Application of a Broad Range Lytic Phage LPST94 for Biological Control of Salmonella in Foods
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Enrichment, Isolation, Purification, and Preparation of Phages
2.3. Screening of Phages Based on Spot Test and Lytic Capacity
2.3.1. Lytic Spectrum Determination by Spot Test
2.3.2. Lytic Activity of Phage LPST94
2.4. Efficiency of Plating
2.5. Morphological Observation of Phage LPST94
2.6. Genomic Analysis of Phage LPST94
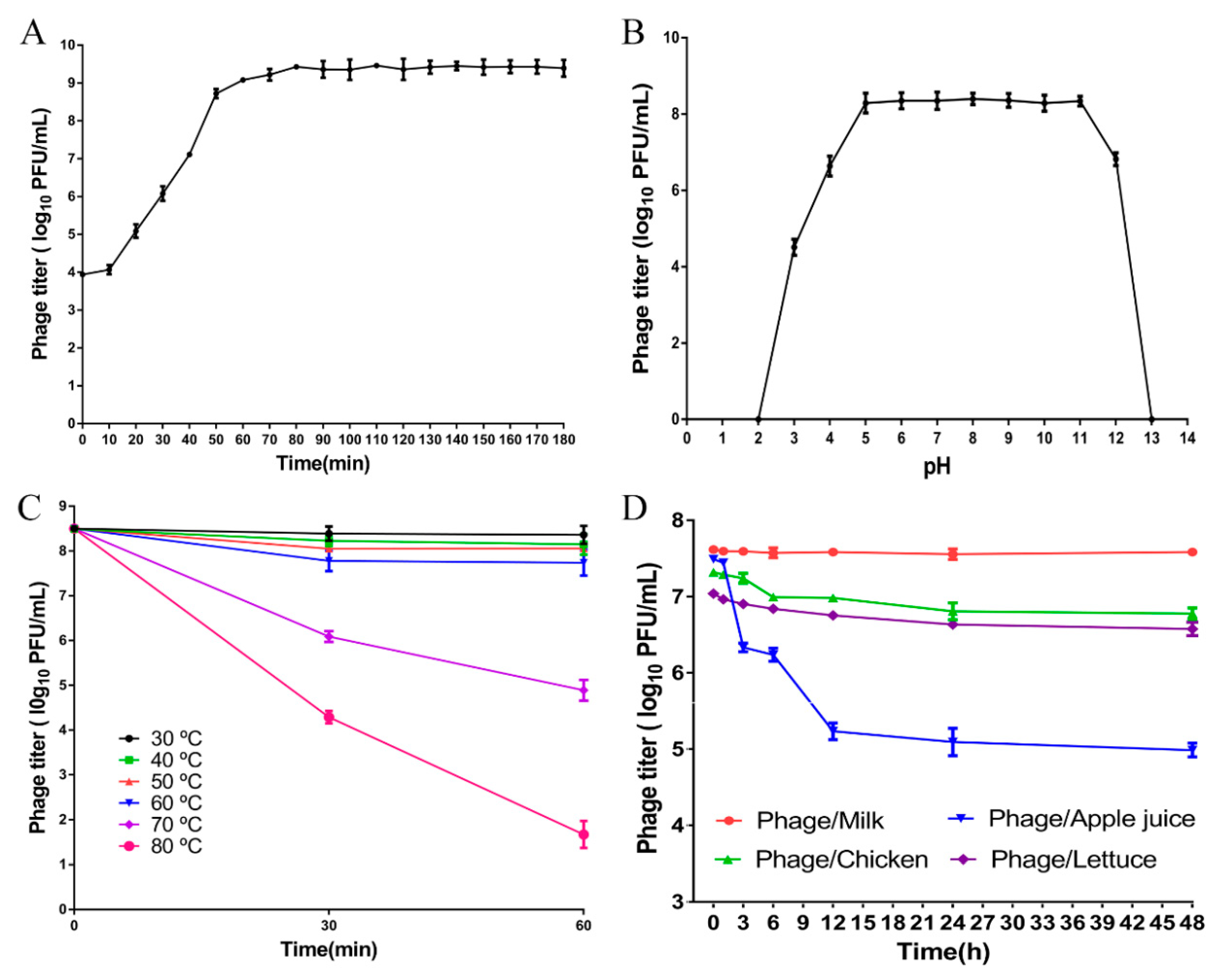
2.7. One-Step Growth Curve
2.8. pH and Thermal Tolerance of the Phage LPST94
2.9. Phage Stability in Diverse Food Samples
2.10. Ability of Lysogenic Formation of the Phage
2.11. Biological Control of Salmonella in Foods Using Phage LPST94
2.11.1. Testing in Milk and Apple Juice
2.11.2. Testing in Chicken Breast and Lettuce
2.12. Statistical Analysis
3. Results
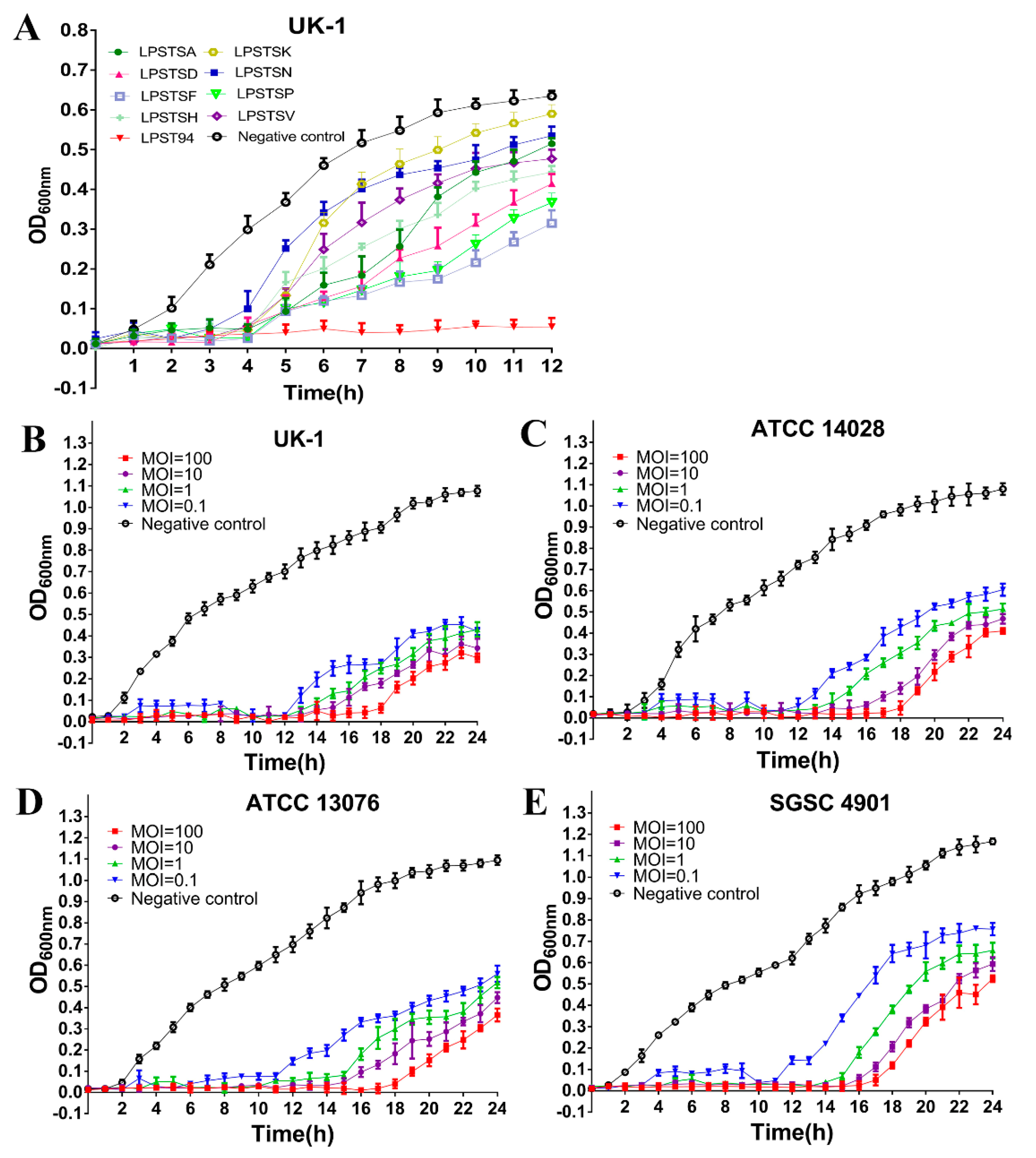
3.1. Isolation and Screening of Phage
3.2. Relative Replication Efficiency of Phage LPST94
3.3. Phage Morphology and Genomic Analysis
3.4. Characteristics of Phage LPST94
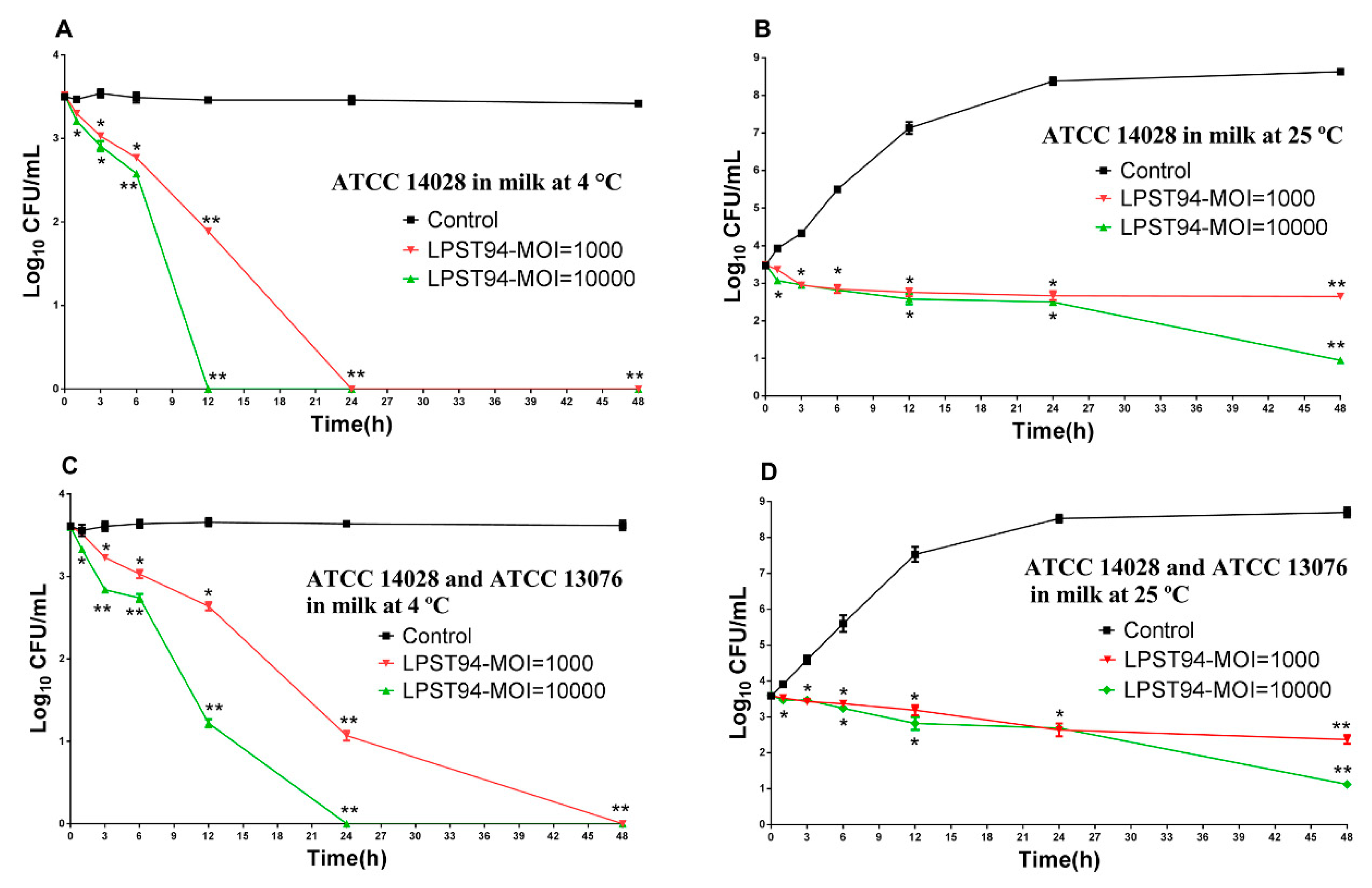
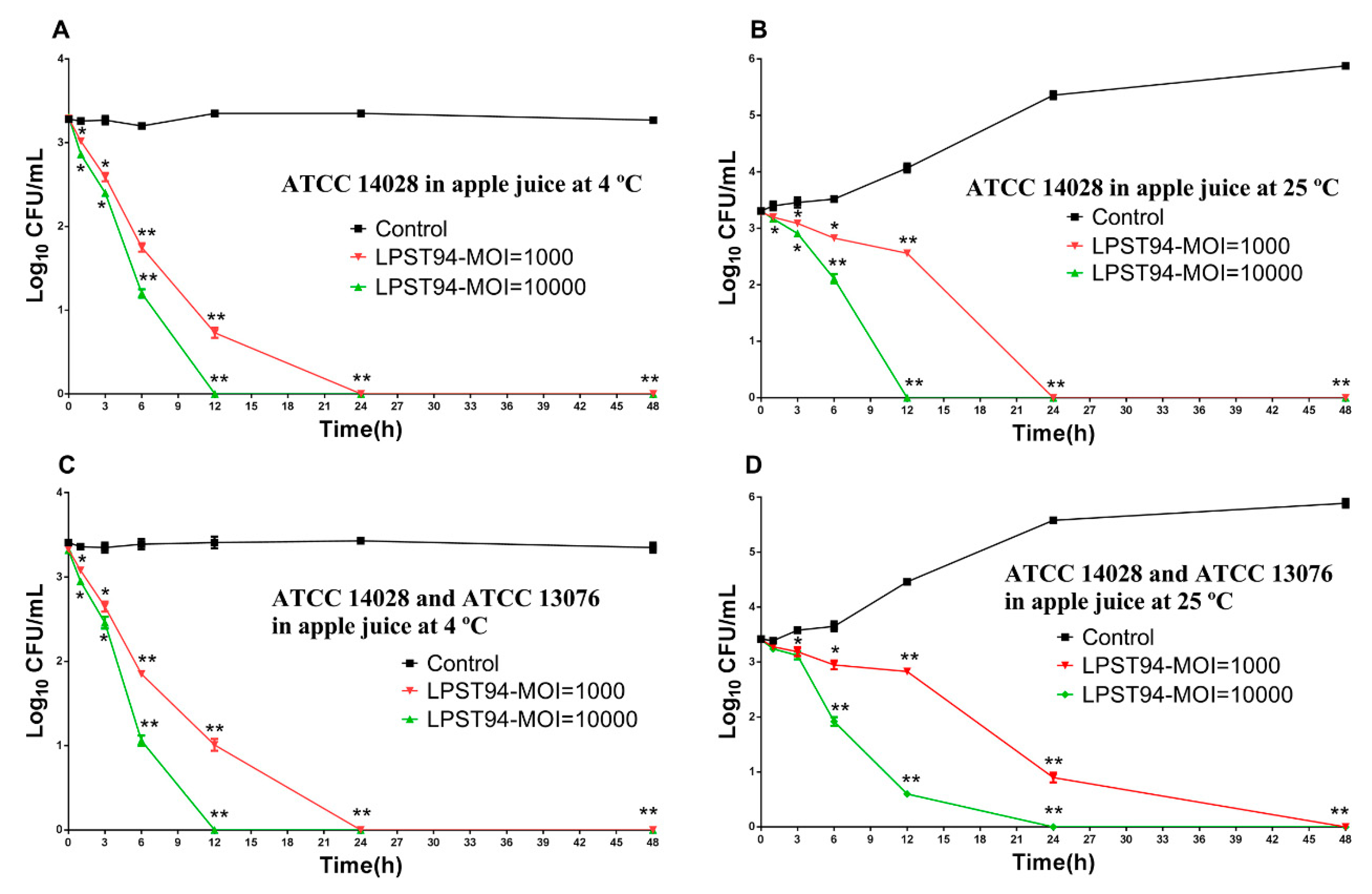
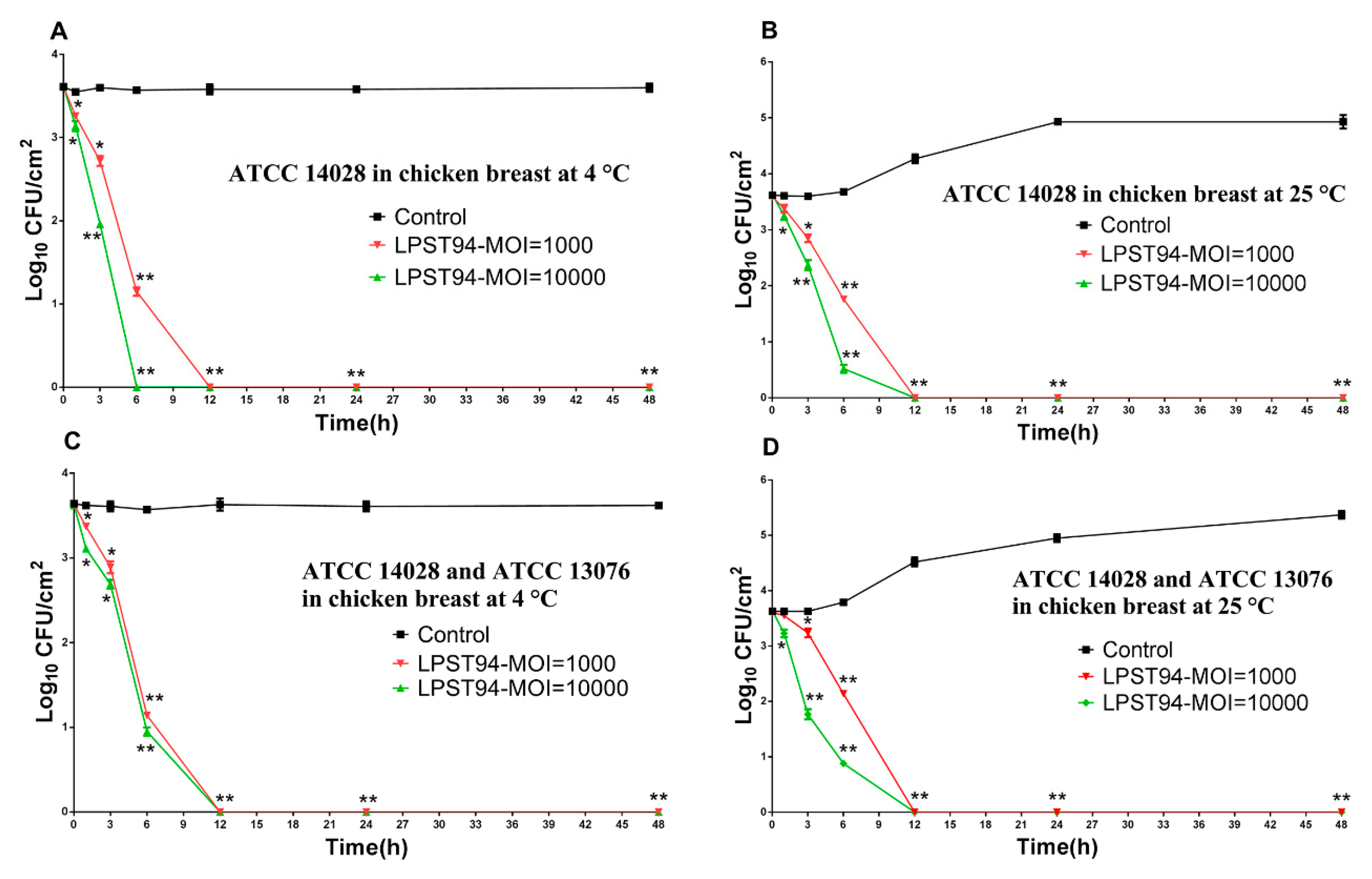
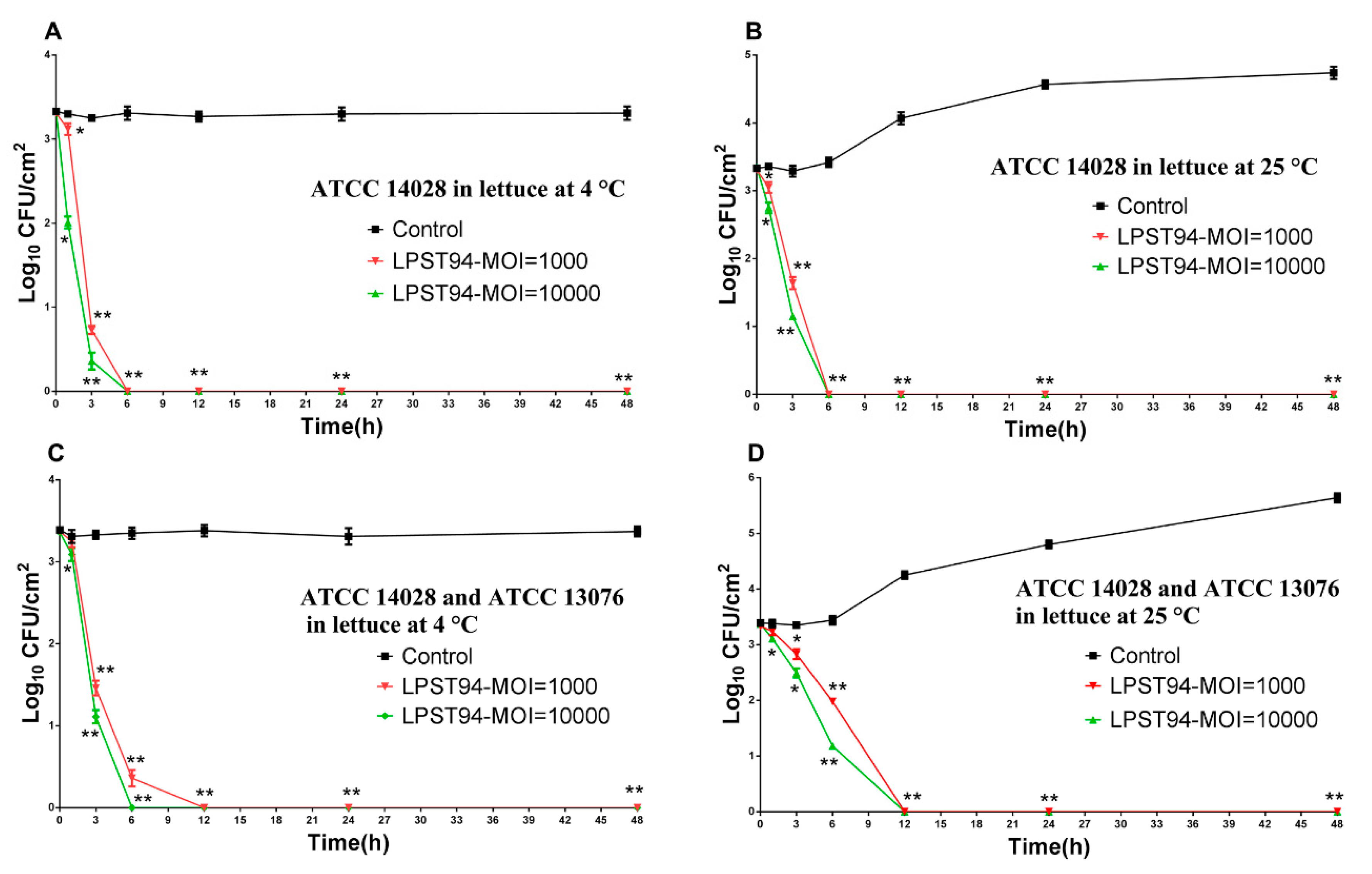
3.5. Application of Phage LPST94 in Controlling Food-Borne S. Typhimurium and S. Enteritidis
3.5.1. Milk
3.5.2. Apple Juice
3.5.3. Chicken Breast
3.5.4. Lettuce
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Balasubramanian, R.; Im, J.; Lee, J.-S.; Jeon, H.J.; Mogeni, O.D.; Kim, J.H.; Rakotozandrindrainy, R.; Baker, S.; Marks, F. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccines Immunother. 2019, 15, 1421–1426. [Google Scholar] [CrossRef]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008, 6, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.F.; Wong, W.C.; Chai, L.C.; Tunung, R.; Jeyaletchumi, P.; Hidayah, M.S.N.; Ubong, A.; Farinazleen, M.G.; Cheah, Y.K. Salmonella: A foodborne pathogen. Int. Food Res. J. 2011, 18, 465–473. [Google Scholar]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 2011, 17, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Multistate Outbreak of Salmonella Typhimurium Linked to Chicken Salad. Centers for Disease Control and Prevention. 2018. Available online: https://www.cdc.gov/salmonella/typhimurium-02-8/index.html (accessed on 11 October 2018).
