Soehngenia longivitae sp. nov., a Fermenting Bacterium Isolated from a Petroleum Reservoir in Azerbaijan, and Emended Description of the Genus Soehngenia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of the Sampling Site
2.2. Isolation of Methanogenic Enrichment and Pure Culture of a Fermenting Bacterium
2.3. DNA Isolation, Amplification and Sequencing of the 16S rRNA Genes from the Methanogenic Enrichment
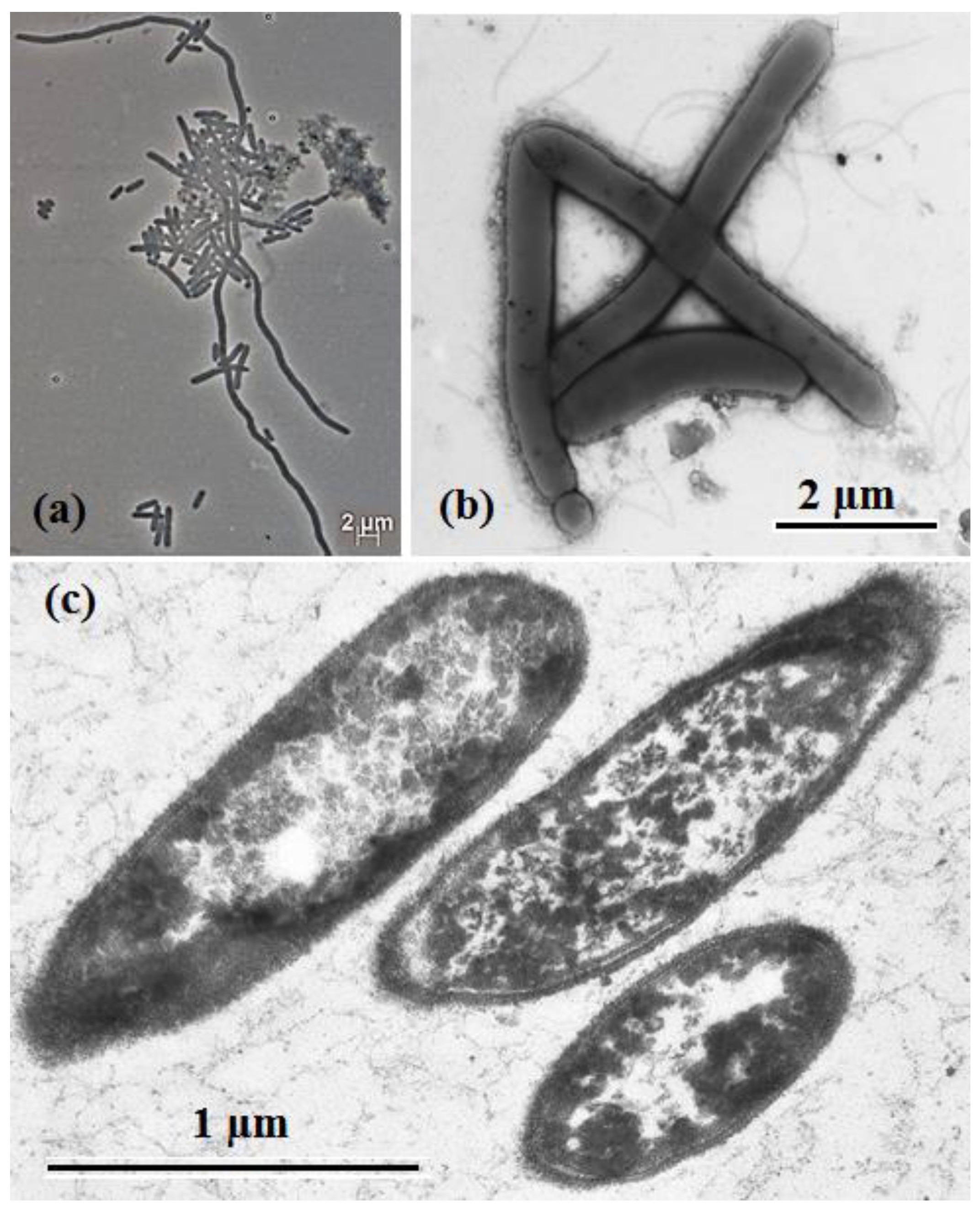
2.4. Morphological, Physiological and Chemotaxonomic Characterization
2.5. 16S rRNA Gene Sequencing and Phylogenetic Analysis
2.6. Genome Analysis
2.7. Nucleotide Sequence Accession Numbers
3. Results and Discussion
3.1. Phylogenetic Diversity of Prokaryotes in Methanogenic Enrichment
3.2. Phenotypic Characterization of Strain 1933PT
3.3. Phylogenetic Analysis of 16S rRNA Gene Sequences
3.4. Whole Genome Sequencing and Phylogenomic Analyses
3.5. Description of Soehngenia Longivitae sp. nov
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, K.; Dai, L.; Tu, B.; Zhang, X.; Zhang, H.; Deng, Y.; Lawson, P.A.; Cheng, L. Gudongella oleilytica gen. nov., sp. nov., an aerotorelant bacterium isolated from Shengli oilfield and validation of family Tissierellaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Alauzet, C.; Marchandin, H.; Courtin, P.; Mory, F.; Lemée, L.; Pons, J.-L.; Chapot-Chartier, M.-P.; Lozniewski, A.; Jumas-Bilak, E. Multilocus analysis reveals diversity in the genus Tissierella: Description of Tissierella carlieri sp. nov. in the new class Tissierellia classis nov. Syst. Appl. Microbiol. 2014, 37, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Parshina, S.N.; Kleerebezem, R.; Sans, J.L.; Lettinga, G.; Nozhevnikova, A.N.; Kostrikina, N.A.; Lysenko, A.M.; Stams, A.J.M. Soehngenia saccharolytica gen. nov., sp. nov., and Clostridium amygdalinum sp. nov., two novel, anaerobic, benzaldehyde-converting bacteria. Int. J. Syst. Evol. Microbiol. 2003, 53, 1791–1799. [Google Scholar] [CrossRef]
- Parshina, S.N.; Kleerebezem, R.; van Kempen, E.; Nozhevnikova, A.N.; Lettinga, G.; Stams, A.J.M. Benzaldehyde conversion by two anaerobic bacteria isolated from an upflow anaerobic sludge bed reactor. Process Biochem. 2000, 36, 423–429. [Google Scholar] [CrossRef]
- Zhang, F.; She, Y.-H.; Chai, L.-J.; Banat, I.M.; Zhang, X.-T.; Shu, F.-C.; Wang, Z.-L.; Yu, L.-J.; Hou, D.-J. Microbial diversity in long-term water-flooded oil reservoirs with different in situ temperatures in China. Sci. Rep. 2012, 2, 760. [Google Scholar] [CrossRef]
- Pavlova-Kostryukova, N.K.; Tourova, T.P.; Poltaraus, A.B.; Feng, Q.; Nazina, T.N. Microbial diversity in formation water and enrichment cultures from the Gangxi bed of the Dagang terrigenous oilfield (PRC). Microbiology 2014, 83, 616–633. [Google Scholar] [CrossRef]
- Nazina, T.N.; Shestakova, N.M.; Semenova, E.M.; Korshunova, A.V.; Kostrukova, N.K.; Tourova, T.P.; Min, L.; Feng, Q.; Poltaraus, A.B. Diversity of metabolically active Bacteria in water-flooded high-temperature heavy oil reservoir. Front. Microbiol. 2017, 8, 707. [Google Scholar] [CrossRef] [Green Version]
