Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins
Abstract
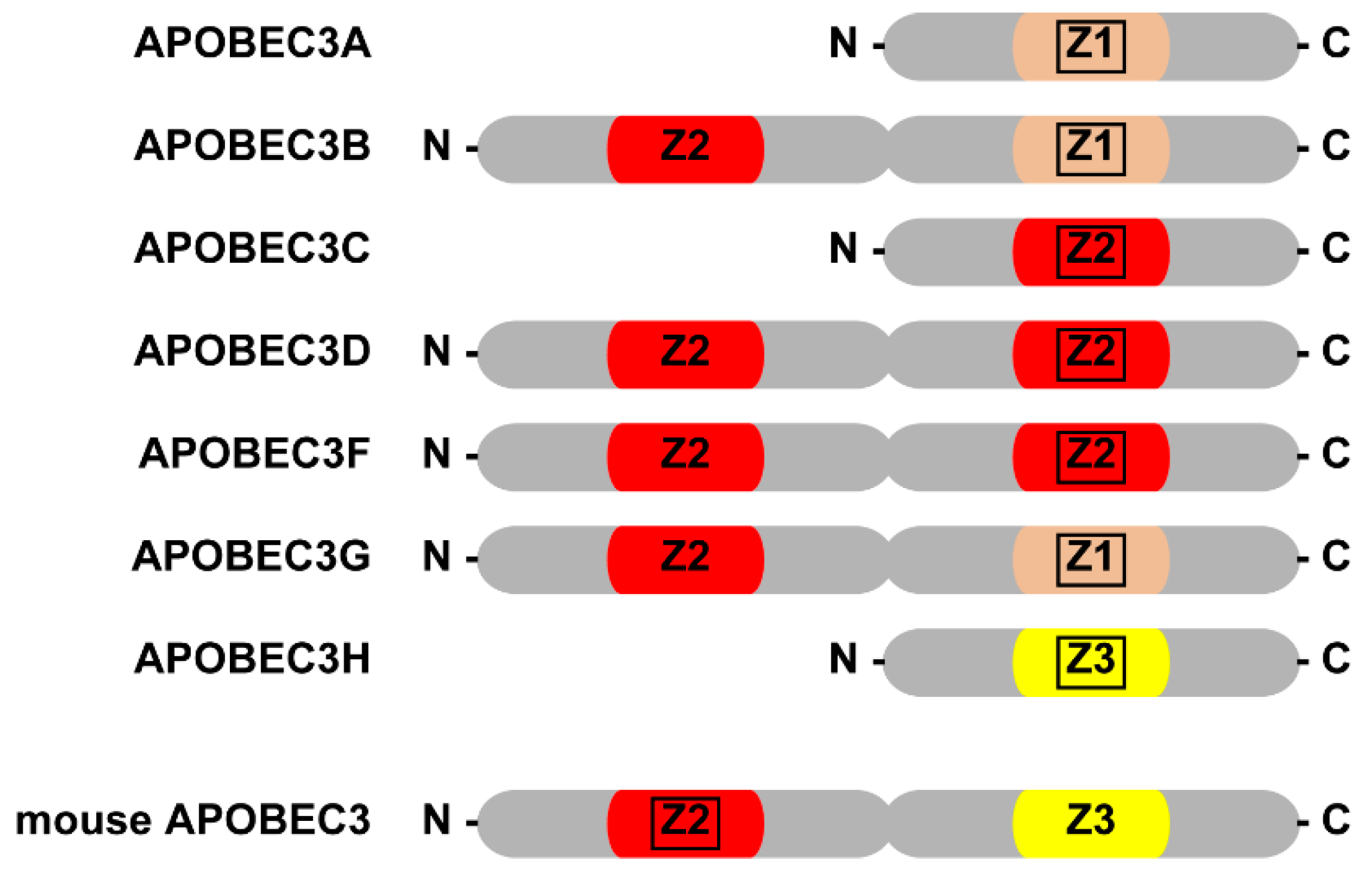
:1. Introduction
2. AID/APOBEC Family
3. Deaminase-Dependent Functions of APOBEC3s
4. Viruses Inhibited by a Deaminase-Independent Function of APOBEC3s
5. Mechanisms of the Deaminase-Independent Antiretroviral Function of APOBEC3s
6. A Deaminase-Independent Cellular Function of APOBEC3B
7. An Alternative Deaminase-Independent Anti-MuLV Mechanism of mA3
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, C.E.; Papavasiliou, F.N.; Rosenberg, B.R. Diverse functions for DNA and RNA editing in the immune system. RNA Biol. 2010, 7, 220–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longerich, S.; Basu, U.; Alt, F.W.; Storb, U. AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 2006, 18, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Stavrou, S.; Ross, S.R. APOBEC3 Proteins in Viral Immunity. J. Immunol. 2015, 195, 4565–4570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knisbacher, B.A.; Gerber, D.; Levanon, E.Y. DNA Editing by APOBECs: A Genomic Preserver and Transformer. Trends Genet. 2016, 32, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nat. Cell Biol. 2013, 494, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Chang, C.-J.; Xie, X.; Xia, W.; Yang, J.-Y.; Wang, S.-C.; Wang, Y.; Xia, J.; Chen, L.; Cai, C.; et al. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J. Clin. Investig. 2011, 121, 4526–4536. [Google Scholar] [CrossRef]
- Taylor, B.J.; Nik-Zainal, S.; Wu, Y.L.; Stebbings, L.; Raine, K.; Campbell, P.J.; Rada, C.; Stratton, M.R.; Neuberger, M.S. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2013, 2, e00534. [Google Scholar] [CrossRef]
- Harris, R.S.; Bishop, K.N.; Sheehy, A.M.; Craig, H.M.; Petersen-Mahrt, S.K.; Watt, I.N.; Neuberger, M.S.; Malim, M.H. DNA Deamination Mediates Innate Immunity to Retroviral Infection. Cell 2003, 113, 803–809. [Google Scholar] [CrossRef] [Green Version]
- Mangeat, B.; Turelli, P.; Caron, G.; Friedli, M.; Perrin, L.; Trono, D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nat. Cell Biol. 2003, 424, 99–103. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Pomerantz, R.J.; Zhang, C.; Arunachalam, S.C.; Gao, L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nat. Cell Biol. 2003, 424, 94–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nik-Zainal, S.; Alexandrov, L.B.; Wedge, D.C.; Van Loo, P.; Greenman, C.D.; Raine, K.; Jones, D.; Hinton, J.; Marshall, J.; Stebbings, L.A.; et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell 2012, 149, 979–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef] [PubMed]
- Chaurasiya, K.R.; McCauley, M.J.; Wang, W.; Qualley, M.F.; Wu, T.; Kitamura, S.; Geertsema, H.; Chan, D.S.B.; Hertz, A.; Iwatani, Y.; et al. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat. Chem. 2013, 6, 28–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwatani, Y.; Chan, D.S.; Wang, F.; Stewart-Maynard, K.; Sugiura, W.; Gronenborn, A.M.; Rouzina, I.; Williams, M.C.; Musier-Forsyth, K.; Levin, J.G. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007, 35, 7096–7108. [Google Scholar] [CrossRef] [PubMed]
- Newman, E.N.; Holmes, R.K.; Craig, H.M.; Klein, K.C.; Lingappa, J.R.; Malim, M.H.; Sheehy, A.M. Antiviral Function of APOBEC3G Can Be Dissociated from Cytidine Deaminase Activity. Curr. Biol. 2005, 15, 166–170. [Google Scholar] [CrossRef] [Green Version]
- Hakata, Y.; Li, J.; Fujino, T.; Tanaka, Y.; Shimizu, R.; Miyazawa, M. Mouse APOBEC3 interferes with autocatalytic cleavage of murine leukemia virus Pr180gag-pol precursor and inhibits Pr65gag processing. PLoS Pathog. 2019, 15, e1008173. [Google Scholar] [CrossRef] [Green Version]
- Muramatsu, M.; Sankaranand, V.S.; Anant, S.; Sugai, M.; Kinoshita, K.; Davidson, N.O.; Honjo, T. Specific Expression of Activation-induced Cytidine Deaminase (AID), a Novel Member of the RNA-editing Deaminase Family in Germinal Center B Cells. J. Biol. Chem. 1999, 274, 18470–18476. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Seija, N.; Di Noia, J.M.; Pezo, R.C. AID in Antibody Diversification: There and Back Again. Trends Immunol. 2020, 41, 586–600. [Google Scholar] [CrossRef]
- Endo, Y.; Marusawa, H.; Kinoshita, K.; Morisawa, T.; Sakurai, T.; Okazaki, I.-M.; Watashi, K.; Shimotohno, K.; Honjo, T.; Chiba, T. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-κB signaling. Oncogene 2007, 26, 5587–5595. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, Y.; Marusawa, H.; Kinoshita, K.; Endo, Y.; Kou, T.; Morisawa, T.; Azuma, T.; Okazaki, I.-M.; Honjo, T.; Chiba, T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 2007, 13, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okazaki, I.-M.; Hiai, H.; Kakazu, N.; Yamada, S.; Muramatsu, M.; Kinoshita, K.; Honjo, T. Constitutive Expression of AID Leads to Tumorigenesis. J. Exp. Med. 2003, 197, 1173–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, S.; Kitamura, K.; Simadu, M.; Koura, M.; Muramatsu, M. Concerted action of activation-induced cytidine deaminase and uracil-DNA glycosylase reduces covalently closed circular DNA of duck hepatitis B virus. FEBS Lett. 2013, 587, 3148–3152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, G.; Kitamura, K.; Wang, Z.; Liu, G.; Chowdhury, S.; Fu, W.; Koura, M.; Wakae, K.; Honjo, T.; Muramatsu, M. RNA editing of hepatitis B virus transcripts by activation-induced cytidine deaminase. Proc. Natl. Acad. Sci. USA 2013, 110, 2246–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, Y.; Han, X.; Guan, G.; Wu, N.; Sun, J.; Pak, V.; Liang, G. TGF-β triggers HBV cccDNA degradation through AID-dependent deamination. FEBS Lett. 2016, 590, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Liu, G.; Kitamura, K.; Wang, Z.; Chowdhury, S.; Monjurul, A.M.; Wakae, K.; Koura, M.; Shimadu, M.; Kinoshita, K.; et al. TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex. PLoS Pathog. 2015, 11, e1004780. [Google Scholar] [CrossRef] [PubMed]
