Impact of Phosphorus Fertilization on Tomato Growth and Arbuscular Mycorrhizal Fungal Communities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Soil Sampling and Measurement of Soil Biochemical Properties before Transplanting Tomatoes
2.3. Estimation of Tomato Growth Parameters
2.4. Root Sampling and Staining
2.5. Extraction of Genomic DNA and Polymerase Chain Reaction (PCR)
2.6. Amplicon Sequencing for AMF Communities in Roots
2.7. Statistical Analysis
3. Results
3.1. Influence of P Fertilizer Level on Soil Biochemical Properties
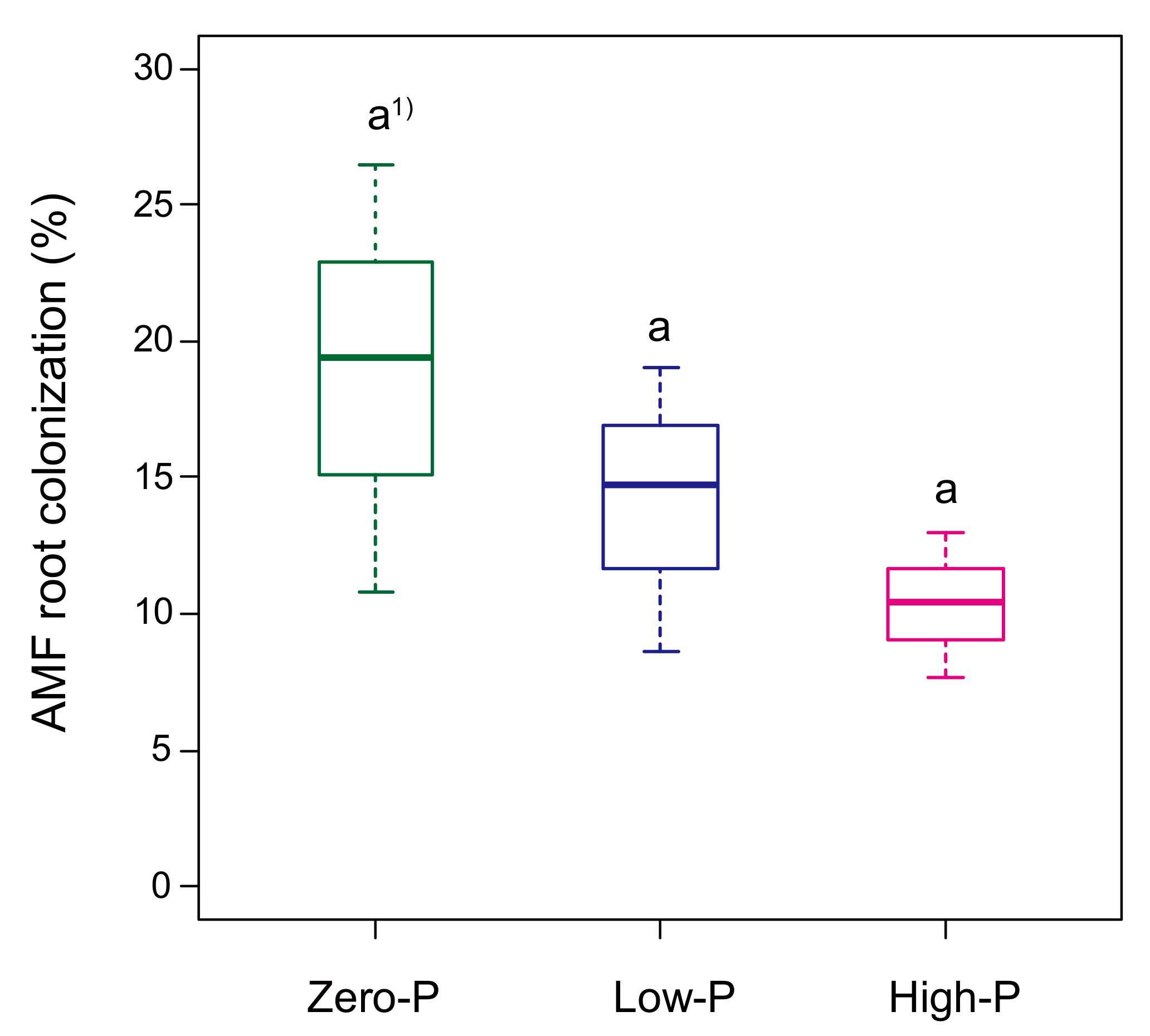
3.2. Influence of P Fertilizer Level on Plant Growth, P Uptake and AMF Colonization of Tomato Plants
3.3. General Sequencing Information and Molecular Diversity of AMF Communities
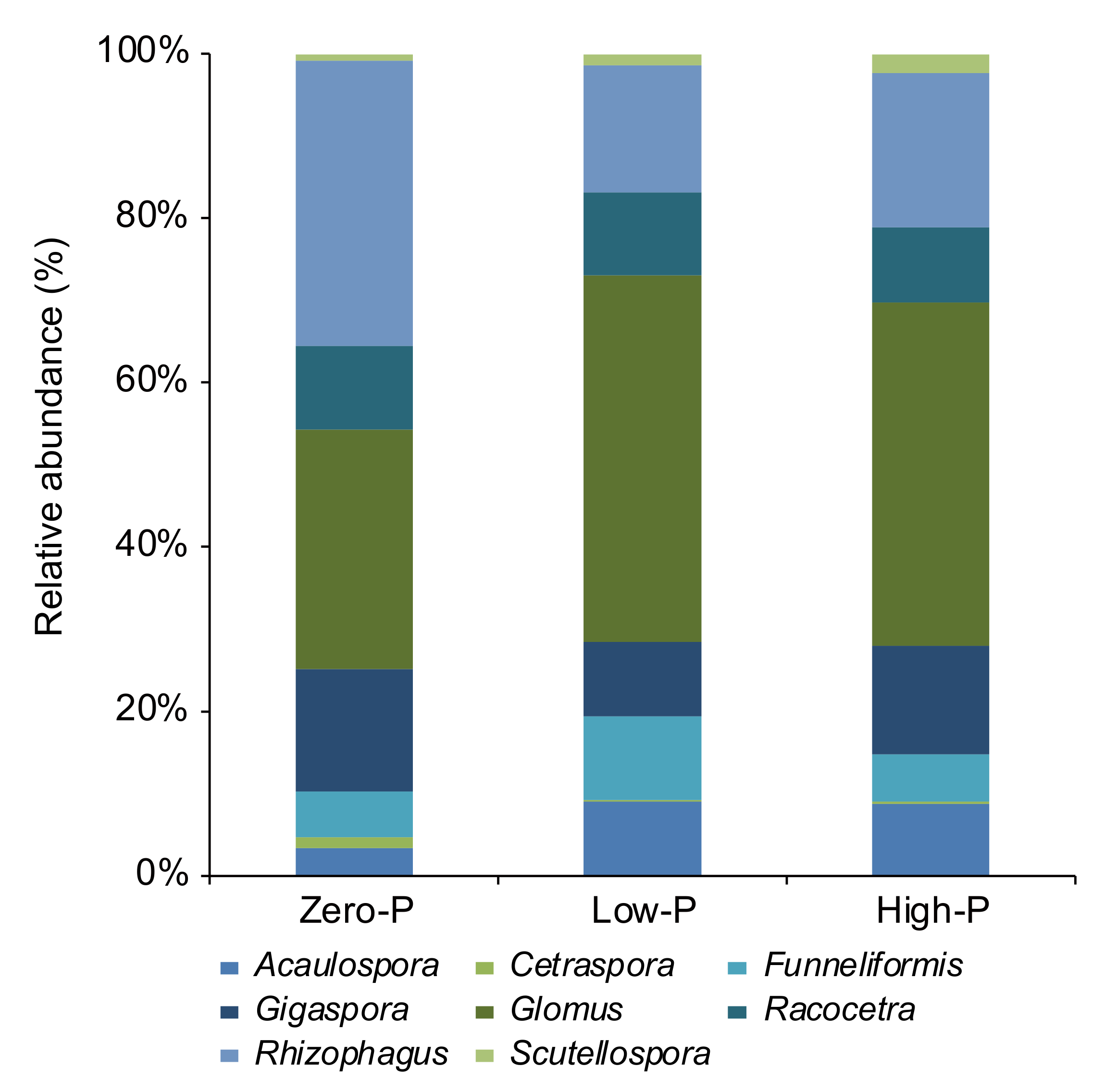
3.4. Influence of P Fertilizer Level on the AMF Communities in Tomato
4. Discussion
4.1. Influence of P Fertilizer Level on Tomato Growth
4.2. Effect of P Fertilizer Level on the AMF Communities in Tomato Roots
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lynch, J.P. Roots of the Second Green Revolution. Aust. J. Bot. 2007, 55, 493–512. [Google Scholar] [CrossRef]
- Mishima, S.; Endo, A.; Kohyama, K. Recent trends in phosphate balance nationally and by region in Japan. Nutr. Cycl. Agroecosyst. 2010, 86, 69–77. [Google Scholar] [CrossRef]
- Menezes-Blackburn, D.; Giles, C.; Darch, T.; George, T.S.; Blackwell, M.; Stutter, M.; Shand, C.; Lumsdond, C.P.; Wendler, R.; Brown, L.; et al. Opportunities for mobilizing recalcitrant phosphorus from agricultural soils: A review. Plant Soil 2018, 427, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Li, M.; Whelan, M. Phosphorus activators contribute to legacy phosphorus availability in agricultural soils: a review. Sci. Total Environ. 2018, 612, 522–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishima, S.; Itahashi, S.; Kimura, R.; Inoue, T. Trends of phosphate fertilizer demand and phosphate balance in farmland soils in Japan. Soil Sci. Plant Nutr. 2003, 49, 39–45. [Google Scholar] [CrossRef] [Green Version]
- Canene-Adams, K.; Campbell, J.K.; Zaripheh, S.; Jeffery, E.H.; Erdman, J.W., Jr. The tomato as a functional food. J. Nutr. 2005, 135, 1226–1230. [Google Scholar] [CrossRef]
- Avio, L.; Turrini, A.; Giovannetti, M.; Sbrana, C. Designing the ideotype mycorrhizal symbionts for the production of healthy food. Front. Plant Sci. 2018, 9, 1089. [Google Scholar]
