Interactions between Kazachstania humilis Yeast Species and Lactic Acid Bacteria in Sourdough
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Inocula
2.2. Laboratory Sourdough Preparation and Indirect Fermentation Record
2.3. Sampling and CFU Count
2.4. Metabolite Analysis
2.5. Acidity Tolerance Analysis
2.6. Statistical Analysis
2.6.1. Repeatability
2.6.2. Single Strain Analysis
2.6.3. Interaction Analysis
2.6.4. Acidity Tolerance Analysis
2.6.5. Multivariate Analysis
3. Results
3.1. Phenotypic Variation between Strains in Monoculture
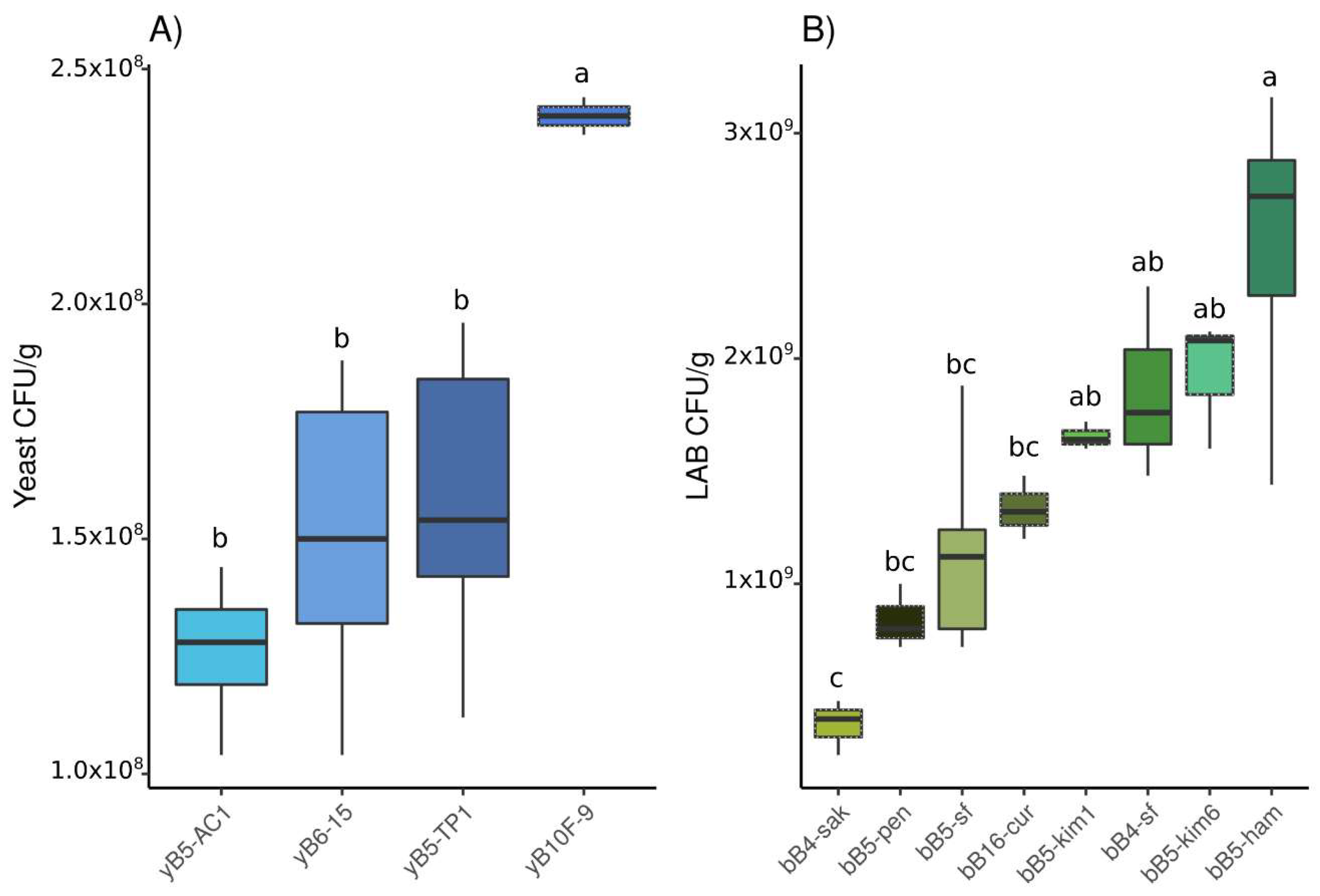
3.1.1. Population Density
3.1.2. Dough Height
3.1.3. Concentration of Metabolites in Dough
3.2. LAB Effect on Yeast
3.2.1. Population Density
3.2.2. Dough Height
3.3. Yeast Effect on LAB
3.3.1. Population Density
3.3.2. Dough Height
3.4. Metabolite Profiles and pH in Cocultures and Monocultures
3.5. Multivariate Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-Fungal Interactions: Hyphens between Agricultural, Clinical, Environmental, and Food Microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furukawa, S.; Watanabe, T.; Toyama, H.; Morinaga, Y. Significance of microbial symbiotic coexistence in traditional fermentation. J. Biosci. Bioeng. 2013, 116, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Calasso, M.; Archetti, G.; Rizzello, C.G. Novel insights on the functional/nutritional features of the sourdough fermentation. Int. J. Food Microbiol. 2019, 302, 103–113. [Google Scholar] [CrossRef]
- Vogel, R.F.; Knorr, R.; Müller, M.R.; Steudel, U.; Gänzle, M.G.; Ehrmann, M.A. Non-dairy lactic fermentations: The cereal world. Antonie Van Leeuwenhoek 1999, 76, 403–411. [Google Scholar] [CrossRef]
- Van Kerrebroeck, S.; Maes, D.; De Vuyst, L. Sourdoughs as a function of their species diversity and process conditions, a meta-analysis. Trends Food Sci. Technol. 2017, 68, 152–159. [Google Scholar] [CrossRef]
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef]
- Lhomme, E.; Lattanzi, A.; Dousset, X.; Minervini, F.; de Angelis, M.; Lacaze, G.; Onno, B.; Gobbetti, M. Lactic acid bacterium and yeast microbiotas of sixteen French traditional sourdoughs. Int. J. Food Microbiol. 2015, 215, 161–170. [Google Scholar] [CrossRef]
- Lhomme, E.; Onno, B.; Chuat, V.; Durand, K.; Orain, S.; Valence, F.; Dousset, X.; Jacques, M.-A. Genotypic diversity of Lactobacillus sanfranciscensis strains isolated from French organic sourdoughs. Int. J. Food Microbiol. 2016, 226, 13–19. [Google Scholar] [CrossRef]
- De Vuyst, L.; Harth, H.; Van Kerrebroeck, S.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef]
- Liu, S.Q. Sourdough. In Bakery Products Science and Technology, 2nd ed.; Zhou, Y.H., Hui, I., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2014; Chapter 29; pp. 511–521. [Google Scholar]
