Genome Sequencing of Pantoea agglomerans C1 Provides Insights into Molecular and Genetic Mechanisms of Plant Growth-Promotion and Tolerance to Heavy Metals
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Extraction, Genome Sequencing, Assembly and Annotation
2.2. Phylogenetic Tree Construction and ANI
2.3. Functional Genome Annotation and Identification of Genomic Islands
2.4. Production of Indole-3-Acetic Acid
2.5. Determination of Siderophore Production
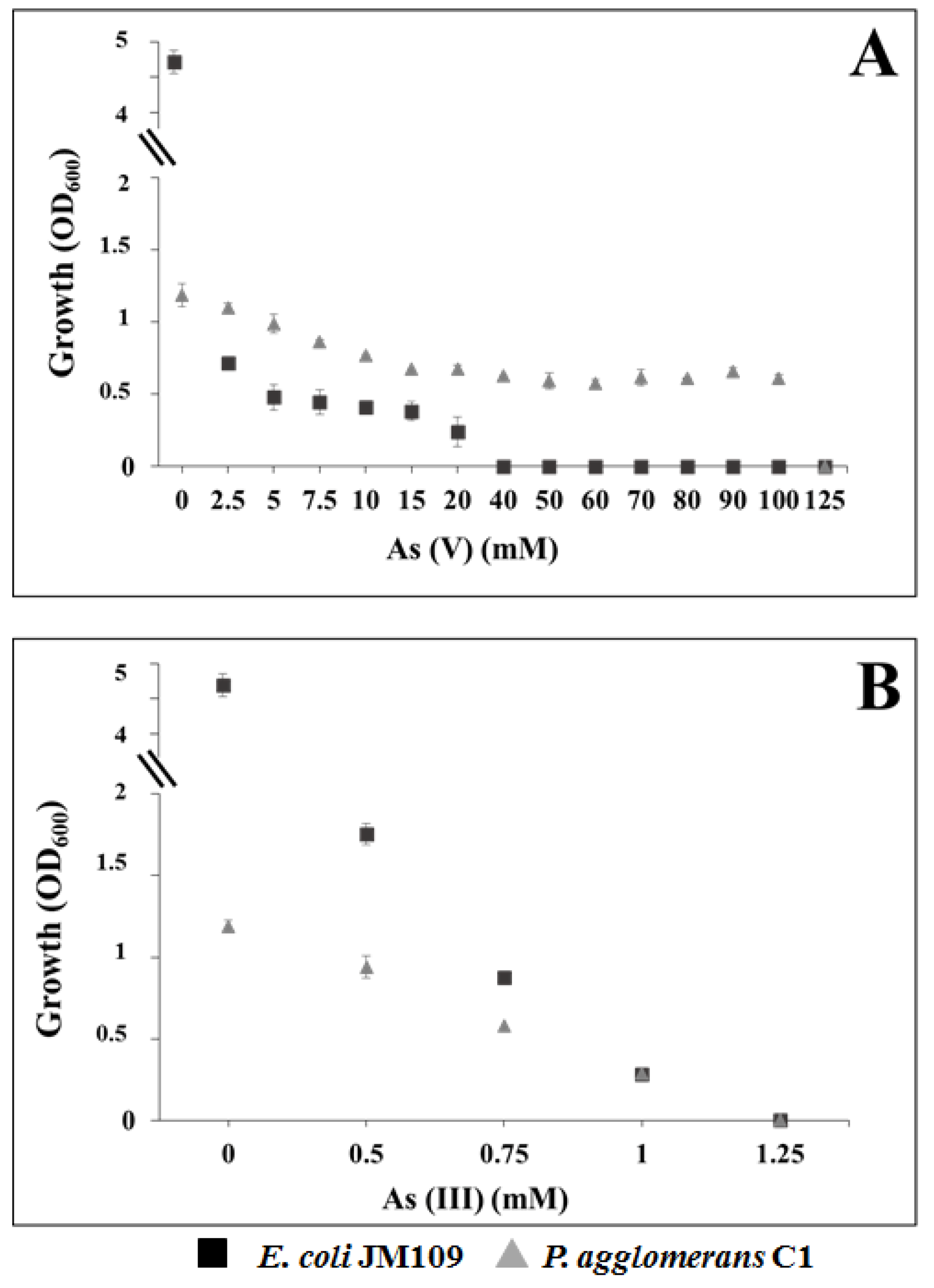
2.6. Determination of Minimal Inhibitory Concentration of Arsenic
2.7. Plant Inoculation
2.8. Statistical Analysis
2.9. Nucleotide Sequence Accession Number
3. Results and Discussion
3.1. Genome Sequencing and Comparison with Pantoea Genomes
3.2. Plant Beneficial Properties of Pantoea agglomerans C1
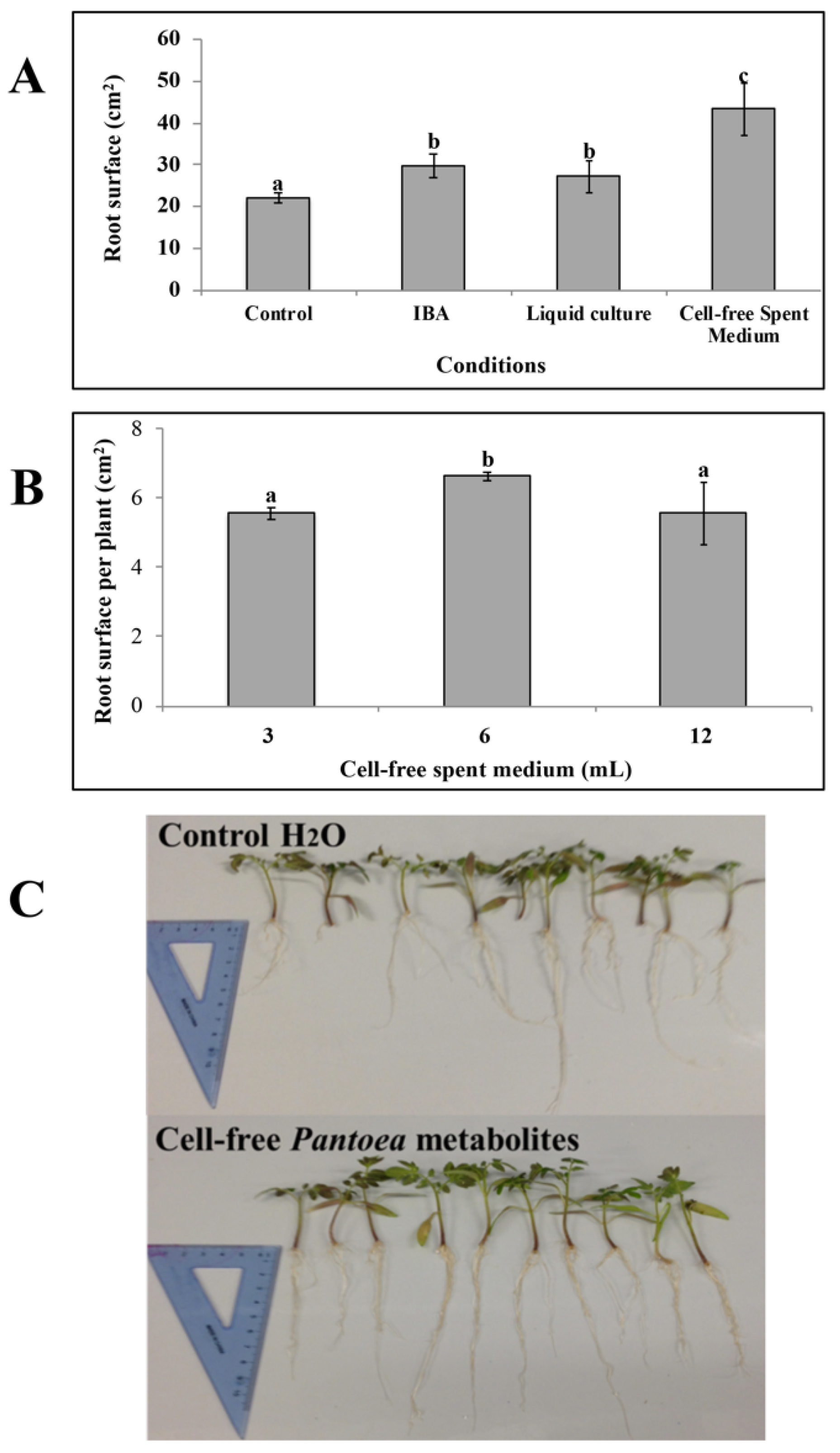