- Ilahy, R.; Siddiqui, M.W.; Tlili, I.; Montefusco, A.; Piro, G.; Hdider, C.; Lenucci, M.S. When color really matters: horticultural performance and functional quality of high-lycopene tomatoes. CRC Crit. Rev. Plant Sci. 2018, 37, 15–53. [Google Scholar] [CrossRef]
- Giovannetti, M.; Avio, L.; Barale, R.; Ceccarelli, N.; Cristofani, R.; Iezzi, A.; Mignolli, F.; Picciarelli, P.; Pinto, B.; Reali, D.; et al. Nutraceutical value and safety of tomato fruits produced by AM plants. Br. J. Nutr. 2012, 107, 242–251. [Google Scholar] [CrossRef] [Green Version]
- Loiudice, R.; Impembo, M.; Laratta, B.; Villari, G.; Lo Voi, A.; Siviero, P.; Castaldo, D. Composition of San Marzano tomato varieties. Food Chem. 1995, 53, 81–89. [Google Scholar] [CrossRef]
- Langlois, D.; Etievant, P.X.; Pierron, P.; Jorrot, A. Sensory and instrumental characterization of commercial tomato varieties. Z. Lebensm. Unters. Forsch. 1996, 203, 534–540. [Google Scholar] [CrossRef]
- Klee, H.J. Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol. 2010, 187, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Goulet, C.; Kamiyoshihara, Y.; Lam, N.B.; Richard, T.; Taylor, M.G.; Tieman, D.M.; Klee, H.J. Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Mol. Plant 2015, 8, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; White, P.J.; Li, C. Colonization and community structure of arbuscular mycorrhizal fungi in maize roots at different depths in the soil profile respond differently to phosphorus inputs on a long-term experimental site. Mycorrhiza 2017, 27, 369–381. [Google Scholar] [CrossRef]
- Higo, M.; Sato, R.; Serizawa, A.; Takahashi, Y.; Gunji, K.; Tatewaki, Y.; Isobe, K. Can phosphorus application and cover cropping alter arbuscular mycorrhizal fungal communities and soybean performance after a five-year phosphorus-unfertilized crop rotational system? PeerJ 2018, 6, e4606. [Google Scholar] [CrossRef] [Green Version]
- Bona, E.; Cantamessa, S.; Massa, N.; Manassero, P.; Marsano, F.; Copetta, A.; Lingua, G.; D’Agostino, G.; Gamalero, E.; Berta, G. Arbuscular mycorrhizal fungi and plant growth-promoting pseudomonads improve yield, quality and nutritional value of tomato: a field study. Mycorrhiza 2017, 27, 1–11. [Google Scholar] [CrossRef]
