Characterization of Ocular Clinical Isolates of Pseudomonas aeruginosa from Non-Contact Lens Related Keratitis Patients from South India
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Culture
2.2. Antibiotic Susceptibility Test
2.3. Genotyping of Virulence Factors
2.4. Biofilm Assay
2.5. Swarming Assay
2.6. Pyoverdine Estimation
2.7. Culture of HCEC
2.8. Cytotoxicity Assay
2.9. Statistical Analysis
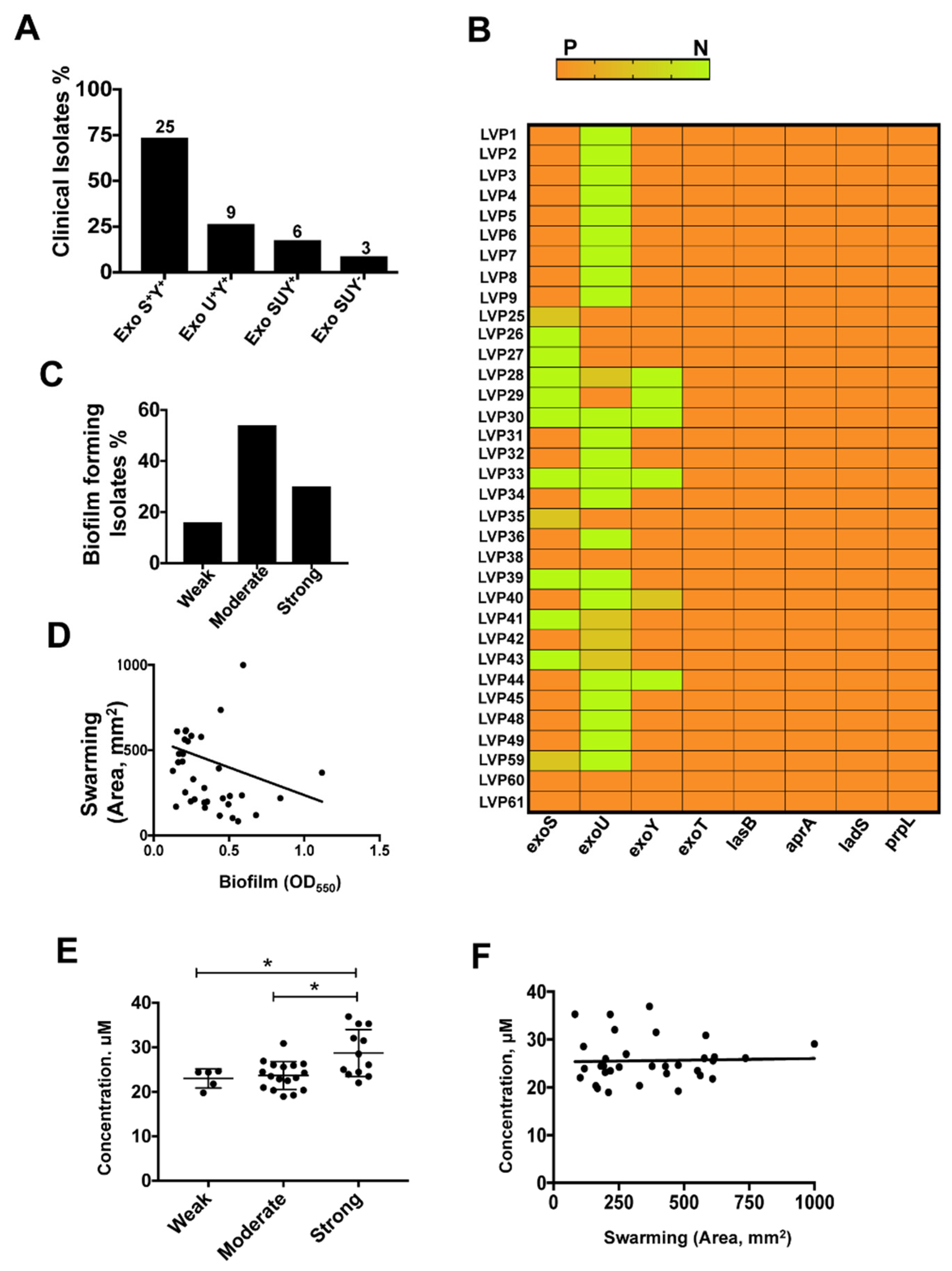
3. Results
3.1. Clinical Features
3.2. Antibiotic Susceptibility of the Clinical Isolates
3.3. Differential Expression of T3SS Genes among the Clinical Isolates of P. aeruginosa
3.4. Biofilm Assay
3.5. Swarming Motility is Linked to Biofilm Formation
3.6. Pyoverdine Secretion among Isolates
3.7. T3SS Positive Isolates Caused Increased Cell Death in HCEC
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chidambaram, J.D.; Venkatesh Prajna, N.; Srikanthi, P.; Lanjewar, S.; Shah, M.; Elakkiya, S.; Lalitha, P.; Burton, M.J. Epidemiology, risk factors, and clinical outcomes in severe microbial keratitis in South India. Ophthalmic Epidemiol 2018, 25, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Rudner, X.L.; Kernacki, K.A.; Barrett, R.P.; Hazlett, L.D. Prolonged elevation of IL-1 in Pseudomonas aeruginosa ocular infection regulates macrophage-inflammatory protein-2 production, polymorphonuclear neutrophil persistence, and corneal perforation. J. Immunol. 2000, 164, 6576–6582. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Karmakar, M.; Roy, S.; Ramadan, R.T.; Williams, S.R.; Howell, S.; Shive, C.L.; Han, Y.; Stopford, C.M.; Rietsch, A.; et al. TLR4 and TLR5 on corneal macrophages regulate Pseudomonas aeruginosa keratitis by signaling through MyD88-dependent and -independent pathways. J. Immunol. 2010, 185, 4272–4283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleiszig, S.M.; Wiener-Kronish, J.P.; Miyazaki, H.; Vallas, V.; Mostov, K.E.; Kanada, D.; Sawa, T.; Yen, T.S.; Frank, D.W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 1997, 65, 579–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ung, L.; Bispo, P.J.M.; Shanbhag, S.S.; Gilmore, M.S.; Chodosh, J. The persistent dilemma of microbial keratitis: Global burden, diagnosis, and antimicrobial resistance. Surv. Ophthalmol. 2019, 64, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef]
- Hauser, A.R. Pseudomonas aeruginosa: So many virulence factors, so little time. Crit. Care Med. 2011, 39, 2193–2194. [Google Scholar] [CrossRef] [Green Version]
- Barbieri, J.T.; Sun, J. Pseudomonas aeruginosa ExoS and ExoT. Rev. Physiol. Biochem. Pharmacol. 2004, 152, 79–92. [Google Scholar] [CrossRef]
- Sato, H.; Frank, D.W.; Hillard, C.J.; Feix, J.B.; Pankhaniya, R.R.; Moriyama, K.; Finck-Barbancon, V.; Buchaklian, A.; Lei, M.; Long, R.M.; et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003, 22, 2959–2969. [Google Scholar] [CrossRef] [Green Version]