3.3. Effects of Pantoea Agglomerans C1 Cells and Metabolites on Root Growth
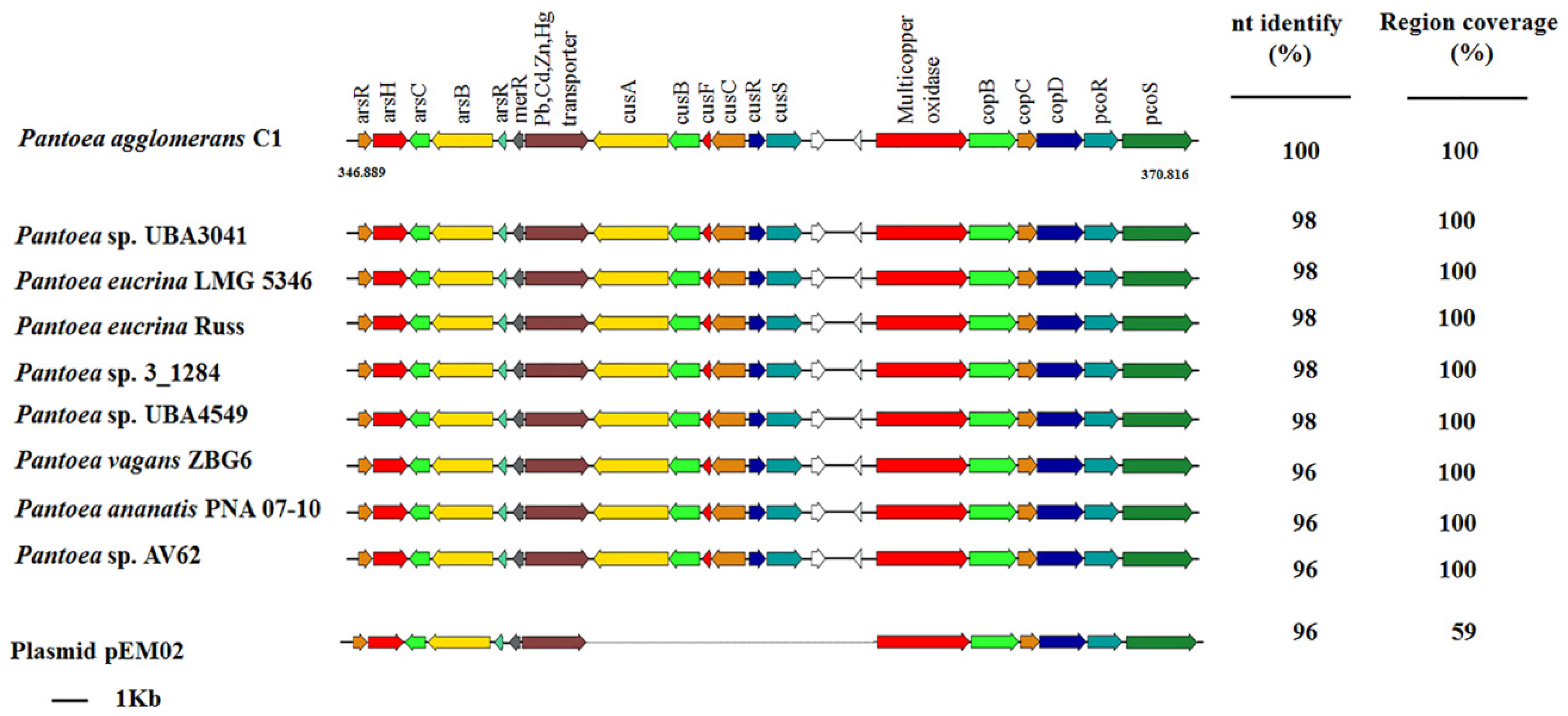
3.4. Tolerance to Heavy Metals in Pantoea Agglomerans C1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chibuike, G.U.; Obiora, S.C. Heavy metal polluted soils: Effect on plants and bioremediation methods. Appl. Environ. Soil Sci. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Glick, B.R. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 2010, 28, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Tak, H.I.; Ahmad, F.; Babalola, O.O. Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. Rev. Environ. Contam. Toxicol. 2013, 223, 33–52. [Google Scholar]
- Ruzzi, M.; Aroca, R. Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 2015, 196, 124–134. [Google Scholar] [CrossRef]
- Rouphael, Y.; Colla, G. Synergistic biostimulatory action: Designing the next generation of plant biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1655. [Google Scholar] [CrossRef] [Green Version]
- Tirry, N.; Tahri Joutey, N.; Sayel, H.; Kouchou, A.; Bahafid, W.; Asri, M.; El Ghachtouli, N. Screening of plant growth promoting traits in heavy metals resistant bacteria: Prospects in phytoremediation. J. Genet. Eng. Biotechnol. 2018, 16, 613–619. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Arsenic and arsenic compounds. In Arsenic, Metals, Fibres, and Dusts; IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 100C; IARC: Lyon, France, 2012; pp. 41–93. [Google Scholar]
- ATSDR. CERCLA Priority List of Hazardous Substances for 2017; Agency for Toxic Substances and Disease Registry, Department of Health and Human Services: Atlanta, GA, USA, 2017. Available online: https://www.atsdr.cdc.gov/SPL/#2017spl (accessed on 16 February 2019).
