP. J. Biotechnol. 2012, 160, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Lu, W.; Rabinowitz, J.D.; Bassler, B.L. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008, 190, 2527–2536. [Google Scholar] [CrossRef] [Green Version]
- Edmunds, A.C.; Castiblanco, L.F.; Sundin, G.W.; Waters, C.M. Cyclic Di-GMP modulates the disease progression of Erwinia amylovora. J. Bacteriol. 2013, 195, 2155–2165. [Google Scholar] [CrossRef] [Green Version]
- Kharadi, R.R.; Castiblanco, L.F.; Waters, C.M.; Sundin, G.W. Phosphodiesterase Genes Regulate Amylovoran Production, Biofilm Formation, and Virulence in Erwinia amylovora. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [Green Version]
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Shevchik, V.E. Bacterial pectate lyases, structural and functional diversity. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef]
- Yi, X.; Yamazaki, A.; Biddle, E.; Zeng, Q.; Yang, C.H. Genetic analysis of two phosphodiesterases reveals cyclic diguanylate regulation of virulence factors in Dickeya dadantii. Mol. Microbiol. 2010, 77, 787–800. [Google Scholar] [CrossRef]
- Yuan, X.; Khokhani, D.; Wu, X.; Yang, F.; Biener, G.; Koestler, B.J.; Raicu, V.; He, C.; Waters, C.M.; Sundin, G.W.; et al. Cross-talk between a regulatory small RNA, cyclic-di-GMP signalling and flagellar regulator FlhDC for virulence and bacterial behaviours. Environ. Microbiol. 2015, 17, 4745–4763. [Google Scholar] [CrossRef]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Tian, F.; He, C.; Severin, G.B.; Waters, C.M.; Zeng, Q.; Liu, F.; Yang, C.H. The diguanylate cyclase GcpA inhibits the production of pectate lyases via the H-NS protein and RsmB regulatory RNA in Dickeya dadantii. Mol. Plant. Pathol. 2018, 19, 1873–1886. [Google Scholar] [CrossRef] [Green Version]
- Nasser, W.; Faelen, M.; Hugouvieux-Cotte-Pattat, N.; Reverchon, S. Role of the nucleoid-associated protein H-NS in the synthesis of virulence factors in the phytopathogenic bacterium Erwinia chrysanthemi. Mol. Plant.-Microbe Interact. 2001, 14, 10–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasser, W.; Reverchon, S. H-NS-dependent activation of pectate lyases synthesis in the phytopathogenic bacterium Erwinia chrysanthemi is mediated by the PecT repressor. Mol. Microbiol. 2002, 43, 733–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouafa, Z.-A.; Reverchon, S.; Lautier, T.; Muskhelishvili, G.; Nasser, W. The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res. 2012, 40, 4306–4319. [Google Scholar] [CrossRef] [PubMed]
- Stauber, J.L.; Loginicheva, E.; Schechter, L.M. Carbon source and cell density-dependent regulation of type III secretion system gene expression in Pseudomonas syringae pathovar tomato DC3000. Res. Microbiol. 2012, 163, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Kim, H.-S.; Delaney, T.P.; Collmer, A. Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in an hrp-dependent manner within tobacco cells. Mol. Plant-Microbe Interact. 1997, 10, 580–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasser, W.; Reverchon, S.; Vedel, R.; Boccara, M. PecS and PecT coregulate the synthesis of HrpN and pectate lyases, two virulence determinants in Erwinia chrysanthemi 3937. Mol. Plant.-Microbe Interact. 2005, 18, 1205–1214. [Google Scholar] [CrossRef]
- Yuan, X.; Zeng, Q.; Xu, J.; Severin, G.B.; Zhou, X.; Waters, C.M.; Sundin, G.W.; Ibekwe, A.M.; Liu, F.; Yang, C.-H. Tricarboxylic Acid (TCA) Cycle Enzymes and Intermediates Modulate Intracellular Cyclic di-GMP Levels and the Production of Plant Cell Wall–Degrading Enzymes in Soft Rot Pathogen Dickeya dadantii. Mol. Plant-Microbe Interact. Mpmi 2020, 33, 296–307. [Google Scholar] [CrossRef]
- Giacalone, D.; Smith, T.J.; Collins, A.J.; Sondermann, H.; Koziol, L.J.; O’Toole, G.A. Ligand-Mediated Biofilm Formation via Enhanced Physical Interaction between a Diguanylate Cyclase and Its Receptor. mBio 2018, 9, e01254-18. [Google Scholar] [CrossRef] [Green Version]