- Michel, E.; Monfort, C.; Deffrasnes, M.; Guezenec, S.; Lhomme, E.; Barret, M.; Sicard, D.; Dousset, X.; Onno, B. Characterization of relative abundance of lactic acid bacteria species in French organic sourdough by cultural, qPCR and MiSeq high-throughput sequencing methods. Int. J. Food Microbiol. 2016, 239, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lhomme, E.; Urien, C.; Legrand, J.; Dousset, X.; Onno, B.; Sicard, D. Sourdough microbial community dynamics: An analysis during French organic bread-making processes. Food Microbiol. 2016, 53, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Urien, C.; Legrand, J.; Montalent, P.; Casaregola, S.; Sicard, D. Fungal Species Diversity in French Bread Sourdoughs Made of Organic Wheat Flour. Front. Microbiol. 2019, 10, 201. [Google Scholar] [CrossRef] [Green Version]
- Carbonetto, B.; Ramsayer, J.; Nidelet, T.; Legrand, J.; Sicard, D. Bakery yeasts, a new model for studies in ecology and evolution. Yeast 2018, 35, 591–603. [Google Scholar] [CrossRef] [Green Version]
- Michel, E.; Masson, E.; Bubbendorf, S.; Lapicque, L.; Legrand, J.; Guézenec, S.; Nidelet, T.; Marlin, T.; Rué, O.; Onno, B.; et al. Artisanal and farmers bread making practices differently shape fungal species diversity in French sourdoughs. bioRxiv 2019, 679472. [Google Scholar] [CrossRef] [Green Version]
- Silla, M.H. The Genera of Lactic Acid Bacteria (Vol. 2); Wood, B.J., Holzapfel, W.H.N., Eds.; Springer Science & Business Media: Boston, MA, USA, 1992; p. XVIII 398. ISBN 07-514-0215. [Google Scholar]
- Vandamme, P.; Pot, B.; Gillis, M.; de Vos, P.; Kersters, K.; Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996, 60, 407–438. [Google Scholar] [CrossRef] [Green Version]
- Hagman, A.; Säll, T.; Compagno, C.; Piskur, J. Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS ONE 2013, 8, e68734. [Google Scholar] [CrossRef]
- Minervini, F.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Ecological parameters influencing microbial diversity and stability of traditional sourdough. Int. J. Food Microbiol. 2014, 171, 136–146. [Google Scholar] [CrossRef]
- Gobbetti, M.; Corsetti, A.; Rossi, J. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: Metabolism of carbohydrates. Appl. Microbiol. Biotechnol. 1994, 41, 456–460. [Google Scholar] [CrossRef]
- De Vuyst, L.; Van Kerrebroeck, S.; Leroy, F. Microbial Ecology and Process Technology of Sourdough Fermentation. Adv. Appl. Microbiol. 2017, 100, 49–160. [Google Scholar]
- Sugihara, T.F.; Kline, L.; Miller, M.W. Microorganisms of the San Francisco Sour Dough Bread Process. Appl. Microbiol. 1971, 21, 456–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuyst, L.D.; De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- De Vuyst, L.; Vrancken, G.; Ravyts, F.; Rimaux, T.; Weckx, S. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 2009, 26, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Venturi, M.; Guerrini, S.; Vincenzini, M. Stable and non-competitive association of Saccharomyces cerevisiae, Candida milleri and Lactobacillus sanfranciscensis during manufacture of two traditional sourdough baked goods. Food Microbiol. 2012, 31, 107–115. [Google Scholar] [CrossRef]
- Gobbetti, M. The sourdough microflora: Interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 1998, 9, 267–274. [Google Scholar] [CrossRef]
- Stolz, P.; Böcker, G.; Vogel, R.F.; Hammes, W.P. Utilisation of maltose and glucose by lactobacilli isolated from sourdough. FEMS Microbiol. Lett 1993, 109, 237–242. [Google Scholar] [CrossRef]