- Shankar, S.; Shanker, U. Arsenic contamination of groundwater: A review of sources, prevalence, health risks, and strategies for mitigation. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef]
- Hughes, M.F.; Beck, B.D.; Chen, Y.; Lewis, A.S.; Thomas, D.J. Arsenic exposure and toxicology: A historical perspective. Toxicol. Sci. 2011, 123, 305–332. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, R.; Rosen, B.P. Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Perspect. 2002, 110 (Suppl. 5), 745–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Páez-Espino, D.; Tamames, J.; De Lorenzo, V.; Cánovas, D. Microbial responses to environmental arsenic. Biometals 2009, 22, 117–130. [Google Scholar]
- Lakshmanan, V.; Cottone, J.; Bais, H.P. Killing two birds with one stone: Natural rice rhizospheric microbes reduce arsenic uptake and blast infections in rice. Front. Plant Sci. 2016, 7, 1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Wang, J.; Jing, C. Comparative genomic analysis reveals organization, function and evolution of ars genes in Pantoea spp. Front. Microbiol. 2017, 8, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Qiu, S. Examining phylogenetic relationships of Erwinia and Pantoea species using whole genome sequence data. Antonie Leeuwenhoek 2015, 108, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 2016, 23, 206–222. [Google Scholar] [CrossRef]
- Luziatelli, F.; Ficca, A.G.; Colla, G.; Švecová, E.; Ruzzi, M. Effects of a protein hydrolysate-based biostimulant and two micronutrient-based fertilizers on plant growth and epiphytic bacterial population of lettuce. Acta Hortic. 2016, 1148, 43–48. [Google Scholar] [CrossRef]
- Luziatelli, F.; Ficca, A.G.; Colla, G.; Baldassarre Švecová, E.; Ruzzi, M. Foliar application of vegetal-derived bioactive compounds stimulates the growth of beneficial bacteria and enhances microbiome biodiversity in lettuce. Front. Plant Sci. 2019, 10, 60. [Google Scholar] [CrossRef] [Green Version]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Luziatelli, F.; Ficca, A.G.; Melini, F.; Ruzzi, M. Genome sequence of the Plant Growth-Promoting Rhizobacterium (PGPR) Pantoea agglomerans C1. Microbiol. Resour. Announc. 2019, 8, e00828-19. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.Y.; Cohoon, M.; De Crecy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R.; et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Zhu, Z.; Fu, L.; Niu, B.; Li, W. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genom. 2011, 12, 444. [Google Scholar] [CrossRef] [Green Version]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef] [Green Version]
- Schwyn, R.; Neilands, J.B. Universal chemical assay for detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Hrynkiewicz, K.; Baum, C.; Leinweber, P. Density, metabolic activity, and identity of cultivable rhizosphere bacteria on Salix viminalis in disturbed arable and landfill soils. J. Plant Nutr. Soil Sci. 2010, 173, 747–756. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Colla, G.; Rouphael, Y.; Canaguier, R.; Svecova, E.; Cardarelli, M. Biostimulant action of a plant-derived protein hydrolysate produced through enzymatic hydrolysis. Front. Plant Sci. 2014, 5, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehman, S.M. Development of a Bacteriophage-Based Biopesticide for Fire Blight. Ph.D. Thesis, Brock University, St. Catharines, ON, Canada, 2007. [Google Scholar]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinf. 2003, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Jin, T.; Kumar Sahu, S.; Xu, J.; Shi, Q.; Liu, H.; Wang, Y. The distribution of tryptophan-dependent indole-3-acetic acid synthesis pathways in bacteria unraveled by large-scale genomic analysis. Molecules 2019, 24, 1411. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.S.; Wu, H.J.; Zang, H.Y.; Wu, L.M.; Zhu, Q.Q.; Gao, X.W. Plant growth-promotion by spermidine-producing Bacillus subtilis OKB105. Mol. Plant Microbe Interact. 2014, 27, 655–663. [Google Scholar] [CrossRef] [Green Version]