- Huang, X.; Huang, Q.; Dun, Z.; Huang, W.; Wu, S.; Liang, J.; Deng, X.; Zhang, Y. Nontyphoidal Salmonella Infection, Guangdong Province, China, 2012. Emerg. Infect. Dis. 2016, 22, 726–729. [Google Scholar] [CrossRef]
- Miao, E.A.; Miller, S.I. Bacteriophages in the evolution of pathogen-host interactions. Proc. Natl. Acad. Sci. USA 1999, 96, 9452. [Google Scholar] [CrossRef]
- Tauxe, R.V.; Doyle, M.P.; Kuchenmã¼Ller, T.; Schlundt, J.; Stein, C.E. Evolving public health approaches to the global challenge of foodborne infections. Int. J. Food Microbiol. 2010, 139, S16–S28. [Google Scholar] [CrossRef]
- Juneja, V.K.; Dwivedi, H.P.; Yan, X. Novel natural food antimicrobials. Annu. Rev. Food Sci. Technol. 2012, 3, 381–403. [Google Scholar] [CrossRef]
- Neetoo, H.; Mahomoodally, F. Use of Antimicrobial Films and Edible Coatings Incorporating Chemical and Biological Preservatives to Control Growth of Listeria monocytogenes on Cold Smoked Salmon. BioMed Res. Int. 2014, 534915, 10. [Google Scholar]
- Musyoka, J.N.; Abong, G.O.; Mbogo, D.M.; Fuchs, R.; Low, J.; Heck, S.; Muzhingi, T. Effects of Acidification and Preservatives on Microbial Growth during Storage of Orange Fleshed Sweet Potato Puree. Int. J. Food Sci. 2018, 2018, 7435–7443. [Google Scholar] [CrossRef]
- Omer Mukhtar, T.; Syed Abdul, A.; Khalid, J.; Askari, B. Study to evaluate the impact of heat treatment on water soluble vitamins in milk. JPMA J. Pak. Med. Assoc. 2010, 60, 909. [Google Scholar]
- Leskova, E.; Kubíková, J.; Kováčiková, E.; Košická, M.; Porubská, J.; Holčíková, K. Vitamin losses: Retention during heat treatment and continual changes expressed by mathematical models. J. Food Compos. Anal. 2006, 19, 252–276. [Google Scholar] [CrossRef]
- Pawlowska, A.M.; Zannini, E.; Coffey, A.; Arendt, E.K. “Green preservatives”: Combating fungi in the food and feed industry by applying antifungal lactic acid bacteria. Adv. Food Nutr. Res. 2012, 66, 217. [Google Scholar] [PubMed]
- Khoshnoud, M.J.; Siavashpour, A.; Bakhshizadeh, M.; Rashedinia, M. Effects of sodium benzoate, a commonly used food preservative, on learning, memory, and oxidative stress in brain of mice. J. Biochem. Mol. Toxicol. 2017, 32, 2. [Google Scholar] [CrossRef]