- Sokolova, D.S.; Semenova, E.M.; Grouzdev, D.S.; Ershov, A.P.; Bidzhieva, S.K.; Ivanova, A.E.; Babich, T.L.; Sissenbayeva, M.R.; Bisenova, M.A.; Nazina, T.N. Microbial diversity and potential sulfide producers in the Karazhanbas oilfield (Kazakhstan). Microbiology 2020, 89, 459–469. [Google Scholar] [CrossRef]
- Pannekens, M.; Voskuhl, L.; Meier, A.; Müller, H.; Haque, S.; Frösler, J.; Brauer, V.S.; Meckenstock, R.U. Densely populated water droplets in heavy-oil seeps. Appl. Environ. Microbiol. 2020, 86, e00164-20. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Shi, S.; Li, Q.; Chen, J.; Zhang, H.; Lu, Y. Progressive degradation of crude oil n-alkanes coupled to methane production under mesophilic and thermophilic conditions. PLoS ONE 2014, 9, e113253. [Google Scholar] [CrossRef]
- Wang, H.Z.; Lv, X.M.; Yi, Y.; Zheng, D.; Gou, M.; Nie, Y.; Hu, B.; Nobu, M.K.; Narihiro, T.; Tang, Y.Q. Using DNA-based stable isotope probing to reveal novel propionate- and acetate-oxidizing bacteria in propionate-fed mesophilic anaerobic chemostats. Sci. Rep. 2019, 9, 17396. [Google Scholar] [CrossRef]
- Quero, G.M.; Cassin, D.; Botter, M.; Perini, L.; Luna, G.M. Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs). Front. Microbiol. 2015, 6, 1053. [Google Scholar] [CrossRef]
- Luque-Almagro, V.M.; Cabello, P.; Sáez, L.P.; Olaya-Abril, A.; Moreno-Vivián, C.; Roldán, M.D. Exploring anaerobic environments for cyanide and cyano-derivatives microbial degradation. Appl. Microbiol. Biotechnol. 2018, 102, 1067–1074. [Google Scholar] [CrossRef] [Green Version]
- Nazina, T.N.; Rozanova, E.P.; Kuznetsov, S.I. Microbial oil transformation processes accompanied by methane and hydrogen-sulfide formation. Geomicrobiol. J. 1985, 4, 103–130. [Google Scholar] [CrossRef]
- Nazina, T.N. Communities of methane-producing bacteria from Apsheron oil formations. Microbiology 1984, 53, 122–127. [Google Scholar]
- Grouzdev, D.S.; Bidzhieva, S.K.; Sokolova, D.S.; Tourova, T.P.; Poltaraus, A.B.; Nazina, T.N. Draft genome sequence of a fermenting bacterium, Soehngenia sp. strain 1933P, isolated from a petroleum reservoir in Azerbaijan. Microbiol. Resour. Announc. 2019, 8, e00689-19. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Zeikus, J.G.; Weimer, P.J.; Nelson, D.R.; Daniels, L. Bacterial methanogenesis: Acetate as a methane precursor in pure culture. Arch. Microbiol. 1975, 104, 129–134. [Google Scholar] [CrossRef]
- Wolin, E.A.; Wolin, M.J.; Wolfe, R.S. Formation of methane by bacterial extracts. J. Biol. Chem. 1963, 238, 2882–2888. [Google Scholar]
- Kevbrin, V.V.; Zavarzin, G.A. The effect of sulfur compounds on growth of halophilic homoacetic bacterium Acetohalobium arabaticum. Mikrobiologiia 1992, 61, 812–817. (In Russian) [Google Scholar]