- Santos-Beneit, F.; Rodríguez-García, A.; Franco-Domínguez, E.; Martín, J.F. Phosphate-dependent regulation of the low- and high-affinity transport systems in the model actinomycete Streptomyces coelicolor. Microbiology 2008, 154, 2356–2370. [Google Scholar] [CrossRef] [Green Version]
- Sashidhar, B.; Podile, A.R. Mineral phosphate solubilization by rhizosphere bacteria and scope for manipulation of the direct oxidation pathway involving glucose dehydrogenase. J. Appl. Microbiol. 2010, 109, 1–12. [Google Scholar] [CrossRef]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Wei, H.X.; Pare, P.W.; Kloepper, J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 8, 4927–4932. [Google Scholar] [CrossRef] [Green Version]
- Lua, X.; Jia, G.; Zonga, H.; Zhugea, B. The role of budABC on 1,3-propanediol and 2,3-butanediol production from glycerol in Klebsiella pneumoniae CICIM B0057. Bioengineered 2016, 7, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis, E.W.I.I.; Lim, C.K.; Shaffer, B.T.; Paulsen, I.T. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef] [PubMed]
- Sayyed, R.Z.; Chincholkar, S.B.; Reddy, M.S.; Gangurde, N.S.; Patel, P.R. Siderophore producing PGPR for crop nutrition and phytopathogen suppression. In Bacteria in Agrobiology: Disease Management; Maheshwari, D., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 449–471. [Google Scholar]
- Cao, J.; Woodhall, M.R.; Alvarez, J.; Cartron, M.L.; Andrews, S.C. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 2007, 65, 857–875. [Google Scholar] [CrossRef] [PubMed]
- Rezzonico, F.; Smits, T.H.; Montesinos, E.; Frey, J.E.; Duffy, B. Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 2009, 9, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.S.; Yoon, I.H.; Kim, K.W. Arsenic biotransformation potential of microbial arsH responses in the biogeochemical cycling of arsenic-contaminated groundwater. Chemosphere 2018, 191, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Ben Fekih, I.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q. Distribution of Arsenic Resistance Genes in Prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef]
- Garbinski, L.D.; Rosen, B.P.; Chen, J. Pathways of arsenic uptake and efflux. Environ. Int. 2019, 126, 585–597. [Google Scholar] [CrossRef]
- Outten, F.W.; Huffman, D.L.; Hale, J.A.; O’Halloran, T.V. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 2001, 276, 30670–30677. [Google Scholar] [CrossRef] [Green Version]
- Silver, S.; Phung, L.T. Bacterial heavy metal resistance: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef]
- Brown, N.L.; Barrett, S.R.; Camakaris, J.; Lee, B.T.; Rouch, D.A. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 1995, 17, 1153–1166. [Google Scholar] [CrossRef]
- Mergeay, M.; Monchy, S.; Vallaeys, T.; Auquier, V.; Benotmane, A.; Bertin, P.; Taghavi, S.; Dunn, J.; Van der Lelie, D.; Wattiez, R. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 2003, 27, 385–410. [Google Scholar] [CrossRef]
- Wu, Q.; Du, J.; Zhuang, G.; Jing, C. Bacillus sp. SXB and Pantoea sp. IMH, aerobic As(V)-reducing bacteria isolated from arsenic-contaminated soil. J. Appl. Microbiol. 2013, 114, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.; Cook, G. Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr. Microbiol. 2004, 48, 341–347. [Google Scholar] [CrossRef] [PubMed]

| Species | Pantoea agglomerans | Source |
|---|---|---|
| Strain | C1 | |
| Assembly level | Contig | |
| No. of sequences | 22 | [21] |
| Genome size (bp) | 4,846,925 | [21] |
| GC content (%) | 55.2 | [21] |
| Gene | 4601 | This work |
| CDS | 4497 | This work |
| RNA | 104 | This work |
| rRNA (5S,16S,23S) | 9, 6, 9 | This work |
| Completed (5S,16S,23S) | 9, 1, 1 | This work |
| Truncated (16S,23S) | 5, 8 | This work |
| tRNA ncRNA | 70 10 | [21] This work |
| Prophage | 2 | This work |
| Genomic island (integrated method) | 11 > 20,000 bp | This work |