- Uribarri, J.; Woodruff, S.; Goodman, S.; Cai, W.; Chen, X.; Pyzik, R.; Yong, A.; Striker, G.E.; Vlassara, H. Advanced glycation end products in foods and a practical guide to their reduction in the diet. J. Am. Diet. Assoc. 2010, 110, 911–916. [Google Scholar] [CrossRef]
- Wei, S.; Chelliah, R.; Rubab, M.; Oh, D.-H.; Uddin, M.J.; Ahn, J. Bacteriophages as Potential Tools for Detection and Control of Salmonella spp. in Food Systems. Microorganisms 2019, 7, 570. [Google Scholar] [CrossRef]
- Svircev, A.; Roach, D.; Castle, A. Framing the Future with Bacteriophages in Agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front. Cell. Infect. Microbiol. 2018, 8, 376. [Google Scholar] [CrossRef]
- Bumunang, E.W.; McAllister, T.A.; Stanford, K.; Anany, H.; Niu, Y.D.; Ateba, C.N. Characterization of Non-O157 STEC Infecting Bacteriophages Isolated from Cattle Faeces in North-West South Africa. Microorganisms 2019, 7, 615. [Google Scholar] [CrossRef]
- Sillankorva, S.M. Bacteriophages and Their Role in Food Safety. Int. J. Microbiol. 2012, 1, 13. [Google Scholar] [CrossRef]
- Blasco, L.; Ambroa, A.; Lopez, M.; Fernandez-Garcia, L. Combined Use of the Ab105-2phiDeltaCI Lytic Mutant Phage and Different Antibiotics in Clinical Isolates of Multi-Resistant Acinetobacter baumannii. Microorg. 2019, 7, 556. [Google Scholar] [CrossRef] [PubMed]
- Mccallin, S.; Alam, S.S.; Barretto, C.; Sultana, S.; Berger, B.; Huq, S.; Krause, L.; Bibiloni, R.; Schmitt, B.; Reuteler, G. Safety analysis of a Russian phage cocktail: From metagenomic analysis to oral application in healthy human subjects. Virology 2013, 443, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Shafiqul Alam, S.; Shawna, M.C.; Caroline, B.; Bernard, B.; Anne-Cécile, P.; Shamima, S.; Lutz, K.; Sayeda, H.; Rodrigo, B.; Anne, B. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 2012, 434, 222–232. [Google Scholar]
- O’Flynn, G.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P. The newly isolated lytic bacteriophages st104a and st104b are highly virulent against Salmonella enterica. J. Appl. Microbiol. 2006, 101, 251–259. [Google Scholar] [CrossRef]
- Spricigo, D.A.; Bardina, C.; Cortés, P.; Llagostera, M. Use of a bacteriophage cocktail to control Salmonella in food and the food industry. Int. J. Food Microbiol. 2013, 165, 169–174. [Google Scholar] [CrossRef]