- Ross, W.; Vrentas, C.E.; Sanchez-Vazquez, P.; Gaal, T.; Gourse, R.L. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 2013, 50, 420–429. [Google Scholar] [CrossRef] [Green Version]
- Hauryliuk, V.; Atkinson, G.C.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015, 13, 298–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.-w.; Yu, M.; Lee, J.H.; Chatnaparat, T.; Zhao, Y. The stringent response regulator (p)ppGpp mediates virulence gene expression and survival in Erwinia amylovora. BMC Genom. 2020, 21, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ancona, V.; Lee, J.H.; Chatnaparat, T.; Oh, J.; Hong, J.-I.; Zhao, Y. The bacterial alarmone (p)ppGpp activates the type III secretion system in Erwinia amylovora. J. Bacteriol. 2015, 197, 1433–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatnaparat, T.; Li, Z.; Korban, S.S.; Zhao, Y. The bacterial alarmone (p)ppGpp is required for virulence and controls cell size and survival of Pseudomonas syringae on plants. Environ. Microbiol. 2015, 17, 4253–4270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatnaparat, T.; Li, Z.; Korban, S.S.; Zhao, Y. The stringent response mediated by (p)ppGpp is required for virulence of Pseudomonas syringae pv. tomato and its survival on tomato. Mol. Plant.-Microbe Interact. 2015, 28, 776–789. [Google Scholar] [CrossRef] [Green Version]
- Young, G.M.; Schmiel, D.H.; Miller, V.L. A new pathway for the secretion of virulence factors by bacteria: The flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 1999, 96, 6456–6461. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Galán, J.E. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 2004, 51, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Pallen, M.J.; Beatson, S.A.; Bailey, C.M. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: A Darwinian perpective. FEMS Microbiol. Rev. 2005, 29, 201–229. [Google Scholar] [CrossRef] [Green Version]
- Erhardt, M.; Namba, K.; Hughes, K.T. Bacterial nanomachines: The flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2010, 2, a000299. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.L.; Chakravarthy, S.; Worley, J.N.; Collmer, A. Consequences of flagellin export through the type III secretion system of Pseudomonas syringae reveal a major difference in the innate immune systems of mammals and the model plant Nicotiana benthamiana. Cell. Microbiol. 2013, 15, 601–618. [Google Scholar] [CrossRef]
- Wang, S.; Fleming, R.T.; Westbrook, E.M.; Matsumura, P.; McKay, D.B. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 2006, 355, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, P.D.; Karlinsey, J.E.; Aldridge, C.; Birchall, C.; Thompson, D.; Yagasaki, J.; Hughes, K.T. The flagellar-specific transcription factor, σ28, is the type III secretion chaperone for the flagellar-specific anti-σ28 factor FlgM. Genes Dev. 2006, 20, 2315–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chilcott, G.S.; Hughes, K.T. Coupling of Flagellar Gene Expression to Flagellar Assembly in Salmonella enterica Serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000, 64, 694–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Chatterjee, A.; Yang, H.; Chatterjee, A.K. Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. J. Bacteriol. 2008, 190, 4610–4623. [Google Scholar] [CrossRef] [Green Version]
- Pesavento, C.; Becker, G.; Sommerfeldt, N.; Possling, A.; Tschowri, N.; Mehlis, A.; Hengge, R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008, 22, 2434–2446. [Google Scholar] [CrossRef] [Green Version]
- Claret, L.; Hughes, C. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J. Mol. Biol. 2002, 321, 185–199. [Google Scholar] [CrossRef]
- Stafford, G.P.; Ogi, T.; Hughes, C. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 2005, 151, 1779. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Gomelsky, M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 2010, 76, 1295–1305. [Google Scholar] [CrossRef]
- Paul, K.; Nieto, V.; Carlquist, W.C.; Blair, D.F.; Harshey, R.M. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 2010, 38, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [Green Version]