- Zhu, Q.; Ozores-Hampton, M.; Li, Y.C.; Morgan, K.T.; Liu, G.; Mylavarapu, R.S. Effect of phosphorus rates on growth, yield, and postharvest quality of tomato in a calcareous soil. Hort. Sci. 2018, 52, 1406–1412. [Google Scholar] [CrossRef]
- Zhu, Q.; Ozores-Hampton, M.; Li, Y.C.; Morgan, K.T. Phosphorus application rates affected phosphorus partitioning and use efficiency in tomato production. Agron. J. 2018, 110, 2050–2058. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.E.; Read, D.J. Arbuscular mycorrhizaes. In Mycorrhizal Symbiosis, 3rd ed.; Smith, S.E., Read, D.J., Eds.; Academic Press: London, UK, 2008; pp. 13–187. [Google Scholar]
- Smith, S.E.; Jakobsen, I.; Grønlund, M.; Smith, F.A. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 2011, 156, 1050–1057. [Google Scholar] [CrossRef] [Green Version]
- Ferrol, N.; Azcón-Aguilar, C.; Pérez-Tienda, J. Arbuscular mycorrhizas as key players in sustainable plant phosphorus acquisition: An overview on the mechanisms involved. Plant Sci. 2019, 280, 441–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hart, M.M.; Forsythe, J.A. Using arbuscular mycorrhizal fungi to improve the nutrient quality of crops; nutritional benefits in addition to phosphorus. Sci. Hortic. 2012, 148, 206–214. [Google Scholar] [CrossRef]
- Sato, T.; Ezawa, T.; Cheng, W.; Tawaraya, K. Release of acid phosphatase from extraradical hyphae of arbuscular mycorrhizal fungus Rhizophagus clarus. Soil Sci. Plant Nutr. 2015, 61, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Aguilera, P.; Borie, F.; Seguel, A.; Cornejo, P. Fluorescence detection of aluminum in arbuscular mycorrhizal fungal structures and glomalin using confocal laser scanning microscopy. Soil Biol. Biochem. 2011, 43, 2427–2431. [Google Scholar] [CrossRef]
- Antunes, P.M.; Koch, A.M.; Morton, J.B.; Rillig, M.C.; Klironomos, J.N. Evidence for functional divergence in arbuscular mycorrhizal fungi from contrasting climatic origins. New Phytol. 2011, 189, 507–514. [Google Scholar] [CrossRef]
- Gosling, P.; Hodge, A.; Goodlass, G.; Bending, G.D. Arbuscular mycorrhizal fungi and organic farming. Agric. Ecosyst. Environ. 2006, 113, 17–35. [Google Scholar] [CrossRef]
- Jansa, J.; Smith, F.A.; Smith, S.E. Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol. 2008, 177, 779–789. [Google Scholar] [CrossRef]
- Verbruggen, E.; van der Heijden, M.G.A.; Rillig, M.C.; Kiers, E.T. Mycorrhizal fungal establishment in agricultural soils: factors determining inoculation success. New Phytol. 2013, 197, 1104–1109. [Google Scholar] [CrossRef] [Green Version]