- Albino, L.A.; Rostagno, M.H.; Húngaro, H.M.; Mendonça, R.C. Isolation, characterization, and application of bacteriophages for Salmonella spp. biocontrol in pigs. Foodborne Pathog. Dis. 2014, 11, 602–609. [Google Scholar] [CrossRef]
- Bao, H.; Zhang, P.; Zhang, H.; Zhou, Y.; Zhang, L.; Wang, R. Bio-Control of Salmonella Enteritidis in Foods Using Bacteriophages. Viruses 2015, 7, 4836–4853. [Google Scholar] [CrossRef]
- Galarce, N.; Escobar, B.; Rojas, V.; Navarro, C.; Turra, G.; Robeson, J.; Borie, C. Application of a virulent bacteriophage cocktail leads to reduction of serovar Enteritidis counts in processed meat products. Biocontrol Sci. Technol. 2016, 26, 1–26. [Google Scholar] [CrossRef]
- Sukumaran, A.T.; Nannapaneni, R.; Kiess, A.; Sharma, C.S. Reduction of Salmonella on chicken meat and chicken skin by combined or sequential application of lytic bacteriophage with chemical antimicrobials. Int. J. Food Microbiol. 2015, 207, 8–15. [Google Scholar] [CrossRef]
- Goodridge, L.D.; Bisha, B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage 2011, 1, 130–137. [Google Scholar] [CrossRef]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018, 359, eaar4120. [Google Scholar] [CrossRef]
- Akhtar, M.; Viazis, S.; Diez-Gonzalez, F. Isolation, identification and characterization of lytic, wide host range bacteriophages from waste effluents against Salmonella enterica serovars. Food Control 2014, 38, 67–74. [Google Scholar] [CrossRef]
- Islam, M.S.; Zhou, Y.; Liang, L.; Nime, I.; Liu, K.; Yan, T.; Wang, X.; Li, J. Application of a Phage Cocktail for Control of Salmonella in Foods and Reducing Biofilms. Viruses 2019, 11, 841. [Google Scholar] [CrossRef]
- Van, T.R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Methods Mol. Biol. 2009, 501, 15–21. [Google Scholar]
- Jin, W.J.; Park, S.C.; Wicklund, A.; Skurnik, M. Bacteriophages reduce Yersinia enterocolitica contamination of food and kitchenware. Int. J. Food Microbiol. 2018, 271, 33–47. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Gomes, A.T.P.C.; Almeida, A. Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella Typhimurium: An In Vitro Preliminary Study. Microorganisms 2019, 7, 94. [Google Scholar] [CrossRef]