- Watashi, K.; Liang, G.; Iwamoto, M.; Marusawa, H.; Uchida, N.; Daito, T.; Kitamura, K.; Muramatsu, M.; Ohashi, H.; Kiyohara, T.; et al. Interleukin-1 and Tumor Necrosis Factor-α Trigger Restriction of Hepatitis B Virus Infection via a Cytidine Deaminase Activation-induced Cytidine Deaminase (AID). J. Biol. Chem. 2013, 288, 31715–31727. [Google Scholar] [CrossRef] [Green Version]
- Navaratnam, N.; Morrison, J.R.; Bhattacharya, S.; Patel, D.; Funahashi, T.; Giannoni, F.; Teng, B.B.; Davidson, N.; Scott, J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 1993, 268, 20709–20712. [Google Scholar]
- Teng, B.; Burant, C.F.; Davidson, N. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 1993, 260, 1816–1819. [Google Scholar] [CrossRef]
- Rosenberg, B.R.; Hamilton, C.E.; Mwangi, M.M.; Dewell, S.; Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 2011, 18, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Skuse, G.R.; Cappione, A.J.; Sowden, M.P.; Metheny, L.J.; Smith, H.C. The Neurofibromatosis Type I Messenger RNA Undergoes Base-Modification RNA Editing. Nucleic Acids Res. 1996, 24, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Anant, S.; Lee, R.M.; Kennedy, S.; Viskochil, D.; Davidson, N.O. C→U Editing of Neurofibromatosis 1 mRNA Occurs in Tumors That Express Both the Type II Transcript and apobec-1, the Catalytic Subunit of the Apolipoprotein B mRNA–Editing Enzyme. Am. J. Hum. Genet. 2002, 70, 38–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc, V.; Park, E.; Schaefer, S.; Miller, M.; Lin, Y.; Kennedy, S.; Billing, A.M.; Ben Hamidane, H.; Graumann, J.; Mortazavi, A.; et al. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 2014, 15, R79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; El Galil, K.H.A.; Tokunaga, K.; Maeda, K.; Sata, T.; Sakaguchi, N.; Heidmann, T.; Koito, A. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res. 2011, 39, 5538–5554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; Ong, E.B.B.; Watanabe, N.; Sakaguchi, N.; Maeda, K.; Koito, A. Creation of chimeric human/rabbit APOBEC1 with HIV-1 restriction and DNA mutation activities. Sci. Rep. 2016, 6, 19035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; Shimoda, M.; Ebrahimi, D.; VandeBerg, J.L.; Harris, R.S.; Koito, A.; Maeda, K. Opossum APOBEC1 is a DNA mutator with retrovirus and retroelement restriction activity. Sci. Rep. 2017, 7, 46719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petit, V.; Guétard, D.; Renard, M.; Keriel, A.; Sitbon, M.; Wain-Hobson, S.; Vartanian, J.-P. Murine APOBEC1 Is a Powerful Mutator of Retroviral and Cellular RNA In Vitro and In Vivo. J. Mol. Biol. 2009, 385, 65–78. [Google Scholar] [CrossRef]
- Petersen-Mahrt, S.K.; Neuberger, M.S. In Vitro Deamination of Cytosine to Uracil in Single-stranded DNA by Apolipoprotein B Editing Complex Catalytic Subunit 1 (APOBEC1). J. Biol. Chem. 2003, 278, 19583–19586. [Google Scholar] [CrossRef] [Green Version]
- Harris, R.S.; Petersen-Mahrt, S.K.; Neuberger, M.S. RNA Editing Enzyme APOBEC1 and Some of Its Homologs Can Act as DNA Mutators. Mol. Cell 2002, 10, 1247–1253. [Google Scholar] [CrossRef]
- Bishop, K.N.; Holmes, R.K.; Sheehy, A.M.; Davidson, N.O.; Cho, S.-J.; Malim, M.H. Cytidine Deamination of Retroviral DNA by Diverse APOBEC Proteins. Curr. Biol. 2004, 14, 1392–1396. [Google Scholar] [CrossRef] [Green Version]
- Bishop, K.N.; Holmes, R.K.; Sheehy, A.M.; Malim, M.H. APOBEC-Mediated Editing of Viral RNA. Science 2004, 305, 645. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Ohsugi, T.; Kimura, T.; Matsushita, S.; Maeda, Y.; Harada, S.; Koito, A. The antiretroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res. 2008, 36, 6859–6871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, M.C.; Suspène, R.; Henry, M.; Guétard, D.; Wain-Hobson, S.; Vartanian, J.-P. Human APOBEC1 cytidine deaminase edits HBV DNA. Retrovirology 2009, 6, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renard, M.; Henry, M.; Guétard, D.; Vartanian, J.-P.; Wain-Hobson, S. APOBEC1 and APOBEC3 Cytidine Deaminases as Restriction Factors for Hepadnaviral Genomes in Non-Humans In Vivo. J. Mol. Biol. 2010, 400, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Ando, Y.; Kitayama, H.; Yamamoto, S.P.; Kanemura, Y.; Ebina, H.; Kawaguchi, Y.; Koyanagi, Y. APOBEC1-Mediated Editing and Attenuation of Herpes Simplex Virus 1 DNA Indicate That Neurons Have an Antiviral Role during Herpes Simplex Encephalitis. J. Virol. 2011, 85, 9726–9736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindič, N.; Budič, M.; Petan, T.; Knisbacher, B.; Levanon, E.Y.; Lovšin, N. Differential inhibition of LINE1 and LINE2 retrotransposition by vertebrate AID/APOBEC proteins. Retrovirology 2013, 10, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, S.; Balestra, M.E.; Ferrell, L.D.; Fan, J.; Arnold, K.S.; Taylor, S.; Taylor, J.M.; Innerarity, T.L. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. USA 1995, 92, 8483–8487. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Probst, H.C.; Tatsumi, R.; Ikeuchi, Y.; Neuberger, M.S.; Rada, C. Deficiency in APOBEC2 Leads to a Shift in Muscle Fiber Type, Diminished Body Mass, and Myopathy. J. Biol. Chem. 2010, 285, 7111–7118. [Google Scholar] [CrossRef] [Green Version]
- Mikl, M.C.; Watt, I.N.; Lu, M.; Reik, W.; Davies, S.L.; Neuberger, M.S.; Rada, C. Mice Deficient in APOBEC2 and APOBEC3. Mol. Cell. Biol. 2005, 25, 7270–7277. [Google Scholar] [CrossRef] [Green Version]
- Lada, A.G.; Krick, C.F.; Kozmin, S.G.; Mayorov, V.I.; Karpova, T.S.; Rogozin, I.B.; Pavlov, Y.I. Mutator effects and mutation signatures of editing deaminases produced in bacteria and yeast. Biochemistry 2011, 76, 131–146. [Google Scholar] [CrossRef] [Green Version]
- Conticello, S.G.; Langlois, M.; Yang, Z.; Neuberger, M.S. DNA Deamination in Immunity: AID in the Context of Its APOBEC Relatives. Adv. Immunol. 2007, 94, 37–73. [Google Scholar] [CrossRef] [PubMed]
- Rogozin, I.B.; Basu, M.K.; Jordan, I.K.; Pavlov, Y.I.; Koonin, E.V. APOBEC4, a New Member of the AID/APOBEC Family of Polynucleotide (Deoxy)Cytidine Deaminases Predicted by Computational Analysis. Cell Cycle 2005, 4, 1281–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino, D.; Perković, M.; Hain, A.; Vasudevan, A.A.J.; Hofmann, H.; Hanschmann, K.-M.; Mühlebach, M.D.; Schumann, G.G.; König, R.; Cichutek, K.; et al. APOBEC4 Enhances the Replication of HIV-1. PLoS ONE 2016, 11, e0155422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, M.; Tan, L.; Zhang, Y.; Meng, C.; Wang, W.; Sun, Y.; Song, C.; Liu, W.; Liao, Y.; Yu, S.; et al. Characterization and functional analysis of chicken APOBEC4. Dev. Comp. Immunol. 2020, 106, 103631. [Google Scholar] [CrossRef]