- Ponomarova, O.; Gabrielli, N.; Sévin, D.C.; Mülleder, M.; Zirngibl, K.; Bulyha, K.; Andrejev, S.; Kafkia, E.; Typas, A.; Sauer, U.; et al. Yeast Creates a Niche for Symbiotic Lactic Acid Bacteria through Nitrogen Overflow. Cell Syst. 2017, 5, 345–357. [Google Scholar] [CrossRef] [Green Version]
- Gobbetti, M.; Corsetti, A.; Rossi, J. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: Metabolism of amino acids. World J. Microbiol. Biotechnol. 1994, 10, 275–279. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y.; Tang, K.; Hu, Y.; Xu, X.; Gänzle, M.G. Effect of Mixed Cultures of Yeast and Lactobacilli on the Quality of Wheat Sourdough Bread. Front. Microbiol. 2019, 10, 2113. [Google Scholar] [CrossRef]
- Minervini, F.; Lattanzi, A.; De Angelis, M.; Celano, G.; Gobbetti, M. House microbiotas as sources of lactic acid bacteria and yeasts in traditional Italian sourdoughs. Food Microbiol. 2015, 52, 66–76. [Google Scholar] [CrossRef]
- Celano, G.; De Angelis, M.; Minervini, F.; Gobbetti, M. Different Flour Microbial Communities Drive to Sourdoughs Characterized by Diverse Bacterial Strains and Free Amino Acid Profiles. Front. Microbiol. 2016, 7, 1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gänzle, M.G.; Zheng, J. Lifestyles of sourdough lactobacilli – Do they matter for microbial ecology and bread quality? Int. J. Food Microbiol. 2019, 302, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Gancedo, J.M. Yeast Carbon Catabolite Repression. Microbiol. Mol. Biol. Rev. 1998, 62, 334–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolland, F.; Winderickx, J.; Thevelein, J.M. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002, 2, 183–201. [Google Scholar] [CrossRef]
- Zaman, S.; Lippman, S.I.; Schneper, L.; Slonim, N.; Broach, J.R. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 2009, 5, 245. [Google Scholar] [CrossRef] [Green Version]
- Broach, J.R. Nutritional Control of Growth and Development in Yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [Green Version]
- De Kroon, R.A.; Koningsberger, V.V. An inducible transport system for alpha-glucosides in protoplasts of Saccharomyces carlsbergensis. Biochim. Biophys. Acta 1970, 204, 590–609. [Google Scholar] [CrossRef]
- Gänzle, M.G.; Follador, R. Metabolism of oligosaccharides and starch in lactobacilli: A review. Front. Microbiol. 2012, 3, 340. [Google Scholar] [CrossRef] [Green Version]
- Elorza, M.V.; Lostau, C.M.; Villanueva, J.R.; Sentandreu, R.; Sanchez, E. Invertase messenger ribonucleic acid in Saccharomyces cerevisiae: Kinetics of formation and decay. Biochim. et Biophys. Acta (BBA)—Nucleic Acids Protein Synth. 1977, 475, 638–651. [Google Scholar] [CrossRef]
- New, A.M.; Cerulus, B.; Govers, S.K.; Perez-Samper, G.; Zhu, B.; Boogmans, S.; Xavier, J.B.; Verstrepen, K.J. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol. 2014, 12, e1001764. [Google Scholar] [CrossRef] [Green Version]