- Ackermann, H.W. Phage classification and characterization. Methods Mol. Biol. 2009, 501, 127. [Google Scholar]
- Islam, M.S.; Raz, A.; Liu, Y.; Elbassiony, K.R.A.; Dong, X.; Zhou, P.; Zhou, Y.; Li, J. Complete Genome Sequence of Aeromonas Phage ZPAH7 with Halo Zones, Isolated in China. Microbiol. Resour. Announc. 2019, 8, e01678-18. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Mount, D.W. Using the basic local alignment search tool (BLAST). C.S.H. Protoc. 2007, 7, pdb–top17. [Google Scholar] [CrossRef]
- Son, H.M.; Duc, H.M.; Masuda, Y.; Honjoh, K.I.; Miyamoto, T. Application of bacteriophages in simultaneously controlling Escherichia coli O157:H7 and extended-spectrum beta-lactamase producing Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 102, 10259–10271. [Google Scholar] [CrossRef]
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of Phage φ6 for Biocontrol of Pseudomonas syringae pv. syringae: An in Vitro Preliminary Study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef]
- Tseng, C.-C.; Chang, D.C.; Chang, K.-C. Development of a Biocontrol Method Applying Bacteriophage-Containing Aerosol against Mycobacterium tuberculosis Using the Bacteriophage BTCU-1 and M. smegmatis as Models. Microorganisms 2019, 7, 237. [Google Scholar] [CrossRef]
- Lópezcuevas, O.; Castrodel, C.N.; Leónfélix, J.; Gonzálezrobles, A.; Chaidez, C. Characterization of bacteriophages with a lytic effect on various Salmonella serotypes and Escherichia coli O157:H7. Can. J. Microbiol. 2011, 57, 1042. [Google Scholar] [CrossRef]
- Guenther, S.; Herzig, O.; Fieseler, L.; Klumpp, J.; Loessner, M.J. Biocontrol of Salmonella Typhimurium in RTE foods with the virulent bacteriophage FO1-E2. Int. J. Food Microbiol. 2012, 154, 66–72. [Google Scholar] [CrossRef]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Cunha, Â.; Delgadillo, I.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Bacteriophages with potential to inactivate Salmonella Typhimurium: Use of single phage suspensions and phage cocktails. Virus Res. 2016, 220, 179–192. [Google Scholar] [CrossRef]
- Laguerre, O.; Derens, E.; Palagos, B. Study of domestic refrigerator temperature and analysis of factors affecting temperature: A French survey. Int. J. Refrig. 2002, 25, 653–659. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Mouttotou, N.; Ahmad, S.; Kamran, Z.; Koutoulis, K.C. Prevalence, Risks and Antibiotic Resistance of Salmonella in Poultry Production Chain. Curr. Top. Salmonella Salmonellosis 2017. [Google Scholar] [CrossRef]