| CODE | STRAIN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Strain C1 | * | 98.7 | 98.7 | 98.7 | 98.6 | 97.8 | 97.3 | 84.0 | 84.1 | 84.2 | 84.3 | 84.1 | 91.3 | 91.3 | 98.7 |
| 2 | Pantoea agglomerans RIT273 | 98.7 | * | 98.7 | 98.7 | 98.7 | 97.9 | 97.3 | 84.0 | 84.0 | 84.0 | 84.1 | 84.1 | 91.3 | 91.3 | 98.8 |
| 3 | Pantoea agglomerans DSM3463T | 98.7 | 98.7 | * | 98.7 | 98.7 | 97.9 | 97.3 | 84.1 | 84.0 | 84.0 | 83.9 | 84.1 | 91.3 | 91.4 | 98.7 |
| 4 | Pantoea agglomerans JM1 | 98.7 | 98.7 | 98.7 | * | 98.7 | 97.8 | 97.3 | 84.0 | 84.0 | 84.0 | 83.9 | 84.1 | 91.3 | 91.3 | 98.8 |
| 5 | Pantoea agglomerans IG1 | 98.6 | 98.7 | 98.7 | 98.7 | * | 97.8 | 97.2 | 83.9 | 83.9 | 83.9 | 83.9 | 84.0 | 91.3 | 91.3 | 98.7 |
| 6 | Pantoea agglomerans C410P1 | 97.8 | 97.9 | 97.9 | 97.8 | 97.8 | * | 97.5 | 84.0 | 84.1 | 83.9 | 83.9 | 84.1 | 91.8 | 91.8 | 97.9 |
| 7 | Pantoea agglomerans P5 | 97.3 | 97.3 | 97.3 | 97.3 | 97.2 | 97.5 | * | 83.9 | 84.0 | 84.0 | 83.9 | 84.0 | 91.2 | 91.3 | 97.3 |
| 8 | Pantoea ananatis LMG2665T | 84.0 | 83.9 | 84.1 | 84.0 | 83.9 | 84.0 | 83.9 | * | 99.3 | 99.2 | 83.9 | 86.0 | 84.1 | 84.2 | 84.0 |
| 9 | Pantoea ananatis LMG20103 | 84.0 | 84.0 | 84.0 | 84.0 | 83.9 | 84.1 | 84.0 | 99.3 | * | 99.2 | 83.8 | 85.9 | 84.2 | 84.2 | 83.9 |
| 10 | Pantoea ananatis PNA 07-10 | 84.2 | 84.0 | 84.0 | 84.0 | 83.9 | 83.9 | 84.0 | 99.2 | 99.2 | * | 84.2 | 85.9 | 84.2 | 84.0 | 84.2 |
| 11 | Pantoea eucrina LMG5346T | 84.3 | 84.1 | 84.0 | 83.9 | 83.9 | 83.9 | 83.9 | 83.9 | 83.8 | 84.2 | * | 84.0 | 83.8 | 83.8 | 84.2 |
| 12 | Pantoea stewartii sub. stewartii DC283T | 84.1 | 84.1 | 84.1 | 84.1 | 84.0 | 84.1 | 84.0 | 86.0 | 85.9 | 85.9 | 84.0 | * | 84.2 | 84.1 | 84.0 |
| 13 | Pantoea vagans C9-1 | 91.3 | 91.3 | 91.3 | 91.3 | 91.3 | 91.8 | 91.2 | 84.1 | 84.2 | 84.2 | 83.8 | 84.2 | * | 96.9 | 91.3 |
| 14 | Pantoea vagans MP7 | 91.3 | 91.3 | 91.4 | 91.3 | 91.3 | 91.8 | 91.3 | 84.1 | 84.2 | 84.0 | 83.8 | 84.1 | 96.9 | * | 91.3 |
| 15 | Pantoea vagans ZBG6 | 98.7 | 98.8 | 98.7 | 98.8 | 98.7 | 97.9 | 97.3 | 84.0 | 83.9 | 84.3 | 84.2 | 84.0 | 91.3 | 91.3 | * |
| Function | Code | Value | %age | Description |
|---|---|---|---|---|
| CELLULAR PROCESSES AND SIGNALING | D | 62 | 1.32 | Cell cycle control, cell division, chromosome partitioning |
| M | 255 | 5.43 | Cell wall/membrane/envelope biogenesis | |
| N | 95 | 2.02 | Cell motility | |
| O | 107 | 2.28 | Post-translational modification, protein turnover, and chaperones | |
| T | 106 | 2.26 | Signal transduction mechanisms | |
| U | 54 | 1.15 | Intracellular trafficking, secretion, and vesicular transport | |
| V | 47 | 1.00 | Defense mechanisms | |
| INFORMATION STORAGE AND PROCESSING | A | 0 | 0.00 | RNA processing and modification |
| B | 0 | 0.00 | Chromatin structure and dynamics | |
| J | 193 | 4.11 | Translation, ribosomal structure and biogenesis | |
| K | 409 | 8.71 | Transcription | |
| L | 158 | 3.36 | Replication, recombination and repair | |
| METABOLISM | C | 234 | 4.98 | Energy production and conversion |
| E | 391 | 8.33 | Amino acid transport and metabolism | |
| F | 106 | 2.26 | Nucleotide transport and metabolism | |
| G | 276 | 5.88 | Carbohydrate transport and metabolism | |
| H | 176 | 3.75 | Coenzyme transport and metabolism | |
| I | 113 | 2.41 | Lipid transport and metabolism | |
| P | 287 | 6.11 | Inorganic ion transport and metabolism | |
| Q | 39 | 0.83 | Secondary metabolites biosynthesis, transport, and catabolism | |
| POORLY CHARACTERIZED | R | 0 | 0.00 | General function prediction only |
| S | 937 | 19.95 | Function unknown | |
| - | 651 | 13.86 | Not in COGs |
| Direct Plant Growth-Promotion | |||
| Gene | EC No. | Annotation | Gene Location, Coding Strand (+/−) |
| IAA production | |||
| ipdC | 4.1.1.74 | Indole-3-pyruvate decarboxylase | Contig1: 2029913-2028261, − |
| amiE | 3.5.1.4 | Aliphatic amidase | Contig1: 254208-254999, + |
| aec | Auxin efflux carrier family protein | Contig1: 1779607-1780566, + | |
| Spermidine biosynthesis | |||
| speA | 3.5.3.11 | Agmatinase | Contig4: 165937-1659017, − |
| speB | 4.1.1.19 | Biosynthetic arginine decarboxylase | Contig4: 168083-1686107, − |