- Duggal, N.K.; Fu, W.; Akey, J.M.; Emerman, M. Identification and antiviral activity of common polymorphisms in the APOBEC3 locus in human populations. Virology 2013, 443, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Henry, M.; Terzian, C.; Peeters, M.; Wain-Hobson, S.; Vartanian, J.-P. Evolution of the Primate APOBEC3A Cytidine Deaminase Gene and Identification of Related Coding Regions. PLoS ONE 2012, 7, e30036. [Google Scholar] [CrossRef]
- Sawyer, S.L.; Emerman, M.; Malik, H.S. Ancient Adaptive Evolution of the Primate Antiviral DNA-Editing Enzyme APOBEC3G. PLoS Biol. 2004, 2, e275. [Google Scholar] [CrossRef]
- Zhang, J. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 2004, 13, 1785–1791. [Google Scholar] [CrossRef]
- Conticello, S.G.; Thomas, C.J.F.; Petersen-Mahrt, S.K.; Neuberger, M.S. Evolution of the AID/APOBEC Family of Polynucleotide (Deoxy)cytidine Deaminases. Mol. Biol. Evol. 2004, 22, 367–377. [Google Scholar] [CrossRef] [Green Version]
- LaRue, R.S.; Andrésdóttir, V.; Blanchard, Y.; Conticello, S.G.; Derse, D.; Emerman, M.; Greene, W.C.; Jónsson, S.R.; Landau, N.R.; Löchelt, M.; et al. Guidelines for Naming Nonprimate APOBEC3 Genes and Proteins. J. Virol. 2008, 83, 494–497. [Google Scholar] [CrossRef] [Green Version]
- Münk, C.; Beck, T.; Zielonka, J.; Hotz-Wagenblatt, A.; Chareza, S.; Battenberg, M.; Thielebein, J.; Cichutek, K.; Bravo, I.G.; O’Brien, S.; et al. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008, 9, R48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaRue, R.S.; Jónsson, S.R.; Silverstein, K.A.T.; Lajoie, M.; Bertrand, D.; El-Mabrouk, N.; Hötzel, I.; Andrésdóttir, V.; Smith, T.P.L.; Harris, R.S. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 2008, 9, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayward, J.; Tachedjian, M.; Cui, J.; Cheng, A.Z.; Johnson, A.; Baker, M.L.; Harris, R.S.; Wang, L.-F.; Tachedjian, G. Differential Evolution of Antiretroviral Restriction Factors in Pteropid Bats as Revealed by APOBEC3 Gene Complexity. Mol. Biol. Evol. 2018, 35, 1626–1637. [Google Scholar] [CrossRef]
- Yang, L.; Emerman, M.; Malik, H.S.; McLaughlin, R.N., Jr. Retrocopying expands the functional repertoire of APOBEC3 antiviral proteins in primates. eLife 2020, 9, e58436. [Google Scholar] [CrossRef]
- Desimmie, B.A.; Burdick, R.C.; Izumi, T.; Doi, H.; Shao, W.; Alvord, W.G.; Sato, K.; Koyanagi, Y.; Jones, S.; Wilson, E.; et al. APOBEC3 proteins can copackage and comutate HIV-1 genomes. Nucleic Acids Res. 2016, 44, 7848–7865. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, S.J.; Liu, C.; Landau, M.; Buzovetsky, O.; Desimmie, B.A.; Zhao, Q.; Sasaki, T.; Burdick, R.C.; Pathak, V.K.; Anderson, K.S.; et al. Insights into DNA substrate selection by APOBEC3G from structural, biochemical, and functional studies. PLoS ONE 2018, 13, e0195048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtz, C.M.; Sadler, H.A.; Mansky, L.M. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 2013, 41, 6139–6148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beale, R.C.; Petersen-Mahrt, S.K.; Watt, I.N.; Harris, R.S.; Rada, C.; Neuberger, M.S. Comparison of the Differential Context-dependence of DNA Deamination by APOBEC Enzymes: Correlation with Mutation Spectra in Vivo. J. Mol. Biol. 2004, 337, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Silvas, T.V.; Hou, S.; Myint, W.; Nalivaika, E.; Somasundaran, M.; Kelch, B.A.; Matsuo, H.; Yilmaz, N.K.; Schiffer, C.A. Substrate sequence selectivity of APOBEC3A implicates intra-DNA interactions. Sci. Rep. 2018, 8, 7511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDaniel, Y.Z.; Wang, D.; Love, R.P.; Adolph, M.B.; Mohammadzadeh, N.; Chelico, L.; Mansky, L.M. Deamination hotspots among APOBEC3 family members are defined by both target site sequence context and ssDNA secondary structure. Nucleic Acids Res. 2020, 48, 1353–1371. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Diaz, M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 2004, 172, 3382–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esnault, C.; Heidmann, O.; Delebecque, F.; Dewannieux, M.; Ribet, D.; Hance, A.J.; Heidmann, T.; Schwartz, O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nat. Cell Biol. 2005, 433, 430–433. [Google Scholar] [CrossRef] [PubMed]
- Suspène, R.; Aynaud, M.-M.; Koch, S.; Pasdeloup, D.; Labetoulle, M.; Gaertner, B.; Vartanian, J.-P.; Meyerhans, A.; Wain-Hobson, S. Genetic Editing of Herpes Simplex Virus 1 and Epstein-Barr Herpesvirus Genomes by Human APOBEC3 Cytidine Deaminases in Culture and In Vivo. J. Virol. 2011, 85, 7594–7602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nat. Cell Biol. 2002, 418, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Okeoma, C.M.; Lovsin, N.; Peterlin, B.M.; Ross, S.R. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nat. Cell Biol. 2007, 445, 927–930. [Google Scholar] [CrossRef]
- Sasada, A.; Takaori-Kondo, A.; Shirakawa, K.; Kobayashi, M.; Abudu, A.; Hishizawa, M.; Imada, K.; Tanaka, Y.; Uchiyama, T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 2005, 2, 32. [Google Scholar] [CrossRef] [Green Version]
- Mahieux, R.; Suspène, R.; Delebecque, F.; Henry, M.; Schwartz, O.; Wain-Hobson, S.; Vartanian, J.-P. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J. Gen. Virol. 2005, 86, 2489–2494. [Google Scholar] [CrossRef]
- Wiegand, H.L.; Cullen, B.R. Inhibition of Alpharetrovirus Replication by a Range of Human APOBEC3 Proteins. J. Virol. 2007, 81, 13694–13699. [Google Scholar] [CrossRef] [Green Version]
- Takeda, E.; Tsuji-Kawahara, S.; Sakamoto, M.; Langlois, M.-A.; Neuberger, M.S.; Rada, C.; Miyazawa, M. Mouse APOBEC3 Restricts Friend Leukemia Virus Infection and Pathogenesis In Vivo. J. Virol. 2008, 82, 10998–11008. [Google Scholar] [CrossRef] [Green Version]
