Haloglomus irregulare gen. nov., sp. nov., a New Halophilic Archaeon Isolated from a Marine Saltern
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Haloarchaeal Strain
2.2. DNA Extraction, Purification, and Sequencing
2.3. Genome Assembly and Annotation
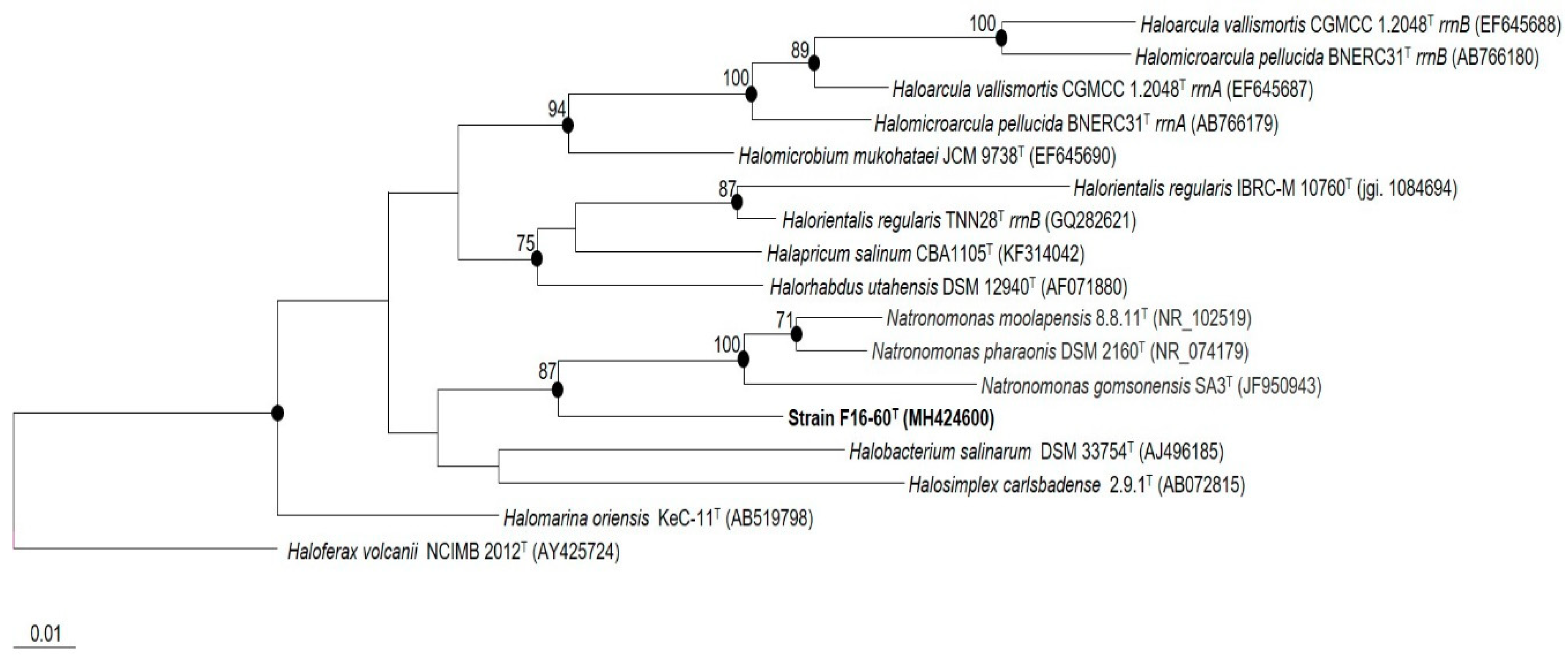
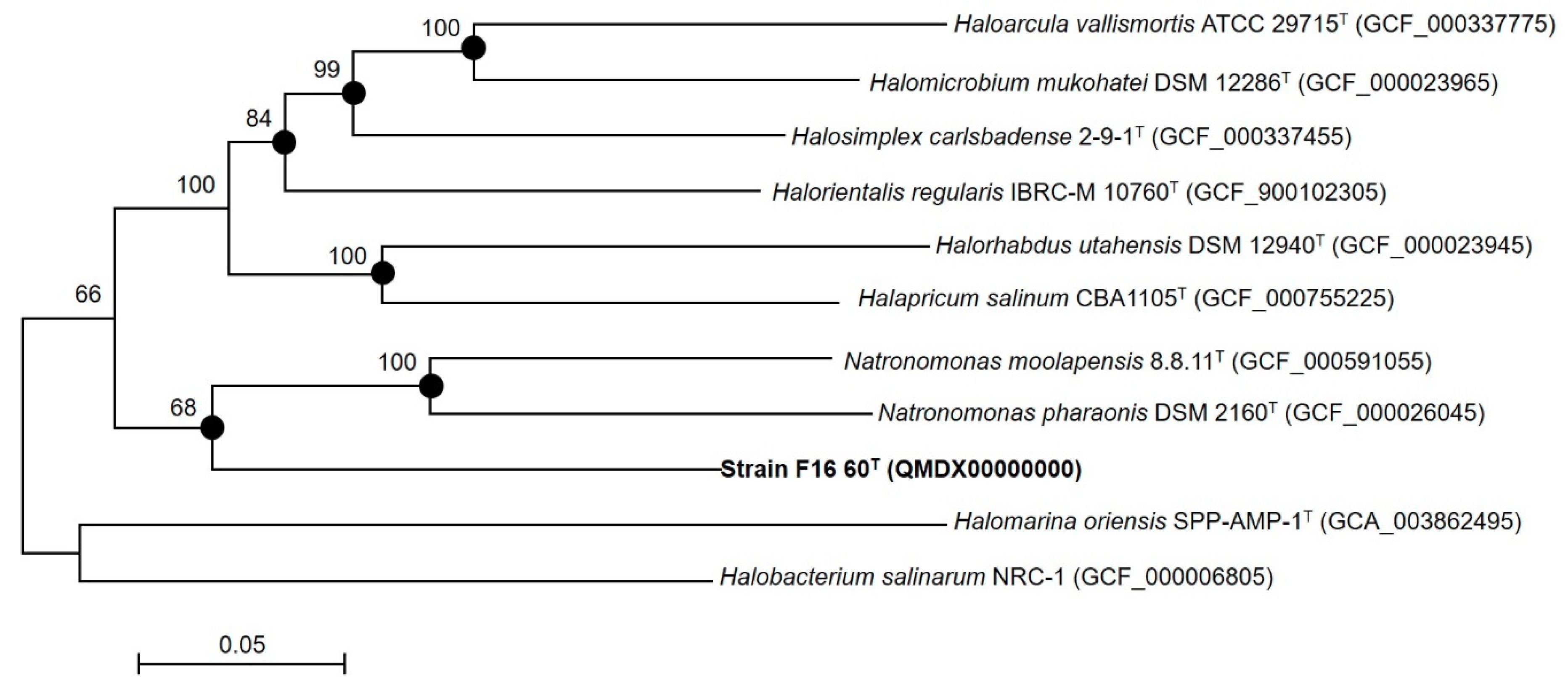
2.4. Phylogenetic Analyses
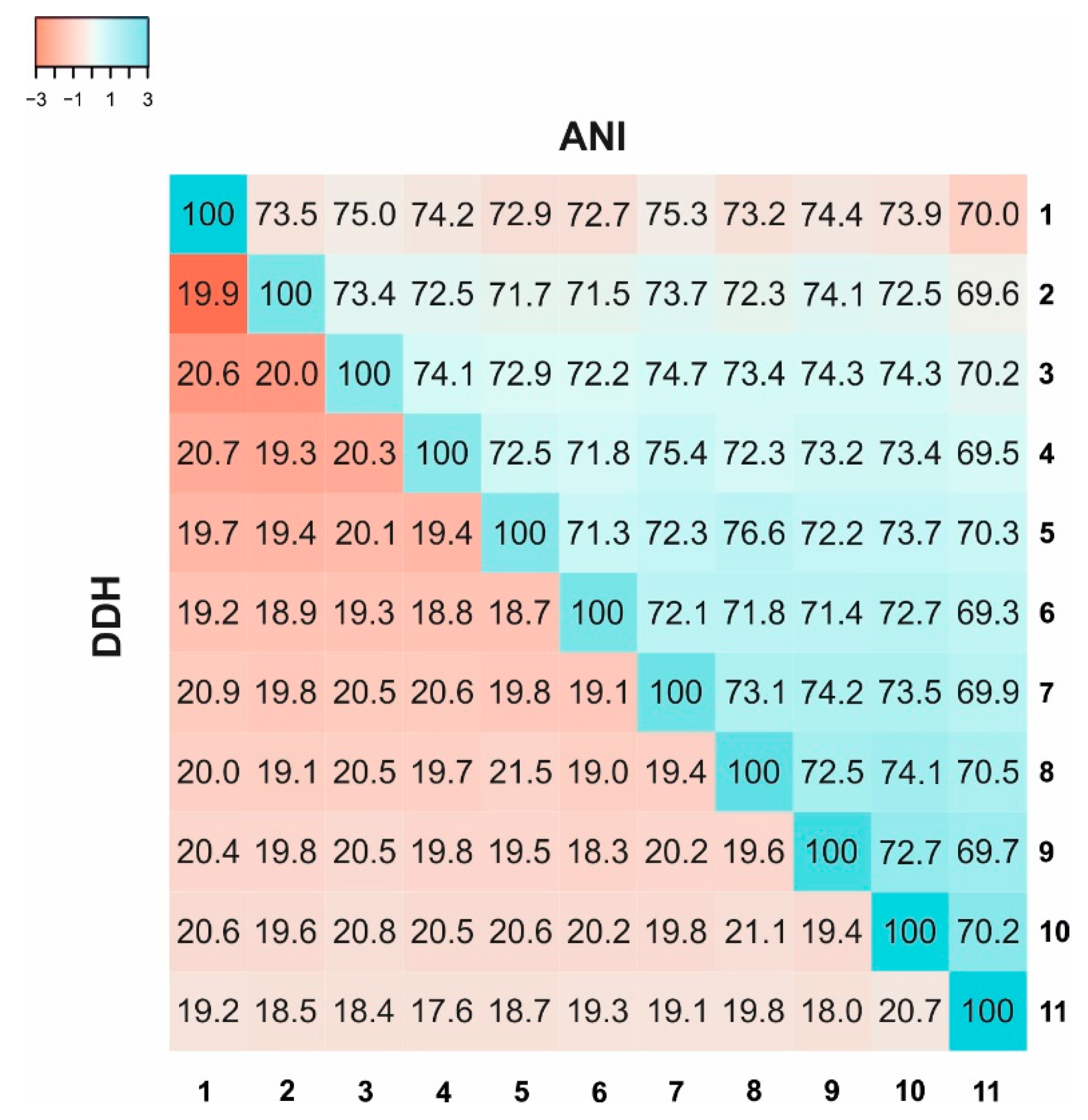
2.5. Average Nucleotide Identity (ANI), Average Amino Acid Identity (AAI), and Digital DNA–DNA Hybridization (DDH)
2.6. Chemotaxonomic Analysis
2.7. Phenotypic Characterization
2.8. Metagenomic Fragment Recruitment Analysis
3. Results and Discussion
3.1. Phylogenetic Analyses
3.2. Chemotaxonomic Characterization
3.3. Phenotypic Characterization
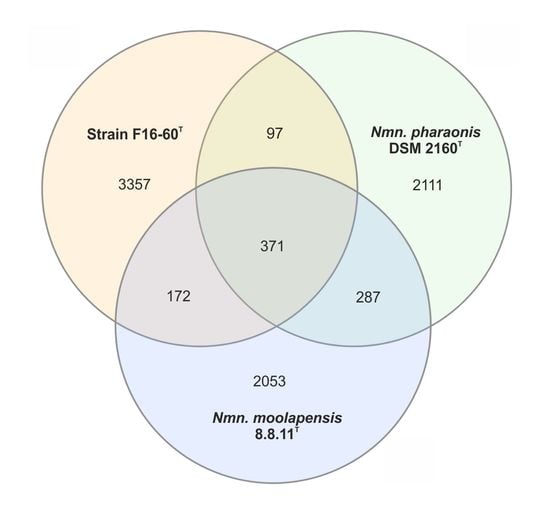
3.4. Genomic Characteristics
3.5. Metagenomic Fragment Recruitment Analysis
3.6. Description of Haloglomus gen. nov.
3.7. Description of Haloglomus irregulare sp. nov.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oren, A.; Ventosa, A.; Kamekura, M. Halobacteria. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: West Sussex, UK, 2017. [Google Scholar]
- Gupta, R.S.; Naushad, S.; Baker, S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: A proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 1050–1069. [Google Scholar] [PubMed]
- Gupta, R.S.; Naushad, S.; Fabros, R.; Adeolu, M. A phylogenomic reappraisal of family-level divisions within the class Halobacteria: Proposal to divide the order Halobacteriales into the families Halobacteriaceae, Haloarculaceae fam. nov., and Halococcaceae fam. nov., and the order Haloferacales into the families, Haloferacaceae and Halorubraceae fam. nov. Antonie van Leeuwenhoek 2016, 109, 565–587. [Google Scholar] [PubMed]
- Amoozegar, M.A.; Siroosi, M.; Atashgahi, S.; Smidt, H.; Ventosa, A. Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology 2017, 163, 623–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oren, A. The Order Halobacteriales. In The Prokaryotes; Springer: Berlin, Germany, 2006; pp. 113–164. [Google Scholar]