- Hungate, R.E. Chapter IV. A roll tube method for cultivation of strict anaerobes. In Methods in Microbiology; Norris, J.R., Ribbons, D.W., Eds.; Academic Press: London, UK, 1969; pp. 117–132. [Google Scholar]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caparoso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv 2016, 081257. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef] [Green Version]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Semenova, E.M.; Sokolova, D.S.; Grouzdev, D.S.; Poltaraus, A.B.; Vinokurova, N.G.; Tourova, T.P.; Nazina, T.N. Geobacillus proteiniphilus sp. nov., a thermophilic bacterium isolated from a high-temperature heavy oil reservoir in China. Int. J. Syst. Evol. Microbiol. 2019, 69, 3001–3008. [Google Scholar] [CrossRef]
- Trüper, H.G.; Schlegel, H.G. Sulfur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek 1964, 30, 321–323. [Google Scholar] [CrossRef]
- Bidzhieva, S.K.; Sokolova, D.S.; Tourova, T.P.; Nazina, T.N. Bacteria of the genus Sphaerochaeta from low-temperature heavy-oil reservoirs (Russia). Microbiology 2018, 87, 757–765. [Google Scholar] [CrossRef]
- Holguin, G.; Guzman, M.A.; Bashan, Y. Two new nitrogen-fixing bacteria from the rhizosphere of mangrove trees: Their isolation, identification and in vitro interaction with rhizosphere Staphylococcus sp. FEMS Microbiol. Ecol. 1992, 101, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Bidzhieva, S.K.; Sokolova, D.S.; Grouzdev, D.S.; Kostrikina, N.A.; Poltaraus, A.B.; Tourova, T.P.; Shcherbakova, V.A.; Troshina, O.Y.; Nazina, T.N. Sphaerochaeta halotolerans sp. nov., a novel spherical halotolerant spirochete from a Russian heavy oil reservoir, emended description of the genus Sphaerochaeta, reclassification of Sphaerochaeta coccoides to a new genus Parasphaerochaeta gen. nov. as Parasphaerochaeta coccoides comb. nov. and proposal of Sphaerochaetaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 4748–4759. [Google Scholar] [CrossRef]
- Minnikin, D.E.; Collins, M.D.; Goodfellow, M. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J. Appl. Bacteriol. 1979, 47, 87–95. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Hoang, D.T.; Vinh, L.S.; Flouri, T.; Stamatakis, A.; von Haeseler, A.; Minh, B.Q. MPBoot: Fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC Evol. Biol. 2018, 18, 11. [Google Scholar] [CrossRef] [Green Version]
- Varghese, N.J.; Mukherjee, S.; Ivanova, N.; Konstantinidis, K.T.; Mavrommatis, K.; Kyrpides, N.C.; Pati, A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015, 43, 6761–6771. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantiukh, K.; Grouzdev, D. POCP-Matrix calculation for a number of genomes. Figshare 2017. [Google Scholar] [CrossRef]
- Grouzdev, D.S.; Rysina, M.S.; Bryantseva, I.A.; Gorlenko, V.M.; Gaisin, V.A. Draft genome sequences of ‘Candidatus Chloroploca asiatica’ and ‘Candidatus Viridilinea mediisalina’, candidate representatives of the Chloroflexales order: Phylogenetic and taxonomic implications. Stand. Genom. Sci. 2018, 13, 24. [Google Scholar] [CrossRef]