- Lange, R.; Hengge-Aronis, R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994, 8, 1600–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Gottesman, S.; Hoskins, J.R.; Maurizi, M.R.; Wickner, S. The RssB response regulator directly targets ςS for degradation by ClpXP. Genes Dev. 2001, 15, 627–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweder, T.; Lee, K.-H.; Lomovskaya, O.; Matin, A. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J. Bacteriol. 1996, 178, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, A.; Cui, Y.; Ma, W.; Liu, Y.; Ishihama, A.; Eisenstark, A.; Chatterjee, A.K. RpoS (sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J. Bacteriol. 1998, 180, 3629–3634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yamazaki, A.; Zou, L.; Biddle, E.; Zeng, Q.; Wang, Y.; Lin, H.; Wang, Q.; Yang, C.-H. ClpXP protease regulates the type III secretion system of Dickeya dadantii 3937 and is essential for the bacterial virulence. Mol. Plant.-Microbe Interact. 2010, 23, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Zhao, Y. ClpXP-Dependent RpoS Degradation Enables Full Activation of Type III Secretion System, Amylovoran Production, and Motility in Erwinia amylovora. Phytopathology 2017, 107, 1346–1352. [Google Scholar] [CrossRef] [Green Version]
- Ellison, D.W.; Miller, V.L. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 2006, 9, 153–159. [Google Scholar] [CrossRef]
- Haque, M.M.; Kabir, M.S.; Aini, L.Q.; Hirata, H.; Tsuyumu, S. SlyA, a MarR family transcriptional regulator, is essential for virulence in Dickeya dadantii 3937. J. Bacteriol. 2009, 191, 5409–5418. [Google Scholar] [CrossRef] [Green Version]
- Linehan, S.A.; Rytkönen, A.; Yu, X.-J.; Liu, M.; Holden, D.W. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 2005, 73, 4354–4362. [Google Scholar] [CrossRef] [Green Version]
- Zou, L.; Zeng, Q.; Lin, H.; Gyaneshwar, P.; Chen, G.; Yang, C.-H. SlyA regulates type III secretion system (T3SS) genes in parallel with the T3SS master regulator HrpL in Dickeya dadantii 3937. Appl. Environ. Microbiol. 2012, 78, 2888–2895. [Google Scholar] [CrossRef] [Green Version]
- Hommais, F.; Oger-Desfeux, C.; Van Gijsegem, F.; Castang, S.; Ligori, S.; Expert, D.; Nasser, W.; Reverchon, S. PecS is a global regulator of the symptomatic phase in the phytopathogenic bacterium Erwinia chrysanthemi 3937. J. Bacteriol. 2008, 190, 7508–7522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reverchon, S.; Rouanet, C.; Expert, D.; Nasser, W. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 2002, 184, 654–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo, T.; Vakulskas, C.A.; Babitzke, P. Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ. Microbiol. 2013, 15, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Van Assche, E.; Van Puyvelde, S.; Vanderleyden, J.; Steenackers, H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015, 6, 141. [Google Scholar] [CrossRef] [Green Version]
- Kudla, J.; Hayes, R.; Gruissem, W. Polyadenylation accelerates degradation of chloroplast mRNA. Embo J. 1996, 15, 7137–7146. [Google Scholar] [CrossRef]
- Leszczyniecka, M.; Kang, D.-c.; Sarkar, D.; Su, Z.-z.; Holmes, M.; Valerie, K.; Fisher, P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPaseold-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. USA 2002, 99, 16636–16641. [Google Scholar] [CrossRef] [Green Version]
- Kinscherf, T.; Apirion, D. Polynucleotide phosphorylase can participate in decay of mRNA in Escherichia coli in the absence of ribonuclease II. Mol. Gen. Genet. MGG. 1975, 139, 357–362. [Google Scholar] [CrossRef]
- Cameron, T.A.; Matz, L.M.; De Lay, N.R. Polynucleotide phosphorylase: Not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet. 2018, 14, e1007654. [Google Scholar] [CrossRef]