| speD | 4.1.1.50 | S-adenosylmethionine decarboxylase proenzyme | Contig3: 73489-74298, + |
| speE | 2.5.1.16 | prokaryotic class 1A Spermidine synthase | Contig3: 73489-74298, + |
| Phosphate solubilization | |||
| gad | 1.1.99.3 | Gluconate 2-dehydrogenase, membrane-bound, cytochrome c | Contig3: 303092-301830, − |
| gad | 1.1.99.3 | Gluconate 2-dehydrogenase, membrane-bound, flavoprotein | Contig3: 304866-303097, − |
| gad | 1.1.99.3 | Gluconate 2-dehydrogenase, membrane-bound, gamma subunit | Contig3: 305631-304903, − |
| gad | 1.1.99.3 | Gluconate 2-dehydrogenase, membrane-bound, cytochrome c | Contig3: 495485-494175, − |
| gad | 1.1.99.3 | Gluconate 2-dehydrogenase, membrane-bound, flavoprotein | Contig3: 497280-495496, − |
| gad | 1.1.99.3 | Gluconate 2-dehydrogenase, membrane-bound, gamma subunit | Contig3: 498017-497283, − |
| gcd | 1.1.5.2 | Glucose dehydrogenase pyrroloquinoline quinone (PQQ)-dependent | Contig3: 476263-478653, + |
| pqq | Coenzyme PQQ synthesis protein B,C,D,E,F | Contig1: 1076330-1081693, + | |
| phoU | Phosphate transport system regulatory protein | Contig6: 207107-206373, − | |
| pstB | Phosphate transport ATP-binding protein | Contig6: 207898-207125, − | |
| pstA | Phosphate transport system permease protein | Contig6: 208833-207943, − | |
| pstC | Phosphate transport system permease protein | Contig6: 209792-208830, − | |
| pstS | Phosphate ABC transporter, periplasmic phosphate-binding protein | Contig6: 210923-209880, − | |
| Indirect Plant Growth-Promotion | |||
| Gene | EC No. | Annotation | Gene Location, Coding Strand (+/−) |
| Volatile organic compounds (VOCs) | |||
| alsR | Transcriptional regulator of alpha-acetolactate operon | Contig7: 135886-136791, + | |
| alsD | 4.1.1.5 | Alpha-acetolactate decarboxylase | Contig7: 135781-134999, − |
| alsS | 2.2.1.6 | Acetolactate synthase | Contig7: 134984-133305, − |
| bdh | 1.1.1.41.1.1.304 | 2,3-butanediol dehydrogenase, S-alcohol forming, (R)-acetoin-specific/Acetoin (diacetyl) reductase | Contig7: 133283-132510, − |
| GABA production | |||
| gabD | 1.2.1.16 | Succinate-semialdehyde dehydrogenase [NAD(P)+] | Contig4: 449240-447789, − |
| gabT | 2.6.1.19 | Gamma-aminobutyrate:alpha-ketoglutarate aminotransferase | Contig2: 419393-420679, + |
| Siderophores biogenesis | |||
| fes | Enterobactin esterase | Contig3: 384712-385917, + | |
| entA | 1.3.1.28 | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | Contig3: 399161-399919, + |
| entB | 3.3.2.1 | Isochorismatase | Contig3: 398310-399164, + |
| entC | 5.4.4.2 | Isochorismate synthase | Contig3: 395486-396664, + |
| entE | 2.7.7.58 | 2,3-dihydroxybenzoate-AMP ligase | Contig3: 396675-398291, + |
| entF | 6.3.2.14 | Enterobactin synthetase component F | Contig3: 386228-390157, + |
| fepA | TonB-dependent receptor; Outer membrane receptor for ferric enterobactin and colicins B, D | Contig3: 384461-382194, − | |
| fepB | Ferric enterobactin-binding periplasmic protein | Contig3: 395308-394340, − | |
| fepC | Ferric enterobactin transport ATP-binding protein | Contig3: 390992-390201, − | |
| fepD | Ferric enterobactin transport system permease protein | Contig3: 392922-391966, − | |
| fepG | Ferric enterobactin transport system permease protein | Contig3: 391969-390989, − | |
| entS | Enterobactin exporter | Contig3: 393083-394345, − | |
| ybdZ | Putative cytoplasmic protein YbdZ in enterobactin biosynthesis operon | Contig3: 386017-386235, + | |
| fhuA | Ferric hydroxamate outer membrane receptor | Contig3: 51852-49651, − | |
| fhuC | Ferric hydroxamate ABC transporter, ATP-binding protein | Contig3: 49611-48817, − | |
| fhuD | Ferric hydroxamate ABC transporter, periplasmic substrate binding protein | Contig3: 48806-47928, − | |
| fhuB | Ferric hydroxamate ABC transporter, permease component | Contig3: 47928-45949, − | |
| Ferrous iron transporter (EfeUOB) | |||
| efeU | Ferrous iron transport permease | Contig1: 1504038-1503214, − | |