- Yahr, T.L.; Vallis, A.J.; Hancock, M.K.; Barbieri, J.T.; Frank, D.W. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 1998, 95, 13899–13904. [Google Scholar] [CrossRef] [Green Version]
- Soscia, C.; Hachani, A.; Bernadac, A.; Filloux, A.; Bleves, S. Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 3124–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overhage, J.; Bains, M.; Brazas, M.D.; Hancock, R.E. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 2008, 190, 2671–2679. [Google Scholar] [CrossRef] [Green Version]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Callaghan, R.J.; Engel, L.S.; Hobden, J.A.; Callegan, M.C.; Green, L.C.; Hill, J.M. Pseudomonas keratitis. The role of an uncharacterized exoprotein, protease IV, in corneal virulence. Investig. Ophthalmol. Vis. Sci. 1996, 37, 534–543. [Google Scholar]
- Marquart, M.E.; Caballero, A.R.; Chomnawang, M.; Thibodeaux, B.A.; Twining, S.S.; O’Callaghan, R.J. Identification of a novel secreted protease from Pseudomonas aeruginosa that causes corneal erosions. Investig. Ophthalmol. Vis. Sci. 2005, 46, 3761–3768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, L.S.; Hill, J.M.; Caballero, A.R.; Green, L.C.; O’Callaghan, R.J. Protease IV, a unique extracellular protease and virulence factor from Pseudomonas aeruginosa. J. Biol. Chem. 1998, 273, 16792–16797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laarman, A.J.; Bardoel, B.W.; Ruyken, M.; Fernie, J.; Milder, F.J.; van Strijp, J.A.; Rooijakkers, S.H. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J. Immunol. 2012, 188, 386–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcorn, J.F.; Wright, J.R. Degradation of pulmonary surfactant protein D by Pseudomonas aeruginosa elastase abrogates innate immune function. J. Biol. Chem. 2004, 279, 30871–30879. [Google Scholar] [CrossRef] [Green Version]
- Skariyachan, S.; Sridhar, V.S.; Packirisamy, S.; Kumargowda, S.T.; Challapilli, S.B. Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal. Folia Microbiol. (Praha) 2018, 63, 413–432. [Google Scholar] [CrossRef]
- Broder, U.N.; Jaeger, T.; Jenal, U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2016, 2, 16184. [Google Scholar] [CrossRef]
- Ventre, I.; Goodman, A.L.; Vallet-Gely, I.; Vasseur, P.; Soscia, C.; Molin, S.; Bleves, S.; Lazdunski, A.; Lory, S.; Filloux, A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 171–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Subedi, D.; Vijay, A.K.; Willcox, M. Overview of mechanisms of antibiotic resistance inPseudomonas aeruginosa: An ocular perspective. Clin. Exp. Optom. 2018, 101, 162–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smitha, S.; Lalitha, P.; Prajna, V.N.; Srinivasan, M. Susceptibility trends of pseudomonas species from corneal ulcers. Indian J. Med. Microbiol. 2005, 23, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Samantaray, R.; Mallick, A.; Sahu, S.K.; Sharma, S. Types of organisms and in-vitro susceptibility of bacterial isolates from patients with microbial keratitis: A trend analysis of 8 years. Indian J. Ophthalmol. 2019, 67, 49–53. [Google Scholar] [CrossRef]