- Turelli, P. Inhibition of Hepatitis B Virus Replication by APOBEC3G. Science 2004, 303, 1829. [Google Scholar] [CrossRef]
- Chen, H.; Lilley, C.E.; Yu, Q.; Lee, D.V.; Chou, J.; Narvaiza, I.; Landau, N.R.; Weitzman, M.D. APOBEC3A Is a Potent Inhibitor of Adeno-Associated Virus and Retrotransposons. Curr. Biol. 2006, 16, 480–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milewska, A.; Kindler, E.; V’Kovski, P.; Zeglen, S.; Ochman, M.; Thiel, V.; Rajfur, Z.; Pyrć, K. APOBEC3-mediated restriction of RNA virus replication. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.A.; Wiegand, H.L.; Moore, M.D.; Schäfer, A.; McClure, M.O.; Cullen, B.R. Foamy Virus Bet Proteins Function as Novel Inhibitors of the APOBEC3 Family of Innate Antiretroviral Defense Factors. J. Virol. 2005, 79, 8724–8731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasudevan, A.A.J.; Perković, M.; Bulliard, Y.; Cichutek, K.; Trono, D.; Häussinger, D.; Münk, C. Prototype Foamy Virus Bet Impairs the Dimerization and Cytosolic Solubility of Human APOBEC3G. J. Virol. 2013, 87, 9030–9040. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.Z.; Yockteng-Melgar, J.; Jarvis, M.C.; Malik-Soni, N.; Borozan, I.; Carpenter, M.A.; McCann, J.L.; Ebrahimi, D.; Shaban, N.M.; Marcon, E.; et al. Epstein–Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 2019, 4, 78–88. [Google Scholar] [CrossRef]
- Svarovskaia, E.S.; Xu, H.; Mbisa, J.L.; Barr, R.; Gorelick, R.J.; Ono, A.; Freed, E.; Hu, W.-S.; Pathak, V.K. Human Apolipoprotein B mRNA-editing Enzyme-catalytic Polypeptide-like 3G (APOBEC3G) Is Incorporated into HIV-1 Virions through Interactions with Viral and Nonviral RNAs. J. Biol. Chem. 2004, 279, 35822–35828. [Google Scholar] [CrossRef] [Green Version]
- Burnett, A.; Spearman, P. APOBEC3G Multimers Are Recruited to the Plasma Membrane for Packaging into Human Immunodeficiency Virus Type 1 Virus-Like Particles in an RNA-Dependent Process Requiring the NC Basic Linker. J. Virol. 2007, 81, 5000–5013. [Google Scholar] [CrossRef] [Green Version]
- Apolonia, L.; Schulz, R.; Curk, T.; Rocha, P.; Swanson, C.M.; Schaller, T.; Ule, J.; Malim, M.H. Promiscuous RNA Binding Ensures Effective Encapsidation of APOBEC3 Proteins by HIV-1. PLoS Pathog. 2015, 11, e1004609. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Chertova, E.; Chen, J.; Ott, D.E.; Roser, J.D.; Hu, W.-S.; Pathak, V.K. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology 2007, 360, 247–256. [Google Scholar] [CrossRef] [Green Version]
- Chiu, Y.-L.; Greene, W.C. The APOBEC3 Cytidine Deaminases: An Innate Defensive Network Opposing Exogenous Retroviruses and Endogenous Retroelements. Annu. Rev. Immunol. 2008, 26, 317–353. [Google Scholar] [CrossRef]
- Jäger, S.; Kim, D.Y.; Hultquist, J.F.; Shindo, K.; LaRue, R.S.; Kwon, E.; Li, M.; Anderson, B.D.; Yen, L.; Stanley, D.J.; et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nat. Cell Biol. 2012, 481, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Du, J.; Evans, S.L.; Yu, Y.; Yu, X.-F. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nat. Cell Biol. 2011, 481, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Hultquist, J.F.; Binka, M.; LaRue, R.S.; Simon, V.; Harris, R.S. Vif Proteins of Human and Simian Immunodeficiency Viruses Require Cellular CBF To Degrade APOBEC3 Restriction Factors. J. Virol. 2011, 86, 2874–2877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kane, J.R.; Stanley, D.J.; Hultquist, J.F.; Johnson, J.R.; Mietrach, N.; Binning, J.M.; Jónsson, S.R.; Barelier, S.; Newton, B.W.; Johnson, T.L.; et al. Lineage-Specific Viral Hijacking of Non-canonical E3 Ubiquitin Ligase Cofactors in the Evolution of Vif Anti-APOBEC3 Activity. Cell Rep. 2015, 11, 1236–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariani, R.; Chen, D.; Schröfelbauer, B.; Navarro, F.; König, R.; Bollman, B.; Münk, C.; Nymark-McMahon, H.; Landau, N.R. Species-Specific Exclusion of APOBEC3G from HIV-1 Virions by Vif. Cell 2003, 114, 21–31. [Google Scholar] [CrossRef] [Green Version]
- D’Arc, M.; Ayouba, A.; Esteban, A.; Learn, G.H.; Boué, V.; Liegeois, F.; Etienne, L.; Tagg, N.; Leendertz, F.H.; Boesch, C.; et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc. Natl. Acad. Sci. USA 2015, 112, E1343–E1352. [Google Scholar] [CrossRef] [Green Version]
- Hultquist, J.F.; Lengyel, J.A.; Refsland, E.W.; LaRue, R.S.; Lackey, L.; Brown, W.L.; Harris, R.S. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-Deficient HIV-1. J. Virol. 2011, 85, 11220–11234. [Google Scholar] [CrossRef] [Green Version]
- Refsland, E.W.; Hultquist, J.F.; Harris, R.S. Endogenous Origins of HIV-1 G-to-A Hypermutation and Restriction in the Nonpermissive T Cell Line CEM2n. PLoS Pathog. 2012, 8, e1002800. [Google Scholar] [CrossRef]
- Refsland, E.W.; Hultquist, J.F.; Luengas, E.M.; Ikeda, T.; Shaban, N.M.; Law, E.K.; Brown, W.L.; Reilly, C.; Emerman, M.; Harris, R.S. Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity. PLoS Genet. 2014, 10, e1004761. [Google Scholar] [CrossRef]
- Ikeda, T.; Symeonides, M.; Albin, J.S.; Li, M.; Thali, M.; Harris, R.S. HIV-1 adaptation studies reveal a novel Env-mediated homeostasis mechanism for evading lethal hypermutation by APOBEC3G. PLoS Pathog. 2018, 14, e1007010. [Google Scholar] [CrossRef] [Green Version]
- Hagen, B.; Kraase, M.; Indikova, I.; Indik, S. A high rate of polymerization during synthesis of mouse mammary tumor virus DNA alleviates hypermutation by APOBEC3 proteins. PLoS Pathog. 2019, 15, e1007533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delviks-Frankenberry, K.; Desimmie, B.A.; Pathak, V.K. Structural Insights into APOBEC3-Mediated Lentiviral Restriction. Viruses 2020, 12, 587. [Google Scholar] [CrossRef]
