Effects of Composting Different Types of Organic Fertilizer on the Microbial Community Structure and Antibiotic Resistance Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Manure and Composting
2.2. Analysis of Physicochemical Properties and Antibiotic Residues
2.3. DNA Extraction from Organic Fertilizer
2.4. 16S rRNA Gene Amplification, Illumina Sequencing, and Bioinformatic Analysis
2.5. Real-Time Quantitative PCR and High-Throughput Quantitative PCR Analysis
2.6. Statistical Analysis
3. Results
3.1. Antibiotic Residues in Organic Fertilizer
3.2. Physicochemical Properties of Organic Fertilizer
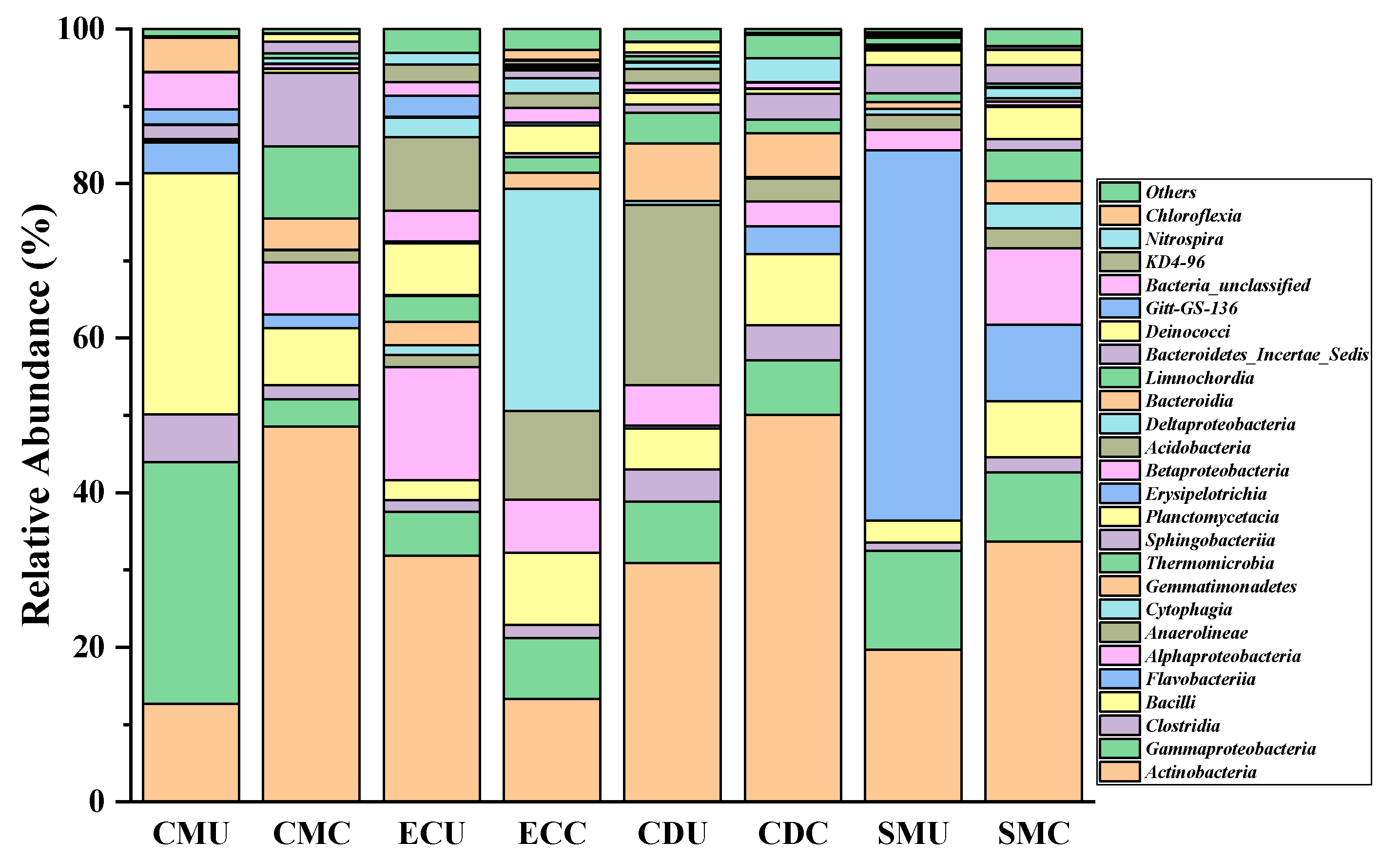
3.3. Structure and Characteristics of Microorganisms in Organic Fertilizer
3.3.1. Diversity of Microorganisms
3.3.2. Composition of Microorganisms