- Qin, Q.L.; Xie, B.B.; Zhang, X.Y.; Chen, X.L.; Zhou, B.C.; Zhou, J.H.; Oren, A.; Zhang, Y.Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef] [Green Version]
- Delmont, T.O.; Eren, E.M. Linking pangenomes and metagenomes: The Prochlorococcus metapangenome. PeerJ 2018, 6, e4320. [Google Scholar] [CrossRef] [Green Version]
- Eren, A.M.; Esen, O.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ’omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Shaffer, M.; Borton, M.A.; McGivern, B.B.; Zayed, A.A.; La Rosa, S.L.; Solden, L.M.; Liu, P.F.; Narrowe, A.B.; Rodríguez-Ramos, J.; Benjamin Bolduc, B.; et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020, 48, 8883–8900. [Google Scholar] [CrossRef]
- Bryant, M.P.; Boone, D.R. Emended description of strain MST (DSM 800T), the type strain of Methanosarcina barkeri. Int. J. Syst. Bacteriol. 1987, 37, 169–170. [Google Scholar] [CrossRef] [Green Version]
- Lomans, B.P.; Maas, R.; Luderer, R.; Op den Camp, H.J.M.; Pol, A.; van der Drift, C.; Vogels, G.D. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl. Environ. Microbiol. 1999, 65, 3641–3650. [Google Scholar] [CrossRef] [Green Version]
- Baena, S.; Fardeau, M.L.; Labat, M.; Ollivier, B.; Garcia, J.L.; Patel, B.K. Desulfovibrio aminophilus sp. nov., a novel amino acid degrading and sulfate reducing bacterium from an anaerobic dairy wastewater lagoon. Syst. Appl. Microbiol. 1998, 21, 498–504. [Google Scholar] [CrossRef]
- Dahle, H.; Birkeland, N.-K. Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int. J. Syst. Evol. Microbiol. 2006, 56, 1539–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabowski, A.; Tindall, B.J.; Bardin, V.; Blanchet, D.; Jeanthon, C. Petrimonas sulfuriphila gen. nov., sp. nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int. J. Syst. Evol. Microbiol. 2005, 55, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Dong, X. Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 2005, 55, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahnke, S.; Langer, T.; Koeck, D.E.; Klocke, M. Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 2016, 66, 1466–1475, Erratum in 2016, 66, 2454. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Kyrpides, N.C.; Woyke, T.; Eisen, J.A.; Garrity, G.; Lilburn, T.G.; Beck, B.J.; Whitman, W.B.; Hugenholtz, P.; Klenk, H.P. Genomic encyclopedia of type strains, phase I: The one thousand microbial genomes (KMG-I) project. Stand. Genom. Sci. 2014, 9, 9031278. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 1, 2399–2406. [Google Scholar] [CrossRef]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Welch, D.M.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedros-Alió, C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006, 14, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Lennon, J.T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. USA 2010, 107, 5881–5886. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Characteristics | 1933PT | S. saccharolytica DSM 12858T | Gudongella oleilytica W6T |
|---|---|---|---|
| Cell size, µM | 0.5 × 2–5 | 0.5–0.7 × 2–11 | 2–4 × 9–21 |