- Iwatani, Y.; Takeuchi, H.; Strebel, K.; Levin, J.G. Biochemical Activities of Highly Purified, Catalytically Active Human APOBEC3G: Correlation with Antiviral Effect. J. Virol. 2006, 80, 5992–6002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, F.; Bollman, B.; Chen, H.; König, R.; Yu, Q.; Chiles, K.; Landau, N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology 2005, 333, 374–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, M.E.; Harris, R.S.; Harki, D.A. APOBEC Enzymes as Targets for Virus and Cancer Therapy. Cell Chem. Biol. 2018, 25, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Hakata, Y.; Landau, N.R. Reversed Functional Organization of Mouse and Human APOBEC3 Cytidine Deaminase Domains. J. Biol. Chem. 2006, 281, 36624–36631. [Google Scholar] [CrossRef] [Green Version]
- Bohn, J.A.; DaSilva, J.; Kharytonchyk, S.; Mercedes, M.; Vosters, J.; Telesnitsky, A.; Hatziioannou, T.; Smith, J.L. Flexibility in Nucleic Acid Binding Is Central to APOBEC3H Antiviral Activity. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Vasudevan, A.A.J.; Hofmann, H.; Willbold, D.; Häussinger, D.; Koenig, B.W.; Münk, C. Enhancing the Catalytic Deamination Activity of APOBEC3C Is Insufficient to Inhibit Vif-Deficient HIV-1. J. Mol. Biol. 2017, 429, 1171–1191. [Google Scholar] [CrossRef]
- Yu, Q.; Chen, D.; König, R.; Mariani, R.; Unutmaz, D.; Landau, N.R. APOBEC3B and APOBEC3C Are Potent Inhibitors of Simian Immunodeficiency Virus Replication. J. Biol. Chem. 2004, 279, 53379–53386. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, S.; Ode, H.; Nakashima, M.; Imahashi, M.; Naganawa, Y.; Kurosawa, T.; Yokomaku, Y.; Yamane, T.; Watanabe, N.; Suzuki, A.; et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 2012, 19, 1005–1010. [Google Scholar] [CrossRef]
- Vasudevan, A.A.J.; Balakrishnan, K.; Gertzen, C.G.W.; Borvető, F.; Zhang, Z.; Sangwiman, A.; Held, U.; Küstermann, C.; Banerjee, S.; Schumann, G.G.; et al. Loop 1 of APOBEC3C Regulates its Antiviral Activity against HIV-1. J. Mol. Biol. 2020, 432, 6200–6227. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Initiative, A.P.C.G.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; et al. Signatures of mutational processes in human cancer. Nat. Cell Biol. 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.; Roberts, S.A.; Klimczak, L.J.; Sterling, J.F.; Saini, N.; Malc, E.P.; Kim, J.; Kwiatkowski, D.J.; Fargo, D.C.; Mieczkowski, P.A.; et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015, 47, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Temiz, N.A.; Harris, R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013, 45, 977–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, B.; Hart, S.N.; Burns, M.B.; Carpenter, M.A.; Temiz, N.A.; Rathore, A.; Vogel, R.I.; Nikas, J.B.; Law, E.K.; Brown, W.L.; et al. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res. 2013, 73, 7222–7231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, L.M.; Brown, A.L.; Dennis, M.A.; Collins, C.D.; Brown, A.J.; Mitchell, D.; Mertz, T.M.; Roberts, S.A. APOBEC3A is a prominent cytidine deaminase in breast cancer. PLoS Genet. 2019, 15, e1008545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, E.K.; Levin-Klein, R.; Jarvis, M.C.; Kim, H.; Argyris, P.P.; Carpenter, M.A.; Starrett, G.J.; Temiz, N.; Larson, L.K.; Durfee, C.; et al. APOBEC3A catalyzes mutation and drives carcinogenesis in vivo. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Asaoka, M.; Ishikawa, T.; Takabe, K.; Patnaik, S.K. APOBEC3-Mediated RNA Editing in Breast Cancer is Associated with Heightened Immune Activity and Improved Survival. Int. J. Mol. Sci. 2019, 20, 5621. [Google Scholar] [CrossRef] [Green Version]
- Bishop, K.N.; Verma, M.; Kim, E.-Y.; Wolinsky, S.M.; Malim, M.H. APOBEC3G Inhibits Elongation of HIV-1 Reverse Transcripts. PLoS Pathog. 2008, 4, e1000231. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Cen, S.; Niu, M.; Saadatmand, J.; Kleiman, L. Inhibition of nhibition of tRNA3(Lys)-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 2006, 80, 11710–11722. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Cen, S.; Niu, M.; Yang, Y.; Gorelick, R.J.; Kleiman, L. The Interaction of APOBEC3G with Human Immunodeficiency Virus Type 1 Nucleocapsid Inhibits tRNA3Lys Annealing to Viral RNA. J. Virol. 2007, 81, 11322–11331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, K.; Wang, T.; Liu, B.; Tian, C.; Xiao, Z.; Kappes, J.C.; Yu, X.-F. Cytidine Deaminases APOBEC3G and APOBEC3F Interact with Human Immunodeficiency Virus Type 1 Integrase and Inhibit Proviral DNA Formation. J. Virol. 2007, 81, 7238–7248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bélanger, K.; Savoie, M.; Gerpe, M.C.R.; Couture, J.-F.; Langlois, M.-A. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res. 2013, 41, 7438–7452. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.K.; Koning, F.A.; Bishop, K.N.; Malim, M.H. APOBEC3F Can Inhibit the Accumulation of HIV-1 Reverse Transcription Products in the Absence of Hypermutation. J. Biol. Chem. 2007, 282, 2587–2595. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-Y.; Guo, F.; Zhang, L.; Kleiman, L.; Cen, S. APOBEC3G Inhibits DNA Strand Transfer during HIV-1 Reverse Transcription. J. Biol. Chem. 2007, 282, 32065–32074. [Google Scholar] [CrossRef] [Green Version]
- Bishop, K.N.; Holmes, R.K.; Malim, M.H. Antiviral Potency of APOBEC Proteins Does Not Correlate with Cytidine Deamination. J. Virol. 2006, 80, 8450–8458. [Google Scholar] [CrossRef] [Green Version]
- Shindo, K.; Takaori-Kondo, A.; Kobayashi, M.; Abudu, A.; Fukunaga, K.; Uchiyama, T. The Enzymatic Activity of CEM15/Apobec-3G Is Essential for the Regulation of the Infectivity of HIV-1 Virion but Not a Sole Determinant of Its Antiviral Activity. J. Biol. Chem. 2003, 278, 44412–44416. [Google Scholar] [CrossRef] [Green Version]
- Gillick, K.; Pollpeter, D.; Phalora, P.; Kim, E.-Y.; Wolinsky, S.M.; Malim, M.H. Suppression of HIV-1 Infection by APOBEC3 Proteins in Primary Human CD4+T Cells Is Associated with Inhibition of Processive Reverse Transcription as Well as Excessive Cytidine Deamination. J. Virol. 2013, 87, 1508–1517. [Google Scholar] [CrossRef] [Green Version]
- Jónsson, S.R.; Haché, G.; Stenglein, M.D.; Fahrenkrug, S.C.; Andrésdóttir, V.; Harris, R.S. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006, 34, 5683–5694. [Google Scholar] [CrossRef]
- Ara, A.; Love, R.P.; Follack, T.B.; Ahmed, K.A.; Adolph, M.B.; Chelico, L. Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G. J. Virol. 2016, 91. [Google Scholar] [CrossRef] [Green Version]