- Foster, K.R.; Bell, T. Competition, Not Cooperation, Dominates Interactions among Culturable Microbial Species. Curr. Biol. 2012, 22, 1845–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, R.R.; Bucci, V.; Toussaint, N.C.; Buffie, C.G.; Rätsch, G.; Pamer, E.G.; Sander, C.; Xavier, J.B. Ecological modeling from time-series inference: Insight into dynamics and stability of intestinal microbiota. PLoS Comput. Biol. 2013, 9, e1003388. [Google Scholar] [CrossRef] [PubMed]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Zelezniak, A.; Andrejev, S.; Ponomarova, O.; Mende, D.R.; Bork, P.; Patil, K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. USA 2015, 112, 6449–6454. [Google Scholar] [CrossRef] [Green Version]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]





| Species | Strain’s Code | Lab Code | Baker | French Region | Main Physiological Characteristic |
|---|---|---|---|---|---|
| Yeast | |||||
| Saccharomyces cerevisiae | yB10F-9 | MTF3947 | B10 | Provence-Alpes Côte d’Azur | maltose-positive |
| Kazachstania humilis | yB6-15 | MTF3948 | B6 | Champagne Ardennes | maltose-negative |
| Kazachstania humilis | yB5-TP1 | MTF3949 | B5 | Ile de France | maltose-negative |
| Kazachstania humilis | yB5-AC1 | MTF4070 | B5 | Ile de France | maltose-negative |
| Bacteria | |||||
| Lactobacilus curvatus | bB16-cur | MTF 4123 | B16 | Ile de France | facultatively heterofermentative |
| Lactobacilus sakei | bB4-sak | MTF 4118 | B4 | Pays de Loire | facultatively heterofermentative |
| Lactobacilus sanfranciscensis | bB4-sf | MTF 3946 | B4 | Pays de Loire | obligately heterofermentative |
| Lactobacilus sanfranciscensis | bB5-sf | MTF 3945 | B5 | Ile de France | obligately heterofermentative |
| Lactobacilus hammesii | bB5-ham | MTF 3944 | B5 | Ile de France | obligately heterofermentative |
| Lactobacilus pentosus | bB5-pen | MTF 4114 | B5 | Ile de France | facultatively heterofermentative |
| Lactobacilus kimchi | bB5-kim1 | MTF 4117 | B5 | Ile de France | facultatively heterofermentative |
| Lactobacilus kimchi | bB5-kim6 | MTF 4116 | B5 | Ile de France | facultatively heterofermentative |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carbonetto, B.; Nidelet, T.; Guezenec, S.; Perez, M.; Segond, D.; Sicard, D. Interactions between Kazachstania humilis Yeast Species and Lactic Acid Bacteria in Sourdough. Microorganisms 2020, 8, 240. https://doi.org/10.3390/microorganisms8020240
Carbonetto B, Nidelet T, Guezenec S, Perez M, Segond D, Sicard D. Interactions between Kazachstania humilis Yeast Species and Lactic Acid Bacteria in Sourdough. Microorganisms. 2020; 8(2):240. https://doi.org/10.3390/microorganisms8020240
Chicago/Turabian StyleCarbonetto, Belén, Thibault Nidelet, Stéphane Guezenec, Marc Perez, Diego Segond, and Delphine Sicard. 2020. "Interactions between Kazachstania humilis Yeast Species and Lactic Acid Bacteria in Sourdough" Microorganisms 8, no. 2: 240. https://doi.org/10.3390/microorganisms8020240
APA StyleCarbonetto, B., Nidelet, T., Guezenec, S., Perez, M., Segond, D., & Sicard, D. (2020). Interactions between Kazachstania humilis Yeast Species and Lactic Acid Bacteria in Sourdough. Microorganisms, 8(2), 240. https://doi.org/10.3390/microorganisms8020240