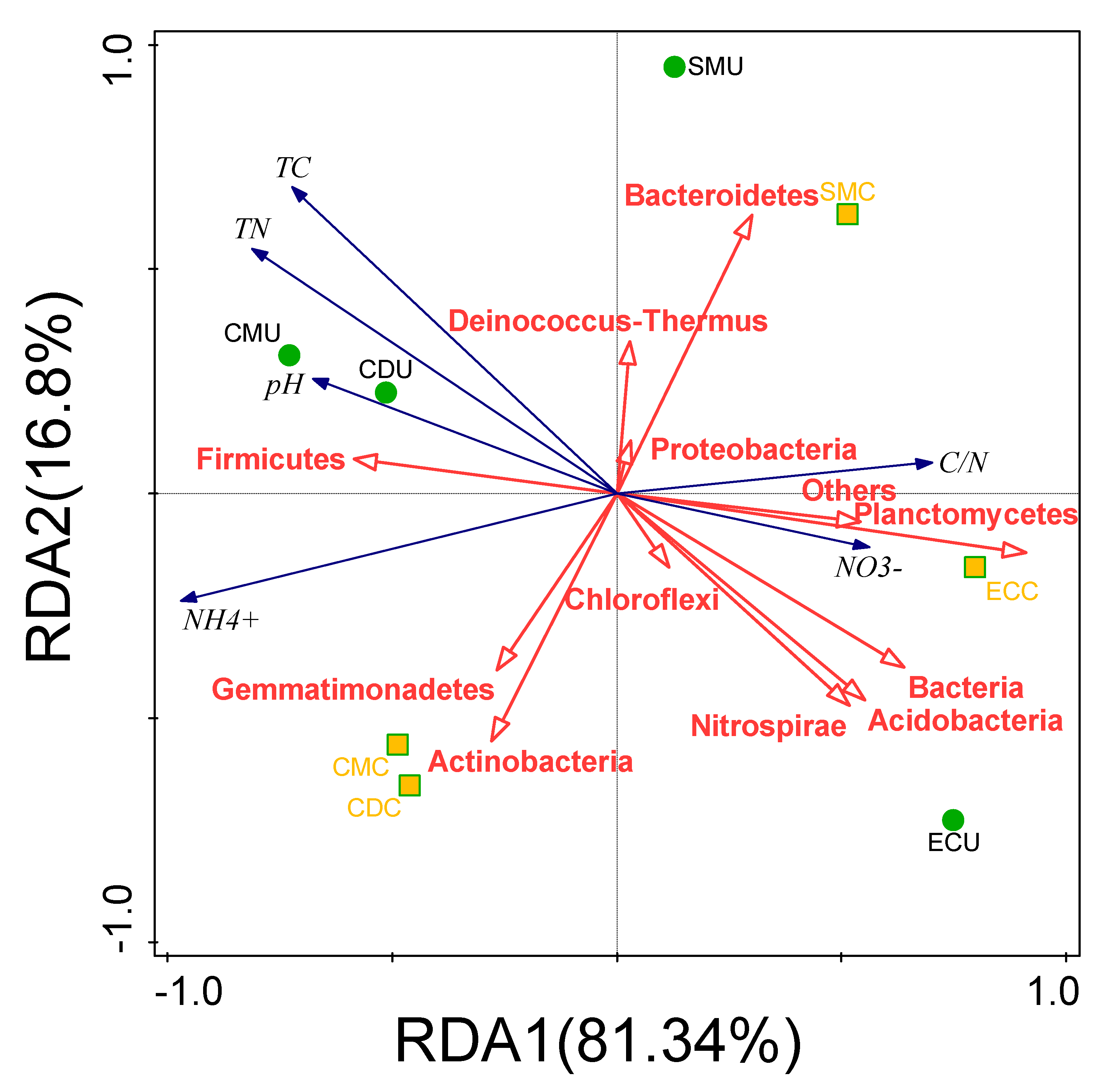
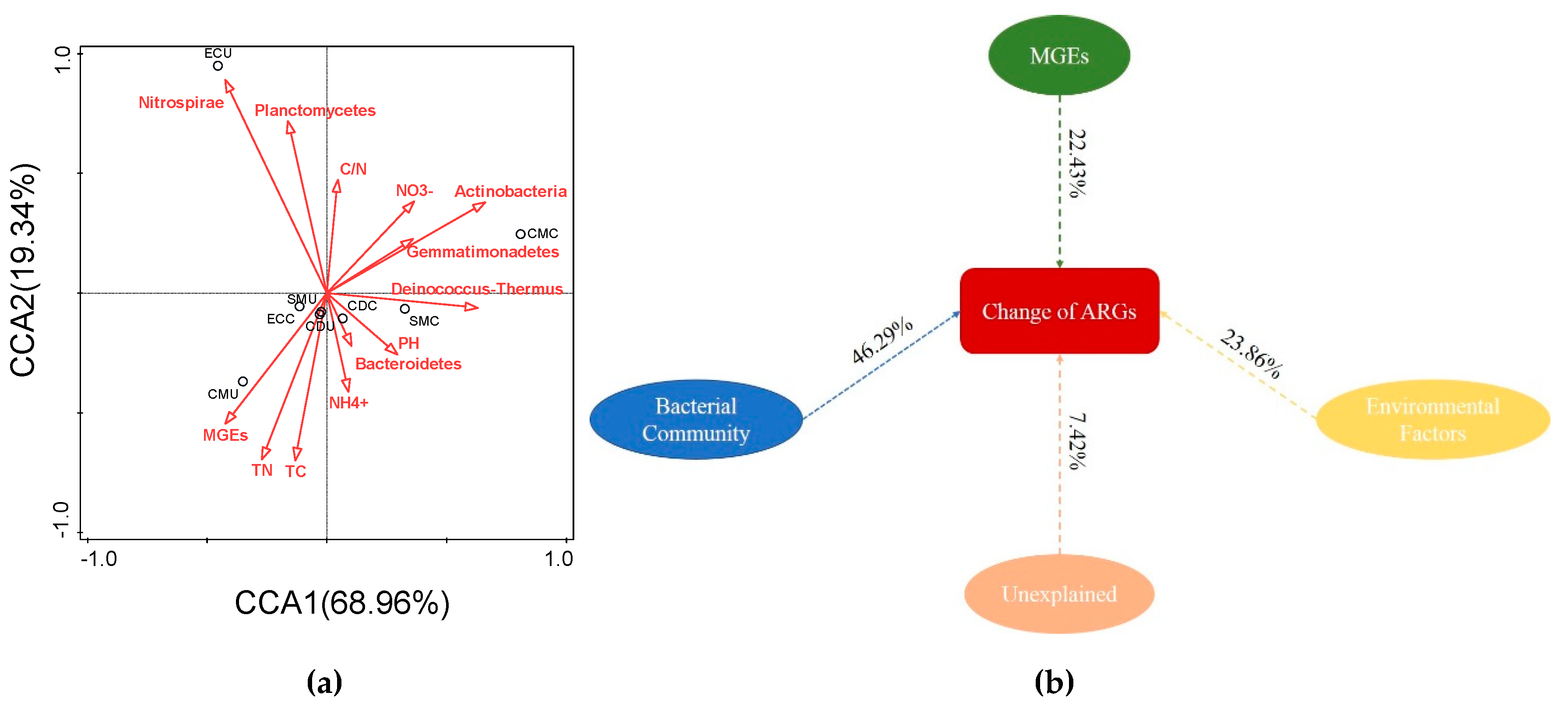
3.3.3. Correlation between Physicochemical Properties and Microorganisms
3.4. ARGs in Organic Fertilizer
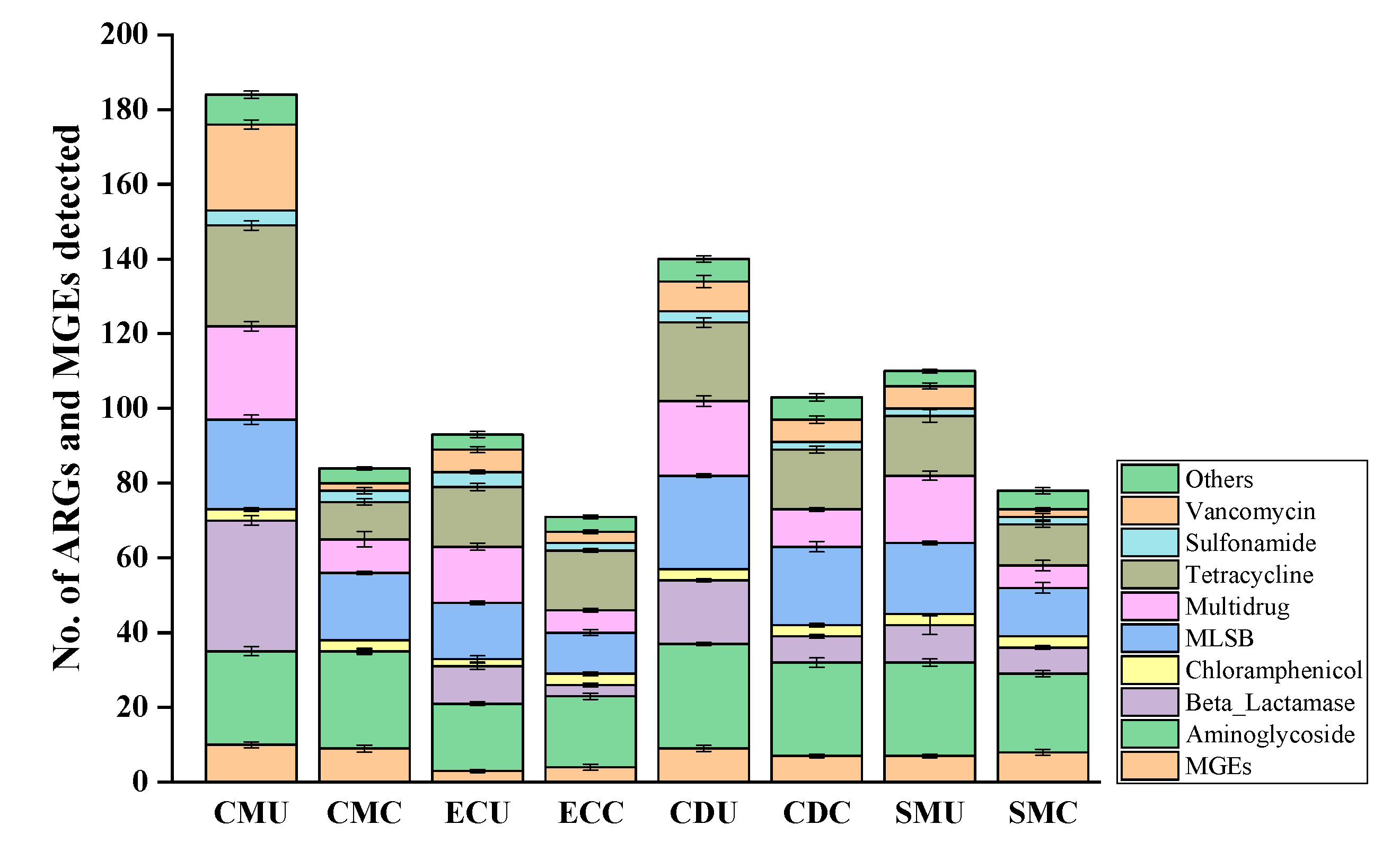
3.4.1. Diversity of ARGs and MGEs in Organic Fertilizer