- Dang, Y.; Siew, L.M.; Wang, X.; Han, Y.; Lampen, R.; Zheng, Y.-H. Human Cytidine Deaminase APOBEC3H Restricts HIV-1 Replication. J. Biol. Chem. 2008, 283, 11606–11614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, M.; Singer, D.; Mano, Y.; Hritz, J.; Nam, G.; Gorelick, R.J.; Byeon, I.-J.L.; Gronenborn, A.M.; Iwatani, Y.; Levin, J.G. Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities. Retrovirology 2015, 12, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derse, D.; Hill, S.A.; Princler, G.; Lloyd, P.; Heidecker, G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. USA 2007, 104, 2915–2920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ooms, M.; Krikoni, A.; Kress, A.K.; Simon, V.; Münk, C. APOBEC3A, APOBEC3B, and APOBEC3H Haplotype 2 Restrict Human T-Lymphotropic Virus Type 1. J. Virol. 2012, 86, 6097–6108. [Google Scholar] [CrossRef] [Green Version]
- Kuramitsu, M.; Sekizuka, T.; Yamochi, T.; Firouzi, S.; Sato, T.; Umeki, K.; Sasaki, D.; Hasegawa, H.; Kubota, R.; Sobata, R.; et al. Proviral Features of Human T Cell Leukemia Virus Type 1 in Carriers with Indeterminate Western Blot Analysis Results. J. Clin. Microbiol. 2017, 55, 2838–2849. [Google Scholar] [CrossRef] [Green Version]
- Köck, J.; Blum, H.E. Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J. Gen. Virol. 2008, 89, 1184–1191. [Google Scholar] [CrossRef]
- Rösler, C.; Köck, J.; Kann, M.; Malim, M.H.; Blum, H.E.; Baumert, T.F.; Von Weizsäcker, F. APOBEC-mediated interference with hepadnavirus production. Hepatology 2005, 42, 301–309. [Google Scholar] [CrossRef]
- Suspène, R.; Guétard, D.; Henry, M.; Sommer, P.; Wain-Hobson, S.; Vartanian, J.-P. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 8321–8326. [Google Scholar] [CrossRef] [Green Version]
- Bonvin, M.; Achermann, F.; Greeve, I.; Stroka, D.; Keogh, A.; Inderbitzin, D.; Candinas, D.; Sommer, P.; Wain-Hobson, S.; Vartanian, J.-P.; et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 2006, 43, 1364–1374. [Google Scholar] [CrossRef]
- Günther, S.; Sommer, G.; Plikat, U.; Iwanska, A.; Wain-Hobson, S.; Will, H.; Meyerhans, A. Naturally Occurring Hepatitis B Virus Genomes Bearing the Hallmarks of Retroviral G—A Hypermutation. Virology 1997, 235, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Vartanian, J.P.; Meyerhans, A.; Asjö, B.; Wain-Hobson, S. Selection, recombination, and G—A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 1991, 65, 1779–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.H.; Gummuluru, S.; Hu, J. Deamination-Independent Inhibition of Hepatitis B Virus Reverse Transcription by APOBEC3G. J. Virol. 2007, 81, 4465–4472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazazian, H.H. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodier, J.L.; Kazazian, H.H. Retrotransposons Revisited: The Restraint and Rehabilitation of Parasites. Cell 2008, 135, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J.V. LINE-1 Retrotransposition Activity in Human Genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef] [Green Version]
- Woodcock, D.; Lawler, C.B.; Linsenmeyer, M.E.; Doherty, J.P.; Warren, W.D. Asymmetric Methylation in the Hypermethylated CpG Promoter Region of the Human L1 Retrotransposon. J. Biol. Chem. 1997, 272, 7810–7816. [Google Scholar] [CrossRef] [Green Version]
- Rodić, N.; Sharma, R.; Sharma, R.; Zampella, J.; Dai, L.; Taylor, M.S.; Hruban, R.H.; Iacobuzio-Donahue, C.A.; Maitra, A.; Torbenson, M.S.; et al. Long Interspersed Element-1 Protein Expression Is a Hallmark of Many Human Cancers. Am. J. Pathol. 2014, 184, 1280–1286. [Google Scholar] [CrossRef] [Green Version]
- Bogerd, H.P.; Wiegand, H.L.; Hulme, A.E.; Garcia-Perez, J.L.; O’Shea, K.S.; Moran, J.V.; Cullen, B.R. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 2006, 103, 8780–8785. [Google Scholar] [CrossRef] [Green Version]
- Kinomoto, M.; Kanno, T.; Shimura, M.; Ishizaka, Y.; Kojima, A.; Kurata, T.; Sata, T.; Tokunaga, K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007, 35, 2955–2964. [Google Scholar] [CrossRef]
- Muckenfuss, H.; Hamdorf, M.; Held, U.; Perković, M.; Löwer, J.; Cichutek, K.; Flory, E.; Schumann, G.G.; Münk, C. APOBEC3 Proteins Inhibit Human LINE-1 Retrotransposition. J. Biol. Chem. 2006, 281, 22161–22172. [Google Scholar] [CrossRef] [Green Version]
- Stenglein, M.D.; Harris, R.S. APOBEC3B and APOBEC3F Inhibit L1 Retrotransposition by a DNA Deamination-independent Mechanism. J. Biol. Chem. 2006, 281, 16837–16841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, S.R.; Narvaiza, I.; Planegger, R.; Weitzman, M.D.; Moran, J.V. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife 2014, 3, e02008. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Xu, J.; Yuan, W.; Song, X.; Zhang, J.; Wei, W.; Yu, X.-F.; Yang, Y. APOBEC3DE Inhibits LINE-1 Retrotransposition by Interacting with ORF1p and Influencing LINE Reverse Transcriptase Activity. PLoS ONE 2016, 11, e0157220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, A.V.; Klawitter, S.; Held, U.; Berger, A.; Vasudevan, A.A.J.; Bock, A.; Hofmann, H.; Hanschmann, K.-M.O.; Trösemeier, J.-H.; Flory, E.; et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 2014, 42, 396–416. [Google Scholar] [CrossRef] [Green Version]
- Fehrholz, M.; Kendl, S.; Prifert, C.; Weissbrich, B.; Lemon, K.; Rennick, L.; Duprex, P.W.; Rima, B.K.; Koning, F.A.; Holmes, R.K.; et al. The innate antiviral factor APOBEC3G targets replication of measles, mumps and respiratory syncytial viruses. J. Gen. Virol. 2012, 93, 565–576. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nat. Cell Biol. 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nat. Cell Biol. 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Simmonds, P. Rampant C→U Hypermutation in the Genomes of SARS-CoV-2 and Other Coronaviruses: Causes and Consequences for Their Short- and Long-Term Evolutionary Trajectories. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Meier, A.F.; Fraefel, C.; Seyffert, M. The Interplay between Adeno-Associated Virus and Its Helper Viruses. Viruses 2020, 12, 662. [Google Scholar] [CrossRef] [PubMed]