- Ventosa, A.; Fernández, A.B.; León, M.J.; Sánchez-Porro, C.; Rodriguez-Valera, F. The Santa Pola saltern as a model for studying the microbiota of hypersaline environments. Extremophiles 2014, 18, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.B.; Vera-Gargallo, B.; Sánchez-Porro, C.; Ghai, R.; Papke, R.T.; Rodriguez-Valera, F.; Ventosa, A. Comparison of prokaryotic community structure from Mediterranean and Atlantic saltern concentrator ponds by a metagenomic approach. Front. Microbiol. 2014, 5, 196. [Google Scholar] [CrossRef]
- Ventosa, A.; de la Haba, R.R.; Sánchez-Porro, C.; Papke, R.T. Microbial diversity of hypersaline environments: A metagenomic approach. Curr. Opin. Microbiol. 2015, 25, 80–87. [Google Scholar] [CrossRef]
- León, M.J.; Fernández, A.B.; Ghai, R.; Sánchez-Porro, C.; Rodriguez-Valera, F.; Ventosa, A. From metagenomics to pure culture: Isolation and characterization of the moderately halophilic bacterium Spiribacter salinus gen. nov., sp. nov. Appl. Environ. Microbiol. 2014, 80, 3850–3857. [Google Scholar] [CrossRef] [Green Version]
- León, M.J.; Vera-Gargallo, B.; Sánchez-Porro, C.; Ventosa, A. Spiribacter roseus sp. nov., a moderately halophilic species of the genus Spiribacter from salterns. Int. J. Syst. Evol. Microbiol. 2016, 66, 4218–4224. [Google Scholar] [CrossRef]
- León, M.J.; Martínez-Checa, F.; Ventosa, A.; Sánchez-Porro, C. Idiomarina aquatica sp. nov., a moderately halophilic bacterium isolated from salterns. Int. J. Syst. Evol. Microbiol. 2015, 65, 4595–4600. [Google Scholar] [CrossRef]
- León, M.J.; Sánchez-Porro, C.; Ventosa, A. Marinobacter aquaticus sp. nov., a moderately halophilic bacterium from a solar saltern. Int. J. Syst. Evol. Microbiol. 2017, 67, 2622–2627. [Google Scholar] [CrossRef]