- Careysmith, G.V.; Billington, C.; Cornelius, A.J.; Hudson, J.A.; Heinemann, J.A. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol. Lett. 2006, 258, 182–186. [Google Scholar] [CrossRef]
- Bielke, L.; Higgins, S.; Donoghue, A.; Donoghue, D.; Hargis, B.M. Salmonella Host Range of Bacteriophages That Infect Multiple Genera. Poult. Sci. 2007, 86, 2536–2540. [Google Scholar] [CrossRef]
- Hobbs, Z.; Abedon, S.T. Diversity of phage infection types and associated terminology: The problem with “Lytic or Lysogenic”. FEMS Microbiol. Lett. 2016, 363, fnw047. [Google Scholar] [CrossRef]
- Sunderland, K.; Yang, M.; Mao, C. Phage-Enabled Nanomedicine: From Probes to Therapeutics in Precision Medicine. Angew. Chem. 2016, 56, 1964. [Google Scholar] [CrossRef]
- Zinno, P.; Devirgiliis, C.; Ercolini, D.; Ongeng, D.; Mauriello, G. Bacteriophage P22 to challenge Salmonella in foods. Int. J. Food Microbiol. 2014, 191, 69–74. [Google Scholar] [CrossRef]
- Pao, S.; Rolph, S.P.; Westbrook, E.W.; Shen, H. Use of Bacteriophages to Control Salmonella in Experimentally Contaminated Sprout Seeds. J. Food Sci. 2004, 69, M127–M130. [Google Scholar] [CrossRef]
- Seal, B.S.; Fouts, D.E.; Simmons, M.; Garrish, J.K.; Kuntz, R.L.; Woolsey, R.; Schegg, K.M.; Kropinski, A.M.; Ackermann, H.; Siragusa, G.R. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: Genomic organization and proteomic analysis of the virions. Arch. Virol. 2011, 156, 25–35. [Google Scholar] [CrossRef]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef]
- Liu, Y.; Mi, L.; Mi, Z.; Huang, Y.; Li, P.; Zhang, X.; Tong, Y.; Bai, C. Complete Genome Sequence of IME207, a Novel Bacteriophage Which Can Lyse Multidrug-Resistant Klebsiella pneumoniae and Salmonella. Genome Announc. 2016, 4, e01015–e01016. [Google Scholar] [CrossRef]
- Hooton, S.P.; Timms, A.R.; Rowsell, J.; Wilson, R.; Connerton, I.F. Salmonella Typhimurium-specific bacteriophage ΦSH19 and the origins of species specificity in the Vi01-like phage family. Virol. J. 2011, 8, 1–14. [Google Scholar] [CrossRef]