- Narvaiza, I.; Linfesty, D.C.; Greener, B.N.; Hakata, Y.; Pintel, D.J.; Logue, E.; Landau, N.R.; Weitzman, M.D. Deaminase-Independent Inhibition of Parvoviruses by the APOBEC3A Cytidine Deaminase. PLoS Pathog. 2009, 5, e1000439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browne, E.P.; Littman, D.R. Species-Specific Restriction of Apobec3-Mediated Hypermutation. J. Virol. 2008, 82, 1305–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rulli, S.J.; Mirro, J.; Hill, S.A.; Lloyd, P.; Gorelick, R.J.; Coffin, J.M.; Derse, D.; Rein, A. Interactions of Murine APOBEC3 and Human APOBEC3G with Murine Leukemia Viruses. J. Virol. 2008, 82, 6566–6575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macmillan, A.L.; Kohli, R.M.; Ross, S.R. APOBEC3 Inhibition of Mouse Mammary Tumor Virus Infection: The Role of Cytidine Deamination versus Inhibition of Reverse Transcription. J. Virol. 2013, 87, 4808–4817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salas-Briceno, K.; Zhao, W.; Ross, S.R. Mouse APOBEC3 Restriction of Retroviruses. Viruses 2020, 12, 1217. [Google Scholar] [CrossRef] [PubMed]
- Stavrou, S.; Zhao, W.; Blouch, K.; Ross, S.R. Deaminase-Dead Mouse APOBEC3 Is an In Vivo Retroviral Restriction Factor. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Adolph, M.B.; Webb, J.; Chelico, L. Retroviral Restriction Factor APOBEC3G Delays the Initiation of DNA Synthesis by HIV-1 Reverse Transcriptase. PLoS ONE 2013, 8, e64196. [Google Scholar] [CrossRef] [Green Version]
- Morse, M.; Huo, R.; Feng, Y.; Rouzina, I.; Chelico, L.; Williams, M.C. Dimerization regulates both deaminase-dependent and deaminase-independent HIV-1 restriction by APOBEC3G. Nat. Commun. 2017, 8, 597. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ao, Z.; Chen, L.; Kobinger, G.; Peng, J.; Yao, X. The Cellular Antiviral Protein APOBEC3G Interacts with HIV-1 Reverse Transcriptase and Inhibits Its Function during Viral Replication. J. Virol. 2012, 86, 3777–3786. [Google Scholar] [CrossRef] [Green Version]
- Pollpeter, D.; Parsons, M.; Sobala, A.E.; Coxhead, S.; Lang, R.D.; Bruns, A.M.; Papaioannou, S.; McDonnell, J.M.; Luis, A.; Chowdhury, J.A.; et al. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat. Microbiol. 2018, 3, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Mbisa, J.L.; Bu, W.; Pathak, V.K. APOBEC3F and APOBEC3G Inhibit HIV-1 DNA Integration by Different Mechanisms. J. Virol. 2010, 84, 5250–5259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adolph, M.B.; Ara, A.; Chelico, L. APOBEC3 Host Restriction Factors of HIV-1 Can Change the Template Switching Frequency of Reverse Transcriptase. J. Mol. Biol. 2019, 431, 1339–1352. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martinez, S.; Aloia, A.L.; Harvin, D.; Mirro, J.; Gorelick, R.J.; Jern, P.; Coffin, J.M.; Rein, A. Studies on the Restriction of Murine Leukemia Viruses by Mouse APOBEC3. PLoS ONE 2012, 7, e38190. [Google Scholar] [CrossRef] [Green Version]
- Douaisi, M.; Dussart, S.; Courcoul, M.; Bessou, G.; Vigne, R.; Decroly, E. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 2004, 321, 566–573. [Google Scholar] [CrossRef]
- Bukovsky, A.; Göttlinger, H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 1996, 70, 6820–6825. [Google Scholar] [CrossRef] [Green Version]
- Tachedjian, G.; Moore, K.L.; Goff, S.P.; Sluis-Cremer, N. Efavirenz enhances the proteolytic processing of an HIV-1 pol polyprotein precursor and reverse transcriptase homodimer formation. FEBS Lett. 2004, 579, 379–384. [Google Scholar] [CrossRef] [Green Version]
- Quillent, C.; Borman, A.M.; Paulous, S.; Dauguet, C.; Clavel, F. Extensive Regions ofpolAre Required for Efficient Human Immunodeficiency Virus Polyprotein Processing and Particle Maturation. Virology 1996, 219, 29–36. [Google Scholar] [CrossRef]
- Sudo, S.; Haraguchi, H.; Hirai, Y.; Gatanaga, H.; Sakuragi, J.-I.; Momose, F.; Morikawa, Y. Efavirenz Enhances HIV-1 Gag Processing at the Plasma Membrane through Gag-Pol Dimerization. J. Virol. 2013, 87, 3348–3360. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Akkawi, C.; Mougel, M.; Ross, S.R. Murine Leukemia Virus P50 Protein Counteracts APOBEC3 by Blocking Its Packaging. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Stavrou, S.; Nitta, T.; Kotla, S.; Ha, D.; Nagashima, K.; Rein, A.R.; Fan, H.; Ross, S.R. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc. Natl. Acad. Sci. USA 2013, 110, 9078–9083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Li, X.; Li, J.; Lu, Y.; Zhao, S.; Tang, X.; Chen, X.; Li, J.; Zheng, Y.; Li, S.; et al. APOBEC3B interaction with PRC2 modulates microenvironment to promote HCC progression. Gut 2019, 68, 1846–1857. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Ho, D.W.H.; Sze, K.M.-F.; Tsui, Y.-M.; Chan, L.-K.; Lee, J.M.-F.; Ng, I.O. APOBEC3B promotes hepatocarcinogenesis and metastasis through novel deaminase-independent activity. Mol. Carcinog. 2019, 58, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.L.; Johnson, M.; D’Aquila, R.T. APOBEC3G Complexes Decrease Human Immunodeficiency Virus Type 1 Production. J. Virol. 2011, 85, 9314–9326. [Google Scholar] [CrossRef] [Green Version]
- York, A.; Kutluay, S.B.; Errando, M.; Bieniasz, P.D. The RNA Binding Specificity of Human APOBEC3 Proteins Resembles That of HIV-1 Nucleocapsid. PLoS Pathog. 2016, 12, e1005833. [Google Scholar] [CrossRef]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.P.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [Green Version]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [Green Version]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nat. Cell Biol. 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.M.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nat. Cell Biol. 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nat. Cell Biol. 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roundtree, I.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Lence, T.; Akhtar, J.; Bayer, M.; Schmid, K.; Spindler, L.; Ho, C.H.; Kreim, N.; Andrade-Navarro, M.A.; Poeck, L.S.B.; Helm, K.S.M.; et al. m6A modulates neuronal functions and sex determination in Drosophila. Nat. Cell Biol. 2016, 540, 242–247. [Google Scholar] [CrossRef]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, Z.B.N.A.R.G.; Soller, I.U.H.E.S.-M.M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nat. Cell Biol. 2016, 540, 301–304. [Google Scholar] [CrossRef] [Green Version]