- López-Hermoso, C.; de la Haba, R.R.; Sánchez-Porro, C.; Ventosa, A. Salinivibrio kushneri sp. nov., a moderately halophilic bacterium isolated from salterns. Syst. Appl. Microbiol. 2018, 41, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Durán-Viseras, A.; Ventosa, A.; Sánchez-Porro, C. Halonotius aquaticus sp. nov., a new haloarchaeon isolated from a marine saltern. Int. J. Syst. Evol. Microbiol. 2019, 69, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Durán-Viseras, A.; Andrei, A.-S.; Ghai, R.; Sánchez-Porro, C.; Ventosa, A. new Halonotius species provide genomics-based insights into cobalamin synthesis in Haloarchaea. Front. Microbiol. 2019, 10, 1928. [Google Scholar] [CrossRef] [PubMed]
- Durán-Viseras, A.; Sánchez-Porro, C.; Ventosa, A. Halorientalis pallida sp. nov., an extremely halophilic archaeon isolated from a marine saltern. Int. J. Syst. Evol. Microbiol. 2019, 69, 3636–3643. [Google Scholar] [CrossRef]
- Oren, A.; Ventosa, A.; Grant, W.D. Proposed minimal standards for description of new taxa in the Order Halobacteriales. Int. J. Syst. Bacteriol. 1997, 47, 233–238. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, III; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; ISBN 9781936113415. [Google Scholar]
- DeLong, E.F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 1992, 89, 5685–5689. [Google Scholar] [CrossRef] [Green Version]
- Arahal, D.R.; Dewhirst, F.E.; Paster, B.J.; Volcani, B.E.; Ventosa, A. Phylogenetic analyses of some extremely halophilic archaea isolated from dead sea water, determined on the basis of their 16S rRNA sequences. Appl. Environ. Microbiol. 1996, 62, 3779–3786. [Google Scholar] [CrossRef] [Green Version]
- Fullmer, M.S.; Soucy, S.M.; Swithers, K.S.; Makkay, A.M.; Wheeler, R.; Ventosa, A.; Gogarten, J.P.; Papke, R.T. Population and genomic analysis of the genus Halorubrum. Front. Microbiol. 2014, 5, 140. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Elsevier: Amsterdam, The Netherlands, 1969; pp. 21–132. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Auch, A.F.; Klenk, H.-P.; Göker, M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genomic Sci. 2010, 2, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Corcelli, A.; Lobasso, S. 25 Characterization of lipids of halophilic Archaea. Methods Microbiol. 2006, 35, 585–613. [Google Scholar]