- Olsson, P.A.; Baath, E.; Jakobsen, I. Phosphorus effects on the mycelium and storage structures of an arbuscular mycorrhizal fungus as studied in the soil and roots by analysis of fatty acid signatures. Appl. Environ. Microbiol. 1997, 63, 3531–3538. [Google Scholar] [CrossRef] [Green Version]
- Camenzind, T.; Hempel, S.; Homeier, J.; Horn, S.; Velescu, A.; Wilcke, W.; Rillig, M.C. Nitrogen and phosphorus additions impact arbuscular mycorrhizal abundance and molecular diversity in a tropical montane forest. Glob. Chang. Biol. 2014, 20, 3646–3659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.L.; Zhang, X.; Ye, J.S.; Han, H.Y.; Wan, S.Q.; Chen, B.D. Six-year fertilization modifies the biodiversity of arbuscular mycorrhizal fungi in a temperate steppe in Inner Mongolia. Soil Biol. Biochem. 2014, 69, 371–381. [Google Scholar] [CrossRef]
- Kahiluoto, H.; Ketoja, E.; Vestberg, M.; Saarela, I. Promotion of AM utilization through reduced P fertilization 2. Field studies. Plant Soil 2001, 231, 65–79. [Google Scholar]
- Corkidi, L.; Allen, E.B.; Merhaut, D.; Allen, M.F.; Downer, J.; Bohn, J.; Evans, M. Assessing the infectivity of commercial mycorrhizal inoculants in plant nursery conditions. J. Environ. Hortic. 2004, 22, 149–154. [Google Scholar]
- Faye, A.; Dalpé, Y.; Ndung’u-Magiroi, K.; Jefwa, J.; Ndoye, I.; Diouf, M.; Lesueur, D. Evaluation of commercial arbuscular mycorrhizal inoculants. Can. J. Plant Sci. 2013, 93, 1201–1208. [Google Scholar] [CrossRef]
- Bray, R.H.; Kurtz, L.T. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- Ishii, T.; Hayano, K. A method for the estimation of phosphodiesterse activity in soil. Jpn. J. Soil Sci. Plant Nutr. 1974, 45, 505–508. [Google Scholar]
- Hayano, K. A method for the determination of β-glucosidase activity in soil. Soil Sci. Plant Nutr. 1973, 19, 103–108. [Google Scholar] [CrossRef] [Green Version]
- Cavell, A.J. The colorimetric determination of phosphorus in plant materials. J. Sci. Food Agric. 1955, 6, 479–480. [Google Scholar] [CrossRef]
- Kobae, Y.; Ohtomo, R. An improved method for bright-field imaging of arbuscular mycorrhizal fungi in plant roots. Soil Sci. Plant Nutr. 2016, 62, 27–30. [Google Scholar] [CrossRef] [Green Version]
- Giovannetti, M.; Mosse, B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 1980, 84, 489–500. [Google Scholar] [CrossRef]