- Rosenzweig, J.A.; Chromy, B.; Echeverry, A.; Yang, J.; Adkins, B.; Plano, G.V.; McCutchen-Maloney, S.; Schesser, K. Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett. 2007, 270, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Ygberg, S.E.; Clements, M.O.; Rytkönen, A.; Thompson, A.; Holden, D.W.; Hinton, J.C.; Rhen, M. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 2006, 74, 1243–1254. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; Ibekwe, A.M.; Biddle, E.; Yang, C.-H. Regulatory mechanisms of exoribonuclease PNPase and regulatory small RNA on T3SS of Dickeya dadantii. Mol. Plant.-Microbe Interact. 2010, 23, 1345–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viegas, S.C.; Pfeiffer, V.; Sittka, A.; Silva, I.s.J.; Vogel, J.; Arraiano, C.M. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007, 35, 7651–7664. [Google Scholar] [CrossRef] [Green Version]
- Dressaire, C.; Pobre, V.; Laguerre, S.; Girbal, L.; Arraiano, C.M.; Cocaign-Bousquet, M. PNPase is involved in the coordination of mRNA degradation and expression in stationary phase cells of Escherichia coli. BMC Genom. 2018, 19, 848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, Y.; Vogel, J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010, 13, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Zeng, Q.; Khokhani, D.; Tian, F.; Severin, G.B.; Waters, C.M.; Xu, J.; Zhou, X.; Sundin, G.W.; Ibekwe, A.M.; et al. A Feed-forward signaling circuit controls bacterial virulence through linking cyclic di-GMP and two mechanistically distinct sRNAs; ArcZ and RsmB. Environ. Microbiol. 2019, 21, 2755–2771. [Google Scholar] [CrossRef]
- Castillo, A.; Reverchon, S. Characterization of the pecT control region from Erwinia chrysanthemi 3937. J. Bacteriol. 1997, 179, 4909–4918. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Cui, Y.; Ma, W.; Liu, Y.; Chatterjee, A.K. hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Environ. Microbiol. 2000, 2, 203–215. [Google Scholar] [CrossRef]
- Zeng, Q.; Sundin, G.W. Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genom. 2014, 15, 414. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; McNally, R.R.; Sundin, G.W. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J. Bacteriol. 2013, 195, 1706–1717. [Google Scholar] [CrossRef] [Green Version]
- Sundin, G.W.; Castiblanco, L.F.; Yuan, X.; Zeng, Q.; Yang, C.H. Bacterial disease management: Challenges, experience, innovation and future prospects: Challenges in bacterial molecular plant pathology. Mol. Plant. Pathol. 2016, 17, 1506–1518. [Google Scholar] [CrossRef]
- Sundin, G.W.; Wang, N. Antibiotic Resistance in Plant-Pathogenic Bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Khokhani, D.; Zhang, C.; Li, Y.; Wang, Q.; Zeng, Q.; Yamazaki, A.; Hutchins, W.; Zhou, S.-S.; Chen, X.; Yang, C.-H. Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 2013, 79, 5424–5436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, S.; Tian, F.; Li, J.; Hutchins, W.; Chen, H.; Yang, F.; Yuan, X.; Cui, Z.; Yang, C.H.; He, C. Identification of phenolic compounds that suppress the virulence of Xanthomonas oryzae on rice via the type III secretion system. Mol. Plant Pathol. 2017, 18, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Peng, Q.; Selimi, D.; Wang, Q.; Charkowski, A.O.; Chen, X.; Yang, C.-H. The plant phenolic compound p-coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl. Environ. Microbiol. 2009, 75, 1223–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Hutchins, W.; Wu, X.; Liang, C.; Zhang, C.; Yuan, X.; Khokhani, D.; Chen, X.; Che, Y.; Wang, Q. Derivative of plant phenolic compound inhibits the type III secretion system of Dickeya dadantii via HrpX/HrpY two-component signal transduction and Rsm systems. Mol. Plant. Pathol. 2015, 16, 150–163. [Google Scholar] [CrossRef]
- Yang, L.; Ding, W.; Xu, Y.; Wu, D.; Li, S.; Chen, J.; Guo, B. New insights into the antibacterial activity of hydroxycoumarins against Ralstonia solanacearum. Mol. Cells 2016, 21, 468. [Google Scholar] [CrossRef]