- Steinbacher, S.; Baxa, U.; Miller, S.; Weintraub, A.; Seckler, R.; Huber, R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. USA 1996, 93, 10584–10588. [Google Scholar] [CrossRef]

| Bacterial Strains | % of Positive Spot Test Against Salmonella Serovars and Other Bacterial Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| LPSTSA | LPSTSD | LPSTSF | LPST94 | LPSTSH | LPSTSK | LPSTSN | LPSTSP | LPSTSV | |
| Salmonella serovars | |||||||||
| Typhimurium (N = 7) | 85.7 | 100 | 100 | 100 | 85.7 | 71.4 | 71.4 | 85.7 | 85.7 |
| Enteritidis (N = 5) | 80 | 60 | 40 | 100 | 80 | 60 | 80 | 60 | 80 |
| Dublin (N = 2) | 50 | 100 | 100 | 100 | 50 | 100 | 50 | 100 | 50 |
| Choleraesuls (N = 1) | 0 | 100 | 0 | 100 | 100 | 0 | 0 | 100 | 100 |
| Newport (N = 1) | 100 | 0 | 100 | 100 | 0 | 0 | 100 | 0 | 0 |
| Paratyphi B (N = 1) | 100 | 100 | 0 | 100 | 100 | 100 | 0 | 100 | 100 |
| Anatum (N = 1) | 0 | 0 | 100 | 100 | 0 | 0 | 0 | 100 | 0 |
| Pullorum (N = 1) | 0 | 0 | 0 | 100 | 0 | 100 | 100 | 0 | 100 |
| Javiana (N = 1) | 0 | 100 | 100 | 100 | 0 | 0 | 100 | 0 | 100 |
| Kentucky (N = 1) | 100 | 0 | 100 | 100 | 100 | 0 | 100 | 100 | 0 |
| S. arizonae (N = 1) | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 100 | 100 |
| Drug-resistant Salmonella serovars | |||||||||
| Typhimurium (N = 10) | 60 | 80 | 90 | 100 | 80 | 70 | 90 | 80 | 90 |
| Enteritidis (N = 8) | 37.5 | 62.5 | 50.0 | 100 | 62.5 | 50.0 | 62.5 | 87.5 | 37.5 |
| Other bacterial strains | |||||||||
| E. coli (N = 6) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. hydrophila (N = 4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. sakazakii (N = 3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. flexneri (N = 1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| V. parahaemolyticus (N = 2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P. aeruginosa (N = 1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. aureus (N = 3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Listeria (N = 2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. Suis (N = 2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. acidophilus (N = 1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bacterial Strains | EOP of LPST94 | Drug-Resistance Salmonella | EOP of LPST94 |
|---|---|---|---|
| S.Enterica serovar Typhimurium | S.Enterica serovar Typhimurium | ||
| ATCC 14028 | +++ | LST10 | + |