- Higo, M.; Tatewaki, Y.; Gunji, K.; Kaseda, A.; Isobe, K. Cover cropping can be a stronger determinant than host crop identity for arbuscular mycorrhizal fungal communities colonizing maize and soybean. PeerJ 2019, 7, e6403. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Lalonde, M.; Bruns, T.D. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl. Environ. Microbiol. 1992, 58, 291–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helgason, T.; Daniell, T.J.; Husband, R.; Fitter, A.H.; Young, J.P.W. Ploughing up the wood-wide web? Nature 1998, 394, 431. [Google Scholar] [CrossRef]
- Polz, M.F.; Cavanaugh, C.M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 1998, 64, 3724–3730. [Google Scholar] [CrossRef] [Green Version]
- Lumini, E.; Orgiazzi, A.; Borriello, R.; Bonfante, P.; Bianciotto, V. Disclosing arbuscular mycorrhizal fungal biodiversity in soil through a land-use gradient using a pyrosequencing approach. Environ. Microbiol. 2010, 12, 2165–2179. [Google Scholar] [CrossRef]
- Higo, M.; Kang, D.J.; Isobe, K. First report of community dynamics of arbuscular mycorrhizal fungi in radiocesium degradation lands after the Fukushima-Daiichi Nuclear disaster in Japan. Sci. Rep. 2019, 9, 8240. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.5-4. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 20 January 2020).
- Lindahl, B.D.; Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Carlsen, T.; Kjøller, R.; Kõljalg, U.; Pennanen, T.; Rosendahl, S.; Stenlid, J.; et al. Fungal community analysis by high-throughput sequencing of amplified markers—A user’s guide. New Phytol. 2013, 199, 288–299. [Google Scholar] [CrossRef] [Green Version]
- Oliver, A.K.; Brown, S.P.; Callaham, M.A., Jr.; Jumpponen, A. Polymerase matters: Non-proofreading enzymes inflate fungal community richness estimates by up to 15%. Fungal Ecol. 2015, 15, 86–89. [Google Scholar] [CrossRef] [Green Version]
- Lenth, R. Emmean: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.3.4. 2019. Available online: https://cran.r-project.org/package=emmeans (accessed on 20 January 2020).
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, J. eulerr: Area-Proportional Euler and Venn Diagrams with Ellipses. R package version 5.1.0. 2019. Available online: https://cran.r-project.org/package=eulerr (accessed on 20 January 2020).