- Kan, L.; Grozhik, A.V.; Vedanayagam, J.; Patil, D.P.; Pang, N.; Lim, K.-S.; Huang, Y.-C.; Joseph, B.; Lin, C.-J.; Despic, V.; et al. The m6A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017, 8, 15737. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nat. Cell Biol. 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.H.J.; Wu, H.D.Y.Z.H.X.M.L.L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Krug, R.M.; Morgan, M.; Shatkin, A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976, 20, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, S.; Salditt-Georgieff, M.; Bachenheimer, S.; Darnell, J.; Furuichi, Y.; Morgan, M.; Shatkin, A. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 1976, 3, 749–766. [Google Scholar] [CrossRef] [PubMed]
- Beemon, K.; Keith, J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J. Mol. Biol. 1977, 113, 165–179. [Google Scholar] [CrossRef]
- Dimock, K.; Stoltzfus, C.M. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry 1977, 16, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Canaani, D.; Kahana, C.; Lavi, S.; Groner, Y. Identification and mapping of N6-methyladenosine containing sequences in Simian Virus 40 RNA. Nucleic Acids Res. 1979, 6, 2879–2899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moss, B.; Gershowitz, A.; Stringer, J.R.; Holland, L.; Wagner, E.K. 5′-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 1977, 23, 234–239. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.R.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M.; et al. Posttranscriptional m 6 A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016, 19, 675–685. [Google Scholar] [CrossRef] [Green Version]
- Courtney, D.G.; Kennedy, E.M.; Dumm, R.E.; Bogerd, H.P.; Tsai, K.; Heaton, N.S.; Cullen, B.R. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe 2017, 22, 377–386.e5. [Google Scholar] [CrossRef] [Green Version]
- Tsai, K.; Courtney, D.G.; Cullen, B.R. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. 2018, 14, e1006919. [Google Scholar] [CrossRef]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 1–9. [Google Scholar] [CrossRef]
- Hao, H.; Hao, S.; Chen, H.; Chen, Z.; Zhang, Y.; Wang, J.; Wang, H.; Zhang, B.; Qiu, J.; Deng, F.; et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 2019, 47, 362–374. [Google Scholar] [CrossRef] [Green Version]
- Ye, F.; Chen, E.R.; Nilsen, T.W. Kaposi’s Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N6-Adenosine Methylation to Promote Lytic Replication. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Hesser, C.R.; Karijolich, J.; Dominissini, D.; He, C.; Glaunsinger, B.A. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2018, 14, e1006995. [Google Scholar] [CrossRef] [Green Version]
- Courtney, D.G.; Chalem, A.; Bogerd, H.P.; Law, B.A.; Kennedy, E.M.; Holley, C.L.; Cullen, B.R. Extensive Epitranscriptomic Methylation of A and C Residues on Murine Leukemia Virus Transcripts Enhances Viral Gene Expression. mBio 2019, 10, e01209-19. [Google Scholar] [CrossRef] [Green Version]
- Tirumuru, N.; Zhao, B.S.; Lu, W.; Lu, Z.; He, C.; Wu, L. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 2016, 5, e15528. [Google Scholar] [CrossRef]
- Gokhale, N.S.; McIntyre, A.B.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6 -Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Lichinchi, G.; Zhao, B.S.; Wu, Y.; Lu, Z.; Qin, Y.; He, C.; Rana, T.M. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 2016, 20, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, X.; Tang, H.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSUN2-Mediated m5C Methylation and METTL3/METTL14-Mediated m6A Methylation Cooperatively Enhance p21 Translation. J. Cell. Biochem. 2017, 118, 2587–2598. [Google Scholar] [CrossRef]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef]
- Courtney, D.G.; Tsai, K.; Bogerd, H.P.; Kennedy, E.M.; Law, B.A.; Emery, A.; Swanstrom, R.; Holley, C.L.; Cullen, B.R. Epitranscriptomic Addition of m5C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host Microbe 2019, 26, 217–227.e6. [Google Scholar] [CrossRef]
- Eckwahl, M.; Xu, R.; Michalkiewicz, J.; Zhang, W.; Patel, P.; Cai, Z.; Pan, T. 5-Methylcytosine RNA Modifications Promote Retrovirus Replication in an ALYREF Reader Protein-Dependent Manner. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Chen, X.; Li, A.; Sun, B.-F.; Yang, Y.; Han, Y.-N.; Yuan, X.; Chen, R.-X.; Wei, W.-S.; Liu, Y.; Gao, C.-C.; et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Han, X.; Yang, W.-L.; Zhang, M.; Ma, H.-L.; Sun, B.-F.; Li, A.; Xia, J.; Chen, J.; et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol. Cell 2019, 75, 1188–1202.e11. [Google Scholar] [CrossRef]
- Tsai, K.; Vasudevan, A.A.J.; Campos, C.M.; Emery, A.; Swanstrom, R.; Cullen, B.R. Acetylation of Cytidine Residues Boosts HIV-1 Gene Expression by Increasing Viral RNA Stability. Cell Host Microbe 2020, 28, 306–312.e6. [Google Scholar] [CrossRef]
- Manners, O.; Baquero-Perez, B.; Whitehouse, A. m6A: Widespread regulatory control in virus replication. Biochim. Biophys. Acta 2019, 1862, 370–381. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hakata, Y.; Miyazawa, M. Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins. Microorganisms 2020, 8, 1976. https://doi.org/10.3390/microorganisms8121976
Hakata Y, Miyazawa M. Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins. Microorganisms. 2020; 8(12):1976. https://doi.org/10.3390/microorganisms8121976
Chicago/Turabian StyleHakata, Yoshiyuki, and Masaaki Miyazawa. 2020. "Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins" Microorganisms 8, no. 12: 1976. https://doi.org/10.3390/microorganisms8121976
APA StyleHakata, Y., & Miyazawa, M. (2020). Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins. Microorganisms, 8(12), 1976. https://doi.org/10.3390/microorganisms8121976