| ATCC 13311 | +++ | LST11 | ++ |
| UK-1 | +++ | LST12 | ++ |
| ST8 | +++ | LST13 | ++ |
| SGSC 4903 | +++ | LST14 | + |
| SL 1344 | +++ | LST15 | + |
| LT2 | +++ | LST16 | + |
| S.enterica serovar Enteritidis | LST17 | ++ | |
| ATCC 13076 | ++ | LST18 | + |
| SJTUF 10978 | + | LST19 | + |
| SJTUF 10984 | + | S.enterica serovar Enteritidis | |
| LK5-3820 | ++ | LSE6 | + |
| SGSC 4901 | ++ | LSE7 | + |
| S.enterica serovar Dublin | LSE8 | ++ | |
| 3710 | + | LSE9 | ++ |
| 3723 | + | LSE10 | + |
| S. enterica serovar Choleraesuis | LSE11 | + | |
| ATCC 10708 | + | LSE12 | + |
| S. enterica serovar Newport | LSE15 | + | |
| E20002725 | + | ||
| S. enterica serovar Paratyphi B | |||
| CMCC 50094 | ++ | ||
| S. enterica Serovar Pullorum | |||
| CVCC 519 | + | ||
| S. enterica serovar Javiana | |||
| CVM 35943 | + | ||
| S. enterica serovar Anatum | |||
| ATCC 9270 | + | ||
| S. enterica serovar Kentucky | |||
| CVM 29188 | + | ||
| S. enterica Arizonae | |||
| CDC 346-86 | + | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.S.; Zhou, Y.; Liang, L.; Nime, I.; Yan, T.; Willias, S.P.; Mia, M.Z.; Bei, W.; Connerton, I.F.; Fischetti, V.A.; et al. Application of a Broad Range Lytic Phage LPST94 for Biological Control of Salmonella in Foods. Microorganisms 2020, 8, 247. https://doi.org/10.3390/microorganisms8020247
Islam MS, Zhou Y, Liang L, Nime I, Yan T, Willias SP, Mia MZ, Bei W, Connerton IF, Fischetti VA, et al. Application of a Broad Range Lytic Phage LPST94 for Biological Control of Salmonella in Foods. Microorganisms. 2020; 8(2):247. https://doi.org/10.3390/microorganisms8020247
Chicago/Turabian StyleIslam, Md. Sharifull, Yang Zhou, Lu Liang, Ishatur Nime, Ting Yan, Stephan P. Willias, Md. Zakaria Mia, Weicheng Bei, Ian F. Connerton, Vincent A. Fischetti, and et al. 2020. "Application of a Broad Range Lytic Phage LPST94 for Biological Control of Salmonella in Foods" Microorganisms 8, no. 2: 247. https://doi.org/10.3390/microorganisms8020247
APA StyleIslam, M. S., Zhou, Y., Liang, L., Nime, I., Yan, T., Willias, S. P., Mia, M. Z., Bei, W., Connerton, I. F., Fischetti, V. A., & Li, J. (2020). Application of a Broad Range Lytic Phage LPST94 for Biological Control of Salmonella in Foods. Microorganisms, 8(2), 247. https://doi.org/10.3390/microorganisms8020247