| Spore formation | + | + | – |
| Temperature range (optimum), °C | 13–55 (35) | 15–40 (30–37) | 20–45 (40) |
| pH range (optimum) | 6.7–8.0 (7.0) | 6.5–7.5 (7.0) | 6.5–9.0 (7.5) |
| NaCl range (optimum), % w/v | 0–5.0 (0–2.0) | 0–2.0 (0) * | 0–3.5 (0) |
| Genomic G + C content, % | 31.9 | 32.94 | 42.4 |
| Genome size (Mb) | 1.92 | 2.0 | 2.36 |
| Utilization of | |||
| Pyruvate | − | + | W |
| Arabinose | − | + | W |
| Cellobiose | − | + | W |
| Galactose | − | + | − |
| Glucose | − | + | − |
| Fructose | − | + | − |
| Lactose | − | + | − |
| Maltose | + | + | − |
| Mannose | W | + | − |
| Mannitol | − | + | |
| Ribose | − | + | − |
| Sucrose | − | + | − |
| Raffinose | − | − | |
| Xylose | − | + | − |
| Xylan | – | + | ND |
| Peptone | + | − | + |
| Methionine | − | − | + |
| Serine | − | + | ND |
| Hydrolysis of gelatin | W | − | ND |
| Molecular nitrogen fixation | − | – * | + |
| The main products of the yeast extract fermentation | Acetate, H2, CO2 | Acetate, H2, CO2 | ND |
| Electron acceptors: | |||
| Elemental sulfur | + | − | ND |
| Sulfite | − | + | ND |
| Major fatty acids | C14:0, C16:0, iso-C15:0 | C16:0, C18:1, C16:1 * | iso-C15:0, C14:0, C16:0, iso-C13:0 |
| Major polar lipids | ND | PGL, PL, L, DPG * | L, AL |
| Isolation source | Methanogenic enrichment from oilfield | Anaerobic-digester sludge | Oily sludge from oilfield |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazina, T.N.; Bidzhieva, S.K.; Grouzdev, D.S.; Sokolova, D.S.; Tourova, T.P.; Parshina, S.N.; Avtukh, A.N.; Poltaraus, A.B.; Talybly, A.K. Soehngenia longivitae sp. nov., a Fermenting Bacterium Isolated from a Petroleum Reservoir in Azerbaijan, and Emended Description of the Genus Soehngenia. Microorganisms 2020, 8, 1967. https://doi.org/10.3390/microorganisms8121967
Nazina TN, Bidzhieva SK, Grouzdev DS, Sokolova DS, Tourova TP, Parshina SN, Avtukh AN, Poltaraus AB, Talybly AK. Soehngenia longivitae sp. nov., a Fermenting Bacterium Isolated from a Petroleum Reservoir in Azerbaijan, and Emended Description of the Genus Soehngenia. Microorganisms. 2020; 8(12):1967. https://doi.org/10.3390/microorganisms8121967
Chicago/Turabian StyleNazina, Tamara N., Salimat K. Bidzhieva, Denis S. Grouzdev, Diyana S. Sokolova, Tatyana P. Tourova, Sofiya N. Parshina, Alexander N. Avtukh, Andrey B. Poltaraus, and Azhdar K. Talybly. 2020. "Soehngenia longivitae sp. nov., a Fermenting Bacterium Isolated from a Petroleum Reservoir in Azerbaijan, and Emended Description of the Genus Soehngenia" Microorganisms 8, no. 12: 1967. https://doi.org/10.3390/microorganisms8121967
APA StyleNazina, T. N., Bidzhieva, S. K., Grouzdev, D. S., Sokolova, D. S., Tourova, T. P., Parshina, S. N., Avtukh, A. N., Poltaraus, A. B., & Talybly, A. K. (2020). Soehngenia longivitae sp. nov., a Fermenting Bacterium Isolated from a Petroleum Reservoir in Azerbaijan, and Emended Description of the Genus Soehngenia. Microorganisms, 8(12), 1967. https://doi.org/10.3390/microorganisms8121967