- Lakshmi Priya, J.; Prajna, L.; Mohankumar, V. Genotypic and phenotypic characterization of Pseudomonas aeruginosa isolates from post-cataract endophthalmitis patients. Microb. Pathog. 2015, 78, 67–73. [Google Scholar] [CrossRef]
- Oka, N.; Suzuki, T.; Ishikawa, E.; Yamaguchi, S.; Hayashi, N.; Gotoh, N.; Ohashi, Y. Relationship of Virulence Factors and Clinical Features in Keratitis Caused by Pseudomonas aeruginosa. Investig. Ophthalmol. Vis. Sci. 2015, 56, 6892–6898. [Google Scholar] [CrossRef] [Green Version]
- Murray, T.S.; Ledizet, M.; Kazmierczak, B.I. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J. Med. Microbiol. 2010, 59, 511–520. [Google Scholar] [CrossRef]
- Toska, J.; Sun, Y.; Carbonell, D.A.; Foster, A.N.; Jacobs, M.R.; Pearlman, E.; Rietsch, A. Diversity of virulence phenotypes among type III secretion negative Pseudomonas aeruginosa clinical isolates. PLoS ONE 2014, 9, e86829. [Google Scholar] [CrossRef]
- Karthikeyan, R.S.; Priya, J.L.; Leal, S.M., Jr.; Toska, J.; Rietsch, A.; Prajna, V.; Pearlman, E.; Lalitha, P. Host response and bacterial virulence factor expression in Pseudomonas aeruginosa and Streptococcus pneumoniae corneal ulcers. PLoS ONE 2013, 8, e64867. [Google Scholar] [CrossRef]
- Roy, S.; Marla, S.; Praneetha, D.C. Recognition of Corynebacterium pseudodiphtheriticum by Toll-like receptors and up-regulation of antimicrobial peptides in human corneal epithelial cells. Virulence 2015, 6, 716–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopinathan, U.; Sharma, S.; Garg, P.; Rao, G.N. Review of epidemiological features, microbiological diagnosis and treatment outcome of microbial keratitis: Experience of over a decade. Indian J. Ophthalmol. 2009, 57, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Bonfield, T.; Tartakoff, A.M. Non-apoptotic toxicity of Pseudomonas aeruginosa toward murine cells. PLoS ONE 2013, 8, e54245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 25th Informational Supplement; CLSI Document M100-S25; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2005. [Google Scholar] [CrossRef] [Green Version]
- Choy, M.H.; Stapleton, F.; Willcox, M.D.; Zhu, H. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens- and non-contact lens-related keratitis. J. Med. Microbiol. 2008, 57, 1539–1546. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Wilderman, P.J.; Vasil, A.I.; Johnson, Z.; Wilson, M.J.; Cunliffe, H.E.; Lamont, I.L.; Vasil, M.L. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 2001, 69, 5385–5394. [Google Scholar] [CrossRef] [Green Version]