- Yang, L.; Li, S.; Qin, X.; Jiang, G.; Chen, J.; Li, B.; Yao, X.; Liang, P.; Zhang, Y.; Ding, W. Exposure to umbelliferone reduces Ralstonia solanacearum biofilm formation, transcription of type III secretion system regulators and effectors and virulence on tobacco. Front. Microbiol. 2017, 8, 1234. [Google Scholar] [CrossRef]
- Yang, F.; Korban, S.S.; Pusey, P.L.; Elofsson, M.; Sundin, G.W.; Zhao, Y. Small-molecule inhibitors suppress the expression of both type III secretion and amylovoran biosynthesis genes in Erwinia amylovora. Mol. Plant Pathol. 2014, 15, 44–57. [Google Scholar] [CrossRef]
- Puigvert, M.; Solé, M.; López-Garcia, B.; Coll, N.S.; Beattie, K.D.; Davis, R.A.; Elofsson, M.; Valls, M. Type III secretion inhibitors for the management of bacterial plant diseases. Mol. Plant Pathol. 2019, 20, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Velderrain-Rodríguez, G.; Palafox-Carlos, H.; Wall-Medrano, A.; Ayala-Zavala, J.; Chen, C.O.; Robles-Sánchez, M.; Astiazaran-García, H.; Alvarez-Parrilla, E.; González-Aguilar, G. Phenolic compounds: Their journey after intake. Food Funct. 2014, 5, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.O.; Nair, M.G.; Hammerschmidt, R.; Safir, G.R.; Putnam, A.R. Significance of phenolic compounds in plant-soil-microbial systems. Crit. Rev. Plant. Sci. 1991, 10, 63–121. [Google Scholar] [CrossRef]
- Yang, S.; Perna, N.T.; Cooksey, D.A.; Okinaka, Y.; Lindow, S.E.; Ibekwe, A.M.; Keen, N.T.; Yang, C.-H. Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf array. Mol. Plant-Microbe Interact. 2004, 17, 999–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montesano, M.; Brader, G.; Ponce de León, I.; Palva, E.T. Multiple defence signals induced by Erwinia carotovora ssp. carotovora elicitors in potato. Mol. Plant Pathol. 2005, 6, 541–549. [Google Scholar] [CrossRef]
- Vidal, S.; de León, I.P.; Denecke, J.; Palva, E.T. Salicylic acid and the plant pathogen Erwinia carotovora induce defense genes via antagonistic pathways. Plant. J. 1997, 11, 115–123. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085. [Google Scholar] [CrossRef]
- Feys, B.J.; Parker, J.E. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000, 16, 449–455. [Google Scholar] [CrossRef]
- Funk, C.; Brodelius, P.E. Phenylpropanoid metabolism in suspension cultures of Vanilla planifolia Andr.: III. Conversion of 4-methoxycinnamic acids into 4-hydroxybenzoic acids. Plant. Physiol. 1990, 94, 102–108. [Google Scholar] [CrossRef] [Green Version]
- Yalpani, N.; Raskin, I. Salicylic acid: A systemic signal in induced plant disease resistance. Trends Microbiol. 1993, 1, 88–92. [Google Scholar] [CrossRef]
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef]
- Wengelnik, K.; Van den Ackerveken, G.; Bonas, U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria ls homologous to two-component response regulators. Mol. Plant-Microbe Interact. 1996, 9, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Rangaraj, N.; Sonti, R.V. Multiple adhesin-like functions of Xanthomonas oryzae pv. oryzae are involved in promoting leaf attachment, entry, and virulence on rice. Mol. Plant-Microbe Interact. 2009, 22, 73–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Büttner, D.; Bonas, U. Regulation and secretion of Xanthomonas virulence factors. Fems Microbiol. Rev. 2010, 34, 107–133. [Google Scholar] [CrossRef] [Green Version]
- Fan, S.; Tian, F.; Fang, L.; Yang, C.-H.; He, C. Transcriptional responses of Xanthomonas oryzae pv. oryzae to type III secretion system inhibitor ortho-coumaric acid. BMC Microbiol. 2019, 19, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stringlis, I.A.; De Jonge, R.; Pieterse, C.M. The age of coumarins in plant–microbe interactions. Plant. Cell Physiol. 2019, 60, 1405–1419. [Google Scholar] [CrossRef] [Green Version]
- Venugopala, K.N.; Rashmi, V.; Odhav, B. Review on natural coumarin lead compounds for their pharmacological activity. BioMed Res. Int. 2013, 2013, 963248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kauppi, A.M.; Nordfelth, R.; Uvell, H.; Wolf-Watz, H.; Elofsson, M. Targeting bacterial virulence: Inhibitors of type III secretion in Yersinia. Chem. Biol. 2003, 10, 241–249. [Google Scholar] [CrossRef] [Green Version]