- Chase, J.M.; Kraft, N.J.B.; Smith, K.G.; Vellend, M.; Inouye, B.D. Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2011, 2, 1–11. [Google Scholar] [CrossRef]
- Dormann, C.F.; Gruber, B.; Fründ, J. Introducing the bipartite package: analysing ecological networks. R News 2008, 8, 8–11. [Google Scholar]
- Dormann, C.F.; Fründ, J.; Blüthgen, N.; Gruber, B. Indices, graphs and null models: analyzing bipartite ecological networks. Open Ecol. J. 2009, 2, 7–24. [Google Scholar] [CrossRef]
- Dormann, C.F. How to be a specialist? Quantifying specialisation in pollination networks. Netw. Biol. 2011, 1, 1–20. [Google Scholar]
- Almeida-Neto, M.; Guimaraes, P.; Guimaraes, P.R., Jr.; Loyola, R.D.; Ulrich, W. A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos 2008, 117, 1227–1239. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Cavagnaro, T.R. Arbuscular mycorrhizas modify tomato responses to soil zinc and phosphorus addition. Biol. Fertil. Soils 2012, 48, 285–294. [Google Scholar] [CrossRef]
- Xu, P.; Liang, L.Z.; Xiao-Ying, D.O.N.G.; Jing, X.U.; Jiang, P.K.; Ren-Fang, S.H.E.N. Response of soil phosphorus required for maximum growth of Asparagus officinalis L. to inoculation of arbuscular mycorrhizal fungi. Pedosphere 2014, 24, 776–782. [Google Scholar] [CrossRef]
- Wilkinson, S.R. Nutrient interactions in soil and plant nutrition. In Handbook of Soil Science; Sumner, M.E., Ed.; CRC Press: Boca Raton, FL, USA, 2000; pp. 89–112. [Google Scholar]
- Taffouo, V.D.; Ngwene, B.; Akoa, A.; Franken, P. Influence of phosphorus application and arbuscular mycorrhizal inoculation on growth, foliar nitrogen mobilization, and phosphorus partitioning in cowpea plants. Mycorrhiza 2014, 24, 361–368. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Cavagnaro, T.R. Nutrient interactions and arbuscular mycorrhizas: A metaanalysis of a mycorrhiza-defective mutant and wildtype tomato genotype pair. Plant Soil 2014, 384, 79–92. [Google Scholar] [CrossRef]
- Kohler, J.; Caravaca, F.; Azcón, R.; Díaz, G.; Roldán, A. The combination of compost addition and arbuscular mycorrhizal inoculation produced positive and synergistic effects on the phytomanagement of a semiarid mine tailing. Sci. Total Environ. 2015, 514, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Munson, R.D.; Nelson, W.L. Principle and practices in plants analysis. In Soil Testing and Plant Analysis; Westerman, R.L., Ed.; SSSA Book. Ser. 3; SSSA: Madison, WI, USA, 1990; pp. 359–387. [Google Scholar]
- Traina, S.J.; Sposito, G.; Bradford, G.R.; Kafkafi, U. Kinetic Study of Citrate Effects on Orthophosphate Solubility in an Acidic, Montmorillonitic Soil 1. Soil Sci. Soc. Am. J. 1987, 51, 1483–1487. [Google Scholar] [CrossRef]
- Havlin, J.L.; Beaton, J.D.; Tisdale, S.L.; Nelson, W.L. An Introduction to Nutrient Management. In Soil Fertility and Fertilizers, 6th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 1999; p. 499. [Google Scholar]
- Maherali, H.; Klironomos, J.N. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 2007, 316, 1746–1748. [Google Scholar] [CrossRef] [Green Version]
- Xiang, X.; Gibbons, S.M.; He, J.S.; Wang, C.; He, D.; Li, Q.; Ni, Y.; Chu, H. Rapid response of arbuscular mycorrhizal fungal communities to short-term fertilization in an alpine grassland on the Qinghai-Tibet Plateau. PeerJ 2016, 4, e2226. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, C.; Zhang, Z.; Sun, Z.; Chen, Y.; Jiang, J.; Shen, Z. The influence of environmental factors on communities of arbuscular mycorrhizal fungi associated with Chenopodium ambrosioides revealed by MiSeq sequencing investigation. Sci. Rep. 2017, 7, 45134. [Google Scholar] [CrossRef]