- Angelini, R.; Corral, P.; Lopalco, P.; Ventosa, A.; Corcelli, A. Novel ether lipid cardiolipins in archaeal membranes of extreme haloalkaliphiles. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1365–1373. [Google Scholar] [CrossRef] [Green Version]
- Corral, P.; Gutierrez, M.C.; Castillo, A.M.; Dominguez, M.; Lopalco, P.; Corcelli, A.; Ventosa, A. Natronococcus roseus sp. nov., a haloalkaliphilic archaeon from a hypersaline lake. Int. J. Syst. Evol. Microbiol. 2013, 63, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Kates, M. Techniques of Lipidology: Isolation, Analysis, and Identification of Lipids; Elsevier: Amsterdam, The Netherlands, 1986; ISBN 0444807322. [Google Scholar]
- Dussault, H.P. An improved technique for staining red halophilic bacteria. J. Bacteriol. 1955, 70, 484–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrow, G.I.; Feltham, R.K.A. Cowan and Steel’s Manual for the Identification of Medical Bacteria; Cambridge University Press: Cambridge, UK, 2003; ISBN 9780521543. [Google Scholar]
- Kovacs, N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 1956, 178, 703. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, P.; Murray, R.G.; Wood, W.A.; Krieg, N. Methods for General and Molecular Bacteriology; American Society for Microbiology: Washington, DC, USA, 1994. [Google Scholar]
- Clarke, P.H. Hydrogen sulphide production by bacteria. J. Gen. Microbiol. 1953, 8, 397–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smibert, R.M.; Krieg, N.R. General characterization. In Manual of Methods for General Bacteriology; American Society for Microbiology: Washington, DC, USA, 1981; pp. 409–443. [Google Scholar]
- Ventosa, A.; Quesada, E.; Rodriguez-Valera, F.; Ruiz-Berraquero, F.; Ramos-Cormenzana, A. Numerical taxonomy of moderately halophilic gram-negative rods. J. Gen. Microbiol. 1982, 128, 1959–1968. [Google Scholar] [CrossRef] [Green Version]