- Araki-Sasaki, K.; Ohashi, Y.; Sasabe, T.; Hayashi, K.; Watanabe, H.; Tano, Y.; Handa, H. An SV40-immortalized human corneal epithelial cell line and its characterization. Investig. Ophthalmol. Vis. Sci. 1995, 36, 614–621. [Google Scholar]
- Ponsoda, X.; Jover, R.; Castell, J.V.; Gomez-Lechon, M.J. Measurement of intracellular LDH activity in 96-well cultures: A rapid and automated assay for cytotoxicity studies. J. Tissue Cult. Methods 1991, 13, 21–24. [Google Scholar] [CrossRef]
- Sharma, P.; Guha, S.; Garg, P.; Roy, S. Differential expression of antimicrobial peptides in corneal infection and regulation of antimicrobial peptides and reactive oxygen species by type III secretion system of Pseudomonas aeruginosa. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vareechon, C.; Zmina, S.E.; Karmakar, M.; Pearlman, E.; Rietsch, A. Pseudomonas aeruginosa Effector ExoS Inhibits ROS Production in Human Neutrophils. Cell Host Microbe 2017, 21, 611–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anantharajah, A.; Mingeot-Leclercq, M.P.; Van Bambeke, F. Targeting the Type Three Secretion System in Pseudomonas aeruginosa. Trends Pharmacol. Sci. 2016, 37, 734–749. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, R.S.; Roddam, L.F.; Merritt, A.; Reid, D.W.; Champion, A.C. Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 2010, 59, 881–890. [Google Scholar] [CrossRef]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Haussler, S. The LasB Elastase of Pseudomonas aeruginosa Acts in Concert with Alkaline Protease AprA To Prevent Flagellin-Mediated Immune Recognition. Infect. Immun. 2016, 84, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Lomholt, J.A.; Poulsen, K.; Kilian, M. Epidemic population structure of Pseudomonas aeruginosa: Evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect. Immun. 2001, 69, 6284–6295. [Google Scholar] [CrossRef] [Green Version]
- Verstraeten, N.; Braeken, K.; Debkumari, B.; Fauvart, M.; Fransaer, J.; Vermant, J.; Michiels, J. Living on a surface: Swarming and biofilm formation. Trends Microbiol. 2008, 16, 496–506. [Google Scholar] [CrossRef]
- Suzuki, T.; Okamoto, S.; Oka, N.; Hayashi, N.; Gotoh, N.; Shiraishi, A. Role of pvdE Pyoverdine Synthesis in Pseudomonas aeruginosa Keratitis. Cornea 2018, 37 (Suppl. 1), S99–S105. [Google Scholar] [CrossRef]
- Kang, D.; Turner, K.E.; Kirienko, N.V. PqsA Promotes Pyoverdine Production via Biofilm Formation. Pathogens 2017, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Shaver, C.M.; Hauser, A.R. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 2004, 72, 6969–6977. [Google Scholar] [CrossRef] [Green Version]