- Zhao, H.; Li, X.; Zhang, Z.; Zhao, Y.; Yang, J.; Zhu, Y. Species diversity and drivers of arbuscular mycorrhizal fungal communities in a semi-arid mountain in China. PeerJ 2017, 5, e4155. [Google Scholar] [CrossRef] [Green Version]
- Oehl, F.; Sieverding, E.; Ineichen, K.; Mäder, P.; Boller, T.; Wiemken, A. Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of Central Europe. Appl. Environ. Microbiol. 2003, 69, 2816–2824. [Google Scholar] [CrossRef] [Green Version]
- Giovannetti, M.; Azzolini, D.; Citernesi, A.S. Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl. Environ. Microbiol. 1999, 65, 5571–5575. [Google Scholar] [CrossRef] [Green Version]
- Schalamuk, S.; Cabello, M. Arbuscular mycorrhizal fungal propagules from tillage and no-tillage systems: possible effects on Glomeromycota diversity. Mycologia 2010, 102, 261–268. [Google Scholar] [CrossRef]
- Treseder, K.K.; Allen, M.F. Direct nitrogen and phosphorus limitation of arbuscular mycorrhizal fungi: a model and field test. New Phytol. 2002, 155, 507–515. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Feng, Y.; Zhang, H.; Chen, R.; Wang, J.; Zhang, J.; Chu, H. Long-term balanced fertilization decreases arbuscular mycorrhizal fungal diversity in an arable soil in North China revealed by 454 pyrosequencing. Environ. Sci. Technol. 2012, 46, 5764–5771. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Y.; Jiang, S.; Deng, Y.; Christie, P.; Murray, P.J.; Li, X.; Zhang, J. Arbuscular mycorrhizal fungi in soil and roots respond differently to phosphorus inputs in an intensively managed calcareous agricultural soil. Sci. Rep. 2016, 6, 24902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, N.C. Resource stoichiometry elucidates the structure and function of arbuscular mycorrhizas across scales. New Phytol. 2010, 185, 631–647. [Google Scholar] [CrossRef] [PubMed]
- Börstler, B.; Raab, P.A.; Thiery, O.; Morton, J.B.; Redecker, D. Genetic diversity of the arbuscular mycorrhizal fungus Glomus intraradices as determined by mitochondrial large subunit rRNA gene sequences is considerably higher than previously expected. New Phytol. 2008, 180, 452–465. [Google Scholar] [CrossRef] [Green Version]
- Higo, M.; Isobe, K.; Matsuda, Y.; Ichida, M.; Torigoe, Y. Influence of sowing season and host crop identity on the community structure of arbuscular mycorrhizal fungi colonizing roots of two different gramineous and leguminous crop species. Adv. Microbiol. 2015, 5, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Gosling, P.; Mead, A.; Proctor, M.; Hammond, J.P.; Bending, G.D. Contrasting arbuscular mycorrhizal communities colonizing different host plants show a similar response to a soil phosphorus concentration gradient. New Phytol. 2013, 198, 546–556. [Google Scholar] [CrossRef]
- Higo, M.; Isobe, K.; Miyazawa, Y.; Matsuda, Y.; Drijber, R.A.; Torigoe, Y. Molecular diversity and distribution of indigenous arbuscular mycorrhizal communities colonizing roots of two different winter cover crops in response to their root proliferation. J. Microbiol. 2016, 54, 86–97. [Google Scholar] [CrossRef]
- Higo, M.; Isobe, K.; Yamaguchi, M.; Drijber, R.A.; Jeske, E.S.; Ishii, R. Diversity and vertical distribution of indigenous arbuscular mycorrhizal fungi under two soybean rotational systems. Biol. Fertil. Soils 2013, 49, 1085–1096. [Google Scholar] [CrossRef]