- Boucher, Y.; Douady, C.J.; Sharma, A.K.; Kamekura, M.; Doolittle, W.F. Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 2004, 186, 3980–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.-L.; Jiang, X.; Wu, Q.L.; Zhou, N.-Y. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbiol. 2013, 79, 5962–5969. [Google Scholar] [CrossRef] [Green Version]
- Konstantinidis, K.T.; Tiedje, J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 3160–3165. [Google Scholar] [CrossRef] [Green Version]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-R, L.M.; Konstantinidis, K.T. Bypassing cultivation to identify bacterial species. Microbe 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Klappenbach, J.A.; Goris, J.; Vandamme, P.; Coenye, T.; Konstantinidis, K.T.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 11, 2399–2406. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.G.; Janssen, P.H.; Itoh, T.; Minegishi, H.; Usami, R.; Kamekura, M.; Dyall-Smith, M.L. Natronomonas moolapensis sp. nov., non-alkaliphilic isolates recovered from a solar saltern crystallizer pond, and emended description of the genus Natronomonas. Int. J. Syst. Evol. Microbiol. 2010, 60, 1173–1176. [Google Scholar] [CrossRef] [PubMed]


| AAI Values | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | - | 64.0 | 65.2 | 65.2 | 62.8 | 61.6 | 65.9 | 62.4 | 65.1 | 62.5 | 61.3 |
| 2 | - | - | 64.0 | 63.9 | 62.7 | 60.8 | 64.4 | 62.2 | 66.3 | 61.8 | 61.1 |
| 3 | - | - | - | 65.3 | 64.0 | 62.1 | 65.6 | 63.8 | 65.3 | 64.0 | 62.9 |
| 4 | - | - | - | - | 62.5 | 61.5 | 68.7 | 62.9 | 65.4 | 63.1 | 61.7 |
| 5 | - | - | - | - | - | 62.7 | 63.3 | 73.0 | 63.2 | 64.9 | 62.4 |
| 6 | - | - | - | - | - | - | 61.9 | 61.2 | 61.3 | 61.4 | 60.6 |
| 7 | - | - | - | - | - | - | - | 63.4 | 65.9 | 62.9 | 61.9 |
| 8 | - | - | - | - | - | - | - | - | 63.0 | 64.6 | 62.5 |
| 9 | - | - | - | - | - | - | - | - | - | 62.4 | 62.0 |
| 10 | - | - | - | - | - | - | - | - | - | - | 62.0 |
| 11 | - | - | - | - | - | - | - | - | - | - | - |
| Characteristic | 1 | 2 |
|---|---|---|
| Morphology | Irregular, pleomorphic | Rods or pleomorphic * |