- Winstanley, C.; Kaye, S.B.; Neal, T.J.; Chilton, H.J.; Miksch, S.; Hart, C.A.; Microbiology Ophthalmic, G. Genotypic and phenotypic characteristics of Pseudomonas aeruginosa isolates associated with ulcerative keratitis. J. Med. Microbiol. 2005, 54, 519–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowell, B.A.; Weissman, B.A.; Yeung, K.K.; Johnson, L.; Ho, S.; Van, R.; Bruckner, D.; Mondino, B.; Fleiszig, S.M. Phenotype of Pseudomonas aeruginosa isolates causing corneal infection between 1997 and 2000. Cornea 2003, 22, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Karmakar, M.; Taylor, P.R.; Rietsch, A.; Pearlman, E. ExoS and ExoT ADP ribosyltransferase activities mediate Pseudomonas aeruginosa keratitis by promoting neutrophil apoptosis and bacterial survival. J. Immunol. 2012, 188, 1884–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agnello, M.; Wong-Beringer, A. Differentiation in quinolone resistance by virulence genotype in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e42973. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.H.; Kwon, K.C.; Kim, S.; Koo, S.H. Correlation between virulence genotype and fluoroquinolone resistance in carbapenem-resistant Pseudomonas aeruginosa. Ann. Lab. Med. 2014, 34, 286–292. [Google Scholar] [CrossRef] [Green Version]
- Subedi, D.; Vijay, A.K.; Kohli, G.S.; Rice, S.A.; Willcox, M. Association between possession of ExoU and antibiotic resistance in Pseudomonas aeruginosa. PLoS ONE 2018, 13, e0204936. [Google Scholar] [CrossRef] [Green Version]
- Horna, G.; Amaro, C.; Palacios, A.; Guerra, H.; Ruiz, J. High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant “high-risk clones” of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals. Sci. Rep. 2019, 9, 10874. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, J.; Deziel, E. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genom. 2010, 11, 587. [Google Scholar] [CrossRef] [Green Version]
- Lai, S.; Tremblay, J.; Deziel, E. Swarming motility: A multicellular behaviour conferring antimicrobial resistance. Environ. Microbiol. 2009, 11, 126–136. [Google Scholar] [CrossRef]
- Baraquet, C.; Murakami, K.; Parsek, M.R.; Harwood, C.S. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012, 40, 7207–7218. [Google Scholar] [CrossRef]
- Caiazza, N.C.; Merritt, J.H.; Brothers, K.M.; O’Toole, G.A. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 2007, 189, 3603–3612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minandri, F.; Imperi, F.; Frangipani, E.; Bonchi, C.; Visaggio, D.; Facchini, M.; Pasquali, P.; Bragonzi, A.; Visca, P. Role of Iron Uptake Systems in Pseudomonas aeruginosa Virulence and Airway Infection. Infect. Immun. 2016, 84, 2324–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, W.; Yoon, S.S. Virulence Characteristics and an Action Mode of Antibiotic Resistance in Multidrug-Resistant Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 487. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Virulence Genes (Product Length) | Primers (5′-3′) |
|---|---|