- Higo, M.; Isobe, K.; Drijber, R.A.; Kondo, T.; Yamaguchi, M.; Takeyama, S.; Suzuki, Y.; Niijima, D.; Matsuda, Y.; Ishii, R.; et al. Impact of a 5-year winter cover crop rotational system on the molecular diversity of arbuscular mycorrhizal fungi colonizing roots of subsequent soybean. Biol. Fertil. Soils 2014, 50, 913–926. [Google Scholar] [CrossRef]
- Higo, M.; Isobe, K.; Kondo, T.; Yamaguchi, M.; Takeyama, S.; Drijber, R.A.; Torigoe, Y. Temporal variation of the molecular diversity of arbuscular mycorrhizal communities in three different winter cover crop rotational systems. Biol. Fertil. Soils 2015, 51, 21–32. [Google Scholar] [CrossRef]
- Higo, M.; Takahashi, Y.; Gunji, K.; Isobe, K. How are arbuscular mycorrhizal associations related to maize growth performance during short-term cover crop rotation? J. Sci. Food Agric. 2018, 98, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Higo, M.; Tatewaki, Y.; Isobe, K. Amplicon sequencing analysis of arbuscular mycorrhizal fungal communities colonizing maize roots in different cover cropping and tillage systems. Sci. Rep. 2020. [Google Scholar] [CrossRef] [Green Version]




| P Fertilizer Levels | Soil pH | EC | Available Soil P Content | ||||||
| (H2O) | (μS/cm) | (mg/kg) | |||||||
| Zero-P | 5.7 | (0.01) 1 | a 2 | 53.3 | (3.33) | b | 17.0 | (3.9) | b |
| Low-P | 5.7 | (0.03) | a | 74.4 | (2.22) | a | 19.1 | (1.6) | b |
| High-P | 5.7 | (0.07) | a | 84.4 | (7.29) | a | 54.3 | (12.9) | a |
| P fertilizer levels | NO3-N Content | ACP Activity | ALP Activity | ||||||
| (mg/kg) | (mU/g) | (mU/g) | |||||||
| Zero-P | 28.7 | (0.2) | a | 20.1 | (0.3) | a | 53.9 | (2.8) | a |
| Low-P | 25.1 | (0.8) | b | 27.9 | (1.2) | a | 61.8 | (1.2) | a |
| High-P | 22.1 | (0.6) | c | 34.2 | (8.4) | a | 59.1 | (1.0) | a |
| P Fertilizer Levels | Plant Length | SPAD Value | Leaf Area | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (cm) | (cm2/plant) | ||||||||
| Zero-P | 19.5 | (3.5) 1 | b 2 | 48.9 | (1.4) | a | 83.5 | (15.6) | b |
| Low-P | 22.3 | (2.1) | b | 49.1 | (0.8) | a | 364.0 | (85.4) | a |
| High-P | 39.6 | (3.2) | a | 51.5 | (0.3) | a | 350.9 | (24.4) | a |
| P Fertilizer Levels | Shoot Biomass | P Concentration | P Uptake | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (g/m²) | (mg P/g) | (mg P/m²) | |||||||
| Zero-P | 2.9 | (1.2) 1 | b 2 | 1.6 | (0.09) | b | 4.6 | (1.9) | b |
| Low-P | 5.2 | (1.1) | b | 1.7 | (0.06) | b | 8.9 | (2.1) | b |
| High-P | 11.3 | (1.1) | a | 2.2 | (0.08) | a | 24.7 | (3.4) | a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Higo, M.; Azuma, M.; Kamiyoshihara, Y.; Kanda, A.; Tatewaki, Y.; Isobe, K. Impact of Phosphorus Fertilization on Tomato Growth and Arbuscular Mycorrhizal Fungal Communities. Microorganisms 2020, 8, 178. https://doi.org/10.3390/microorganisms8020178
Higo M, Azuma M, Kamiyoshihara Y, Kanda A, Tatewaki Y, Isobe K. Impact of Phosphorus Fertilization on Tomato Growth and Arbuscular Mycorrhizal Fungal Communities. Microorganisms. 2020; 8(2):178. https://doi.org/10.3390/microorganisms8020178
Chicago/Turabian StyleHigo, Masao, Mirai Azuma, Yusuke Kamiyoshihara, Akari Kanda, Yuya Tatewaki, and Katsunori Isobe. 2020. "Impact of Phosphorus Fertilization on Tomato Growth and Arbuscular Mycorrhizal Fungal Communities" Microorganisms 8, no. 2: 178. https://doi.org/10.3390/microorganisms8020178
APA StyleHigo, M., Azuma, M., Kamiyoshihara, Y., Kanda, A., Tatewaki, Y., & Isobe, K. (2020). Impact of Phosphorus Fertilization on Tomato Growth and Arbuscular Mycorrhizal Fungal Communities. Microorganisms, 8(2), 178. https://doi.org/10.3390/microorganisms8020178