| Motility | − | + * |
| Cell size (µm) | 0.6–3.0 | 0.7 × 1.7 * |
| Colony size (mm) | 0.5 | 0.5–1.0 * |
| Colony-pigmentation | Red | Pink * |
| NaCl Range (%, w/v) | 25–35 | 14–36 * |
| NaCl optimum (%, w/v) | 30 | 18–20 * |
| Temperature range for growth (°C) | 25–45 | 25–45 * |
| pH range | 6.5–9.0 | 5.5–8.5 * |
| pH optimum | 7.5 | 7–7.5 * |
| Nitrite reduction | + | − |
| Hydrolisis of gelatin | + | − |
| Utilization as sole carbon and energy source of: | ||
| d-glucose | + | − |
| d-melibiose | + | − |
| d-raffinose | + | − |
| Glycerol | − | + |
| l-cysteine | + | − |
| l-glycine | − | + |
| l-lysine | − | + |
| Isoleucine | − | + |
| Valine | − | + |
| Fumarate | − | + |
| Malate | − | + |
| Pyruvate | + | − |
| Feature | Strain F16-60T |
|---|---|
| Size (bp) | 4019787 |
| Contigs | 54 |
| Completeness (%) | 97.6 |
| G + C (mol%) | 68.0 |
| N50 (bp) | 274728 |
| Total genes | 3922 |
| Protein coding genes | 3673 |
| rRNA | 4 |
| tRNA | 53 |
| Accession number | QMDX00000000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durán-Viseras, A.; Sánchez-Porro, C.; Ventosa, A. Haloglomus irregulare gen. nov., sp. nov., a New Halophilic Archaeon Isolated from a Marine Saltern. Microorganisms 2020, 8, 206. https://doi.org/10.3390/microorganisms8020206
Durán-Viseras A, Sánchez-Porro C, Ventosa A. Haloglomus irregulare gen. nov., sp. nov., a New Halophilic Archaeon Isolated from a Marine Saltern. Microorganisms. 2020; 8(2):206. https://doi.org/10.3390/microorganisms8020206
Chicago/Turabian StyleDurán-Viseras, Ana, Cristina Sánchez-Porro, and Antonio Ventosa. 2020. "Haloglomus irregulare gen. nov., sp. nov., a New Halophilic Archaeon Isolated from a Marine Saltern" Microorganisms 8, no. 2: 206. https://doi.org/10.3390/microorganisms8020206
APA StyleDurán-Viseras, A., Sánchez-Porro, C., & Ventosa, A. (2020). Haloglomus irregulare gen. nov., sp. nov., a New Halophilic Archaeon Isolated from a Marine Saltern. Microorganisms, 8(2), 206. https://doi.org/10.3390/microorganisms8020206