| exoS (235 bp) | FWD: AGAGCGAGGTCAGCAGAGTA REV: GCGGACATACCTTGGTCGAT |
| exoT (219 bp) | FWD: GCATGCGGTAATGGACAAGG REV: GACCGATTCAGGTGCTGGTA |
| exoU (134 bp) | FWD: CGGTACGTGCTGTATCCCTC REV: CGTGTAGCGCGATCTGTAGT |
| exoY (289 bp) | FWD: GCTTCTCGGTGAAGGGGAAA REV: CGAACTCATAGCGTTTGCCG |
| lasB (202 bp) | FWD: ATCGACGTGTCCAAACTCCC REV: CCTTGACTTCGGTGATGGCT |
| aprA (176 bp) | FWD: CTACAGCGCCAACGTCAATC REV: AGCTCATCACCGAATAGGCG |
| ladS (181 bp) | FWD: CCCTGATGGTCCTCGGCTAC REV: GTTCCTGGTTCAGCGCTTCC |
| pscL | FWD: AAAAAAGAATTCGGAGGGCGATGAATGCTTCCATTTGTT REV: AAAAAAAAGCTTTCAACCGGCGTCCCCTTCCTCCT |
| pscU | FWD: AAAAAATCTAGAGGAGGAGACGCCATGAGCGCCGAGAAGA REV: AAAAAAAAGCTTGATAGCGATCAGGGCGTATCCGTCTGCT |
| prpL | FWD: ATCGTATTTCGCCGACTCCC REV: TGAAGACCATCTTCGCCACC |
| Antibiotic | MIC (μg/mL) | % Isolates | ||
|---|---|---|---|---|
| Susceptible (S) | Intermediate (I) | Resistant (R) | ||
| Chloramphenicol (CHL) | 0.016–256 | 6 | 6 | 88 |
| Ciprofloxacin (CIP) | 0.25–4 | 70 | 12 | 18 |
| Moxifloxacin (MXF) | 0.002–32 | 12 | 6 | 82 |
| Gatifloxacin (GAT) | 0.002–32 | 79 | 0 | 21 |
| Ofloxacin (OFX) | 0.002–32 | 79 | 0 | 21 |
| Levofloxacin (LEV) | 0.12–8 | 62 | 0 | 38 |
| Gentamycin (GEM) | 1–16 | 82 | 0 | 18 |
| Amikacin (AKN) | 2–64 | 85 | 0 | 15 |
| Tobramycin (TOB) | 0.016–256 | 82 | 0 | 18 |
| Colistin (CS) | 0.5–16 | 100 | 0 | 0 |
| Ceftazidime (CAZ) | 1–64 | 62 | 0 | 38 |
| Cefepime (CEP) | 1–64 | 67 | 18 | 15 |
| Imipenem (IPM) | 0.25–16 | 85 | 0 | 15 |
| Doripenem (DOR) | 0.12–8 | 79 | 9 | 12 |
| Meropenem (MEM) | 0.25–16 | 85 | 3 | 12 |
| Piperacillin/Tazobactam (TZP) | 4/4/–128/4 | 35 | 15 | 50 |
| Ticarcillin/Clavulanic Acid (TIM) | 8/2–128/2 | 9 | 44 | 47 |
| Cefoperazone/Sublactam (CPZ) | 8–64 | 32 | 44 | 24 |
| Tigercycline (TGC) | 0.5–8 | 0 | 0 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dave, A.; Samarth, A.; Karolia, R.; Sharma, S.; Karunakaran, E.; Partridge, L.; MacNeil, S.; Monk, P.N.; Garg, P.; Roy, S. Characterization of Ocular Clinical Isolates of Pseudomonas aeruginosa from Non-Contact Lens Related Keratitis Patients from South India. Microorganisms 2020, 8, 260. https://doi.org/10.3390/microorganisms8020260
Dave A, Samarth A, Karolia R, Sharma S, Karunakaran E, Partridge L, MacNeil S, Monk PN, Garg P, Roy S. Characterization of Ocular Clinical Isolates of Pseudomonas aeruginosa from Non-Contact Lens Related Keratitis Patients from South India. Microorganisms. 2020; 8(2):260. https://doi.org/10.3390/microorganisms8020260
Chicago/Turabian StyleDave, Alpana, Apurwa Samarth, Roshni Karolia, Savitri Sharma, Esther Karunakaran, Lynda Partridge, Sheila MacNeil, Peter N. Monk, Prashant Garg, and Sanhita Roy. 2020. "Characterization of Ocular Clinical Isolates of Pseudomonas aeruginosa from Non-Contact Lens Related Keratitis Patients from South India" Microorganisms 8, no. 2: 260. https://doi.org/10.3390/microorganisms8020260