- Kauppi, A.M.; Nordfelth, R.; Hägglund, U.; Wolf-Watz, H.; Elofsson, M. Salicylanilides are potent inhibitors of type III secretion in Yersinia. In The Genus Yersinia; Springer: Boston, MA, USA, 2004; pp. 97–100. [Google Scholar]
- Nordfelth, R.; Kauppi, A.M.; Norberg, H.; Wolf-Watz, H.; Elofsson, M. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 2005, 73, 3104–3114. [Google Scholar] [CrossRef] [Green Version]
- Muschiol, S.; Bailey, L.; Gylfe, Å.; Sundin, C.; Hultenby, K.; Bergström, S.; Elofsson, M.; Wolf-Watz, H.; Normark, S.; Henriques-Normark, B. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 2006, 103, 14566–14571. [Google Scholar] [CrossRef] [Green Version]
- Muschiol, S.; Normark, S.; Henriques-Normark, B.; Subtil, A. Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol. 2009, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Veenendaal, A.K.; Sundin, C.; Blocker, A.J. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 2009, 191, 563–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, K.; Betts, H.; Chellas-Gery, B.; Hower, S.; Linton, C.; Fields, K. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 2006, 61, 1543–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, L.; Gylfe, Å.; Sundin, C.; Muschiol, S.; Elofsson, M.; Nordström, P.; Henriques-Normark, B.; Lugert, R.; Waldenström, A.; Wolf-Watz, H. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 2007, 581, 587–595. [Google Scholar] [CrossRef] [Green Version]
- Tree, J.J.; Wang, D.; McInally, C.; Mahajan, A.; Layton, A.; Houghton, I.; Elofsson, M.; Stevens, M.P.; Gally, D.L.; Roe, A.J. Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157: H7. Infect. Immun. 2009, 77, 4209–4220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, D.L.; Layton, A.N.; Field, T.R.; Bowen, A.J.; Wolf-Watz, H.; Elofsson, M.; Stevens, M.P.; Galyov, E.E. Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 2007, 51, 2631–2635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negrea, A.; Bjur, E.; Ygberg, S.E.; Elofsson, M.; Wolf-Watz, H.; Rhen, M. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 2007, 51, 2867–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anantharajah, A.; Buyck, J.M.; Sundin, C.; Tulkens, P.M.; Mingeot-Leclercq, M.-P.; Van Bambeke, F. Salicylidene acylhydrazides and hydroxyquinolines act as inhibitors of type three secretion systems in Pseudomonas aeruginosa by distinct mechanisms. Antimicrob. Agents Chemother. 2017, 61, e02566-16. [Google Scholar] [CrossRef] [Green Version]
- Bellemann, P.; Geider, K. Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biochemical characterization. Microbiology 1992, 138, 931–940. [Google Scholar] [CrossRef] [Green Version]
- Dahlgren, M.K.; Zetterström, C.E.; Gylfe, Å.; Linusson, A.; Elofsson, M. Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram-negative bacterium Yersinia. Bioorganic Med. Chem. 2010, 18, 2686–2703. [Google Scholar] [CrossRef]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Stockwell, V.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Et Tech. Off. Int. Des. Epizoot. 2012, 31, 199–210. [Google Scholar] [CrossRef] [PubMed]
- McGhee, G.C.; Sundin, G.W. Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora. Phytopathology 2011, 101, 192–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benveniste, R.; Davies, J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 1973, 70, 2276–2280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambanthamoorthy, K.; Sloup, R.E.; Parashar, V.; Smith, J.M.; Kim, E.E.; Semmelhack, M.F.; Neiditch, M.B.; Waters, C.M. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob. Agents Chemother. 2012, 56, 5202–5211. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.; Park, J.-S.; Choi, H.-Y.; Yoon, S.S.; Kim, W.-G. Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa: A connection between quorum sensing and c-di-GMP. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Hee, C.-S.; Habazettl, J.; Schmutz, C.; Schirmer, T.; Jenal, U.; Grzesiek, S. Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP. Proc. Natl. Acad. Sci. USA 2020, 117, 17211–17220. [Google Scholar] [CrossRef]