| efeO | Ferrous iron transport periplasmic protein contains peptidase-M75 domain and (frequently) cupredoxin-like domain | Contig1: 1504038-1503214, − | |
| efeB | Ferrous iron transport peroxidase | Contig1: 1503155-1502046, − | |
| Gene | EC No. | Annotation | Gene Location, Coding Strand (+/−) |
|---|---|---|---|
| Arsenic tolerance | |||
| arsRH | |||
| arsR | Arsenical resistance operon repressor | Contig2: 346889-347179, + | |
| arsH | Arsenic resistance protein ArsH | Contig2: 347176-347898, + | |
| arsRBC | |||
| arsR | Arsenical resistance operon repressor | Contig2: 350170-349817, − | |
| arsB | Arsenic efflux pump protein | Contig2: 349720-348437, − | |
| arsC | 1.20.4.1 | Arsenate reductase glutaredoxin-coupled, Glutaredoxin-like family | Contig2: 348387-347959, − |
| arsR-acr3 | |||
| arsR | Arsenical resistance operon repressor | Contig9: 2594-2229, − | |
| acr3 | Arsenical-resistance protein ACR3 | Contig9: 2180-1200, − | |
| Copper tolerance | |||
| cueR-copA | |||
| cueR | Transcriptional regulator, MerR family | Contig1: 198563-198967, + | |
| copA | 3.6.3.3 7.2.2.12 7.2.2.9 | Lead, cadmium, zinc and mercury transporting ATPase Copper-translocating P-type ATPase | Contig1: 195952-198465, − |
| cueR-copA | |||
| cueR | Transcriptional regulator, MerR family | Contig2: 350890-350432, − | |
| copA | 3.6.3.3 7.2.2.12 7.2.2.9 | Lead, cadmium, zinc and mercury transporting ATPase Copper-translocating P-type ATPase | Contig2: 350970-353627, + |
| copABCD_pcoRS | |||
| copA | Multicopper oxidase | Contig2: 364520-366361, + | |
| copB | Copper resistance protein CopB | Contig2: 366396-367349, + | |
| copC | Copper resistance protein CopC | Contig2: 367381-367761, + | |
| copD | Copper resistance protein CopD | Contig2: 367766-368698, + | |
| pcoR | Transcriptional regulator PcoR | Contig2: 368730-369410, + | |
| pcoS | 2.7.13.3 | Sensory protein kinase PcoS | Contig2: 369407-370816, + |
| Cadmium tolerance | |||
| cusCFBA_cusSR | |||
| cusC | Cation efflux system protein CusC | Contig2: 360131-358746, − | |
| cusF | Cation efflux system protein CusF | Contig2: 358717-358364, − | |
| cusB | Cobalt/zinc/cadmium efflux RND transporter, Membrane fusion protein, CzcB family | Contig2: 358250-356958, - | |
| cusA | Cation efflux system protein | Contig2: 356947-357380, − | |
| cusR | Copper-sensing two-component system response regulator CusR | Contig2: 360326-361009, + | |
| cusS | Copper sensory histidine kinase CusS | Contig2: 360999-362453, + | |
| czcAC | |||
| czcA | Cobalt-zinc-cadmium resistance protein CzcA | Contig2: 504650-501588, − | |
| czcC | Probable Co/Zn/Cd efflux system membrane fusion protein | Contig2: 505738-504650, − | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luziatelli, F.; Ficca, A.G.; Cardarelli, M.; Melini, F.; Cavalieri, A.; Ruzzi, M. Genome Sequencing of Pantoea agglomerans C1 Provides Insights into Molecular and Genetic Mechanisms of Plant Growth-Promotion and Tolerance to Heavy Metals. Microorganisms 2020, 8, 153. https://doi.org/10.3390/microorganisms8020153
Luziatelli F, Ficca AG, Cardarelli M, Melini F, Cavalieri A, Ruzzi M. Genome Sequencing of Pantoea agglomerans C1 Provides Insights into Molecular and Genetic Mechanisms of Plant Growth-Promotion and Tolerance to Heavy Metals. Microorganisms. 2020; 8(2):153. https://doi.org/10.3390/microorganisms8020153
Chicago/Turabian StyleLuziatelli, Francesca, Anna Grazia Ficca, Mariateresa Cardarelli, Francesca Melini, Andrea Cavalieri, and Maurizio Ruzzi. 2020. "Genome Sequencing of Pantoea agglomerans C1 Provides Insights into Molecular and Genetic Mechanisms of Plant Growth-Promotion and Tolerance to Heavy Metals" Microorganisms 8, no. 2: 153. https://doi.org/10.3390/microorganisms8020153
APA StyleLuziatelli, F., Ficca, A. G., Cardarelli, M., Melini, F., Cavalieri, A., & Ruzzi, M. (2020). Genome Sequencing of Pantoea agglomerans C1 Provides Insights into Molecular and Genetic Mechanisms of Plant Growth-Promotion and Tolerance to Heavy Metals. Microorganisms, 8(2), 153. https://doi.org/10.3390/microorganisms8020153