- Cho, K.H.; Tryon, R.G.; Kim, J.-H. Screening for Diguanylate Cyclase (DGC) Inhibitors Mitigating Bacterial Biofilm Formation. Front. Chem. 2020, 8, 264. [Google Scholar] [CrossRef]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant.-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Nazir, R.; Mazurier, S.; Yang, P.; Lemanceau, P.; Van Elsas, J.D. The ecological role of type three secretion systems in the interaction of bacteria with fungi in soil and related habitats is diverse and context-dependent. Front. Microbiol. 2017, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Slack, S.; Walters, K.J.; Outwater, C.; Sundin, G.W. Effect of kasugamycin, oxytetracycline, and streptomycin on in-orchard population dynamics of Erwinia amylovora on apple flower stigmas. Plant. Dis. 2020. [Google Scholar] [CrossRef]
| Compound | Structure | Known Modes of Action on T3SS in Plant Pathogens | References |
|---|---|---|---|
| o-coumaric acid |  | Induce T3SS via the RsmA/RsmB-HrpL pathway in D. dadantii; inhibit T3SS in E. amylovora and T3SS in X. oryzae pv. oryzae via the HrpG-HrpX regulatory cascade. | [32,115,116] |
| t-cinnamic acid |  | Induce T3SS in D. dadantii; inhibit T3SS in E. amylovora. | [32,115] |
| p-coumaric acid | 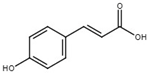 | Inhibit T3SS via the HrpS-HrpL pathway in D. dadantii. | [117] |
| trans-4-hydroxycinnamodydroxamic acid | 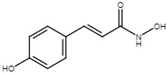 | Inhibit T3SS via the RpoN-HrpL pathway, the HrpX/HrpY-HrpS-HrpL pathway, and the RsmB-HrpL pathway in D. dadantii. | [118] |
| Benzoic acid |  | Inhibit T3SS via targeting HrpS in E. amylovora. | [115] |
| Salicylic acid | 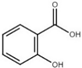 | Inhibit T3SS in E. amylovora. | [115] |
| 4-methoxy-cinnamic acid | 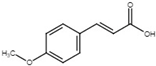 | Inhibit T3SS via targeting HrpL in E. amylovora. | [115] |
| trans-2-methoxycinnamic acid | 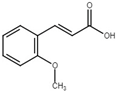 | Inhibit T3SS via the HrpG-HrpX regulatory cascade in X. oryzae pv. oryzae. | [116] |
| trans-2-phenylcyclopropane-1-carboxylic-acid | 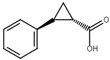 | Inhibit T3SS via the HrpG-HrpX regulatory cascade in X. oryzae pv. oryzae. | [116] |
| trans-2-methylcinnamic acid | 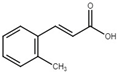 | Inhibit T3SS via the HrpG-HrpX regulatory cascade in X. oryzae pv. oryzae. | [116] |
| Umbelliferone | 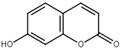 | Inhibit T3SS via targeting HrpG in R. solanacearum. | [119,120] |
| Benzoic acid N′-(2,3,4-trihydroxy-benzylidene)-hydrazide |  | Inhibit multiple T3SS regulon genes in E. amylovora; inhibit T3SS via targeting HrpB in R. solanacearum. | [121,122] |
| 4-nitrobenzoic acid N′-(2,4-dihydroxy-benzylidene)-hydrazide | 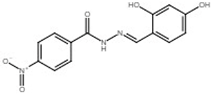 | Inhibit T3SS via targeting HrpB in R. solanacearum; inhibit T3SS via targeting HrpL in E. amylovora. | [121,122] |
| 2-nitro-benzoic acid N′-(3,5-dichloro-2-hydroxy-benzylidene)-hydrazide | 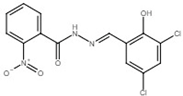 | Inhibit T3SS via targeting HrpB in R. solanacearum. | [122] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Yu, M.; Yang, C.-H. Innovation and Application of the Type III Secretion System Inhibitors in Plant Pathogenic Bacteria. Microorganisms 2020, 8, 1956. https://doi.org/10.3390/microorganisms8121956
Yuan X, Yu M, Yang C-H. Innovation and Application of the Type III Secretion System Inhibitors in Plant Pathogenic Bacteria. Microorganisms. 2020; 8(12):1956. https://doi.org/10.3390/microorganisms8121956
Chicago/Turabian StyleYuan, Xiaochen, Manda Yu, and Ching-Hong Yang. 2020. "Innovation and Application of the Type III Secretion System Inhibitors in Plant Pathogenic Bacteria" Microorganisms 8, no. 12: 1956. https://doi.org/10.3390/microorganisms8121956