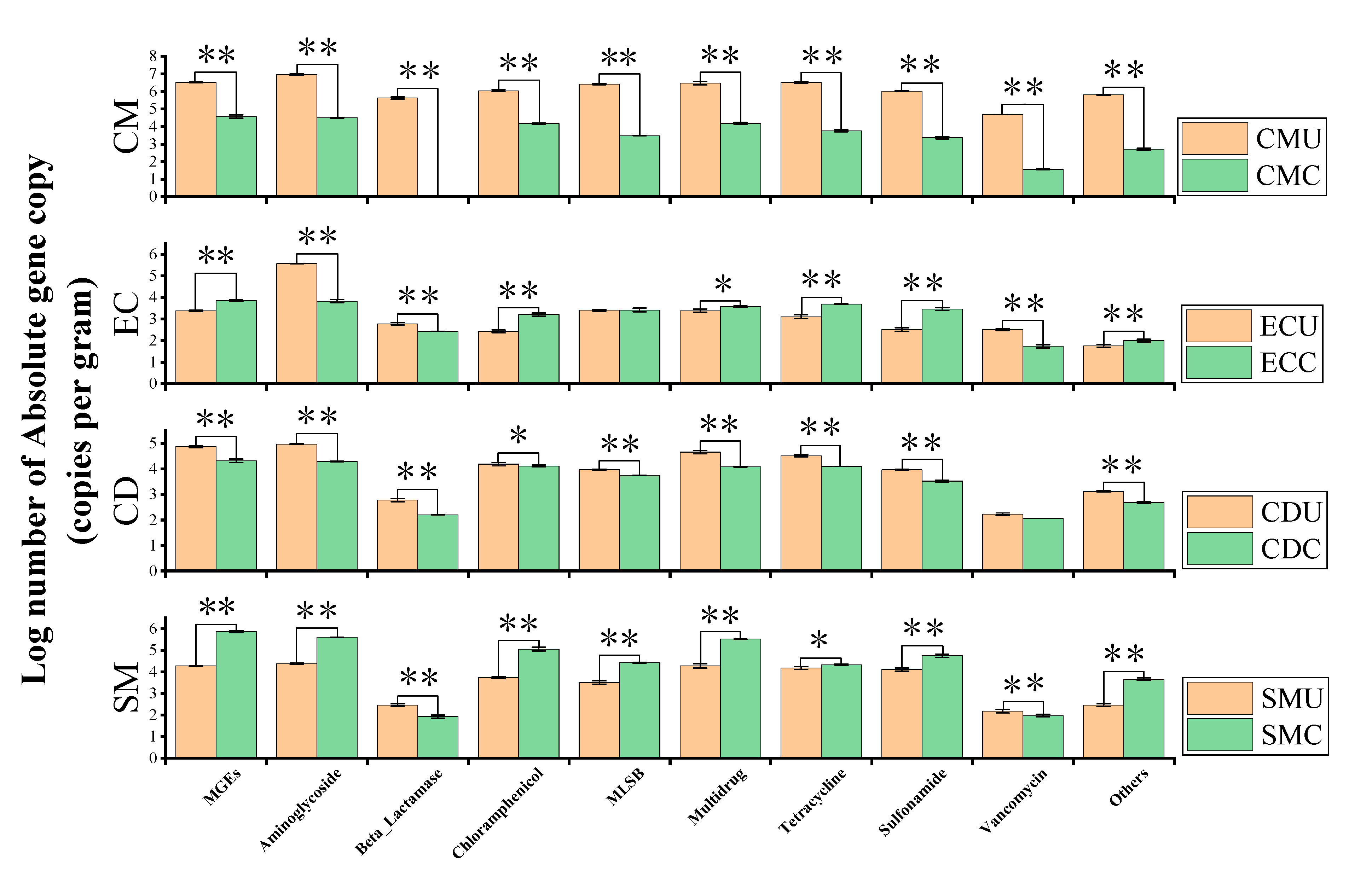
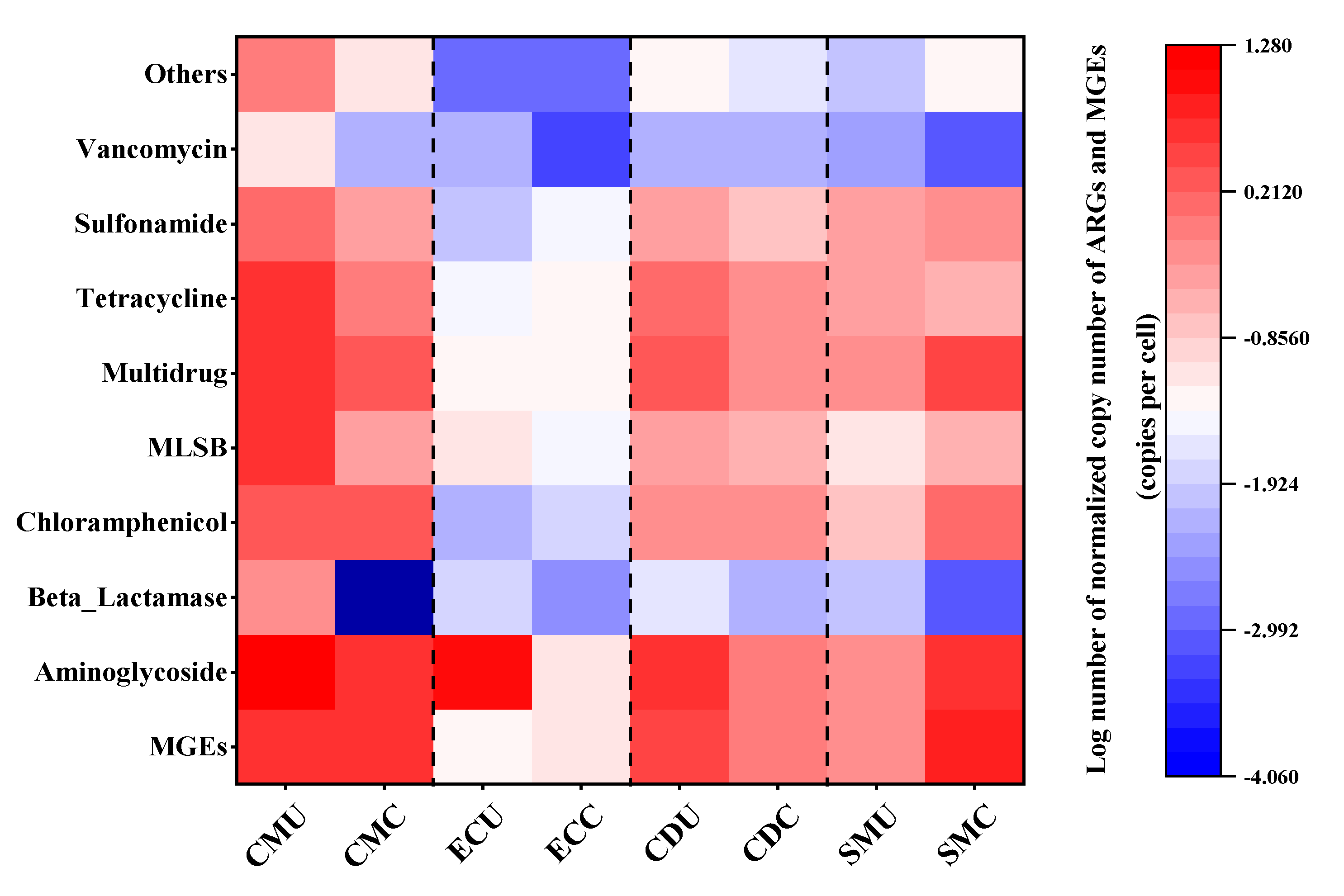
3.4.2. Abundance and Enrichment of ARGs and MGEs in Organic Fertilizer
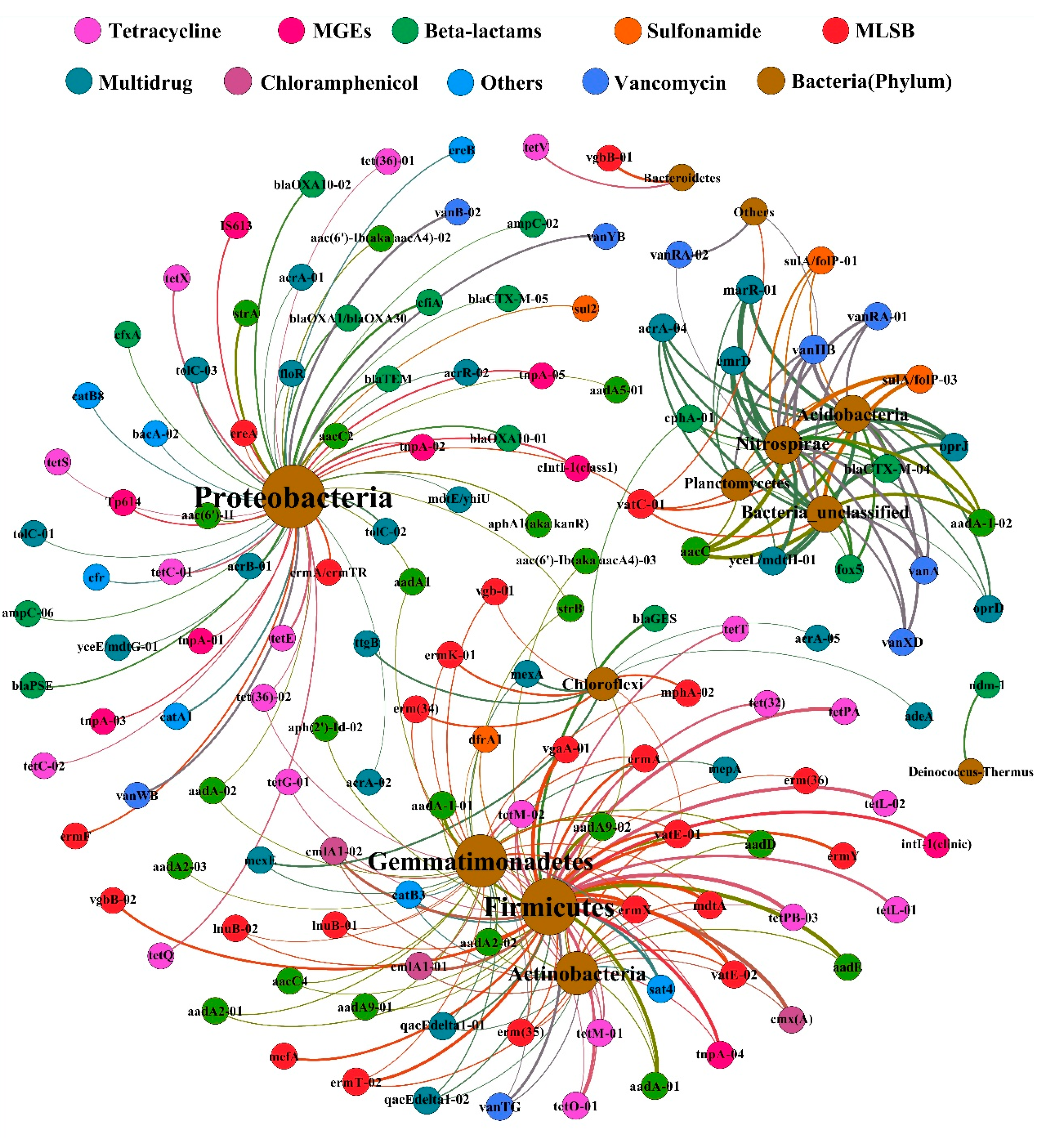
3.4.3. Correlation between ARGs and Microorganisms
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Harada, K.; Suzuki, M.; Kameda, M.; Mitsuhashi, S. On the drug-resistance of enteric bacteria. 2) Transmission of the drug-resistance among Enterobacteriaceae. Jpn. J. Exp. Med. 1960, 30, 289–299. [Google Scholar] [PubMed]
- World Health Organization. Overcoming antimicrobial resistance. Am. Heart J. 2000, 26, 283. [Google Scholar]
- Zuccato, E.; Castiglioni, S.; Bagnati, R.; Melis, M.; Fanelli, R. Source, occurrence and fate of antibiotics in the Italian aquatic environment. J. Hazard. Mater. 2010, 179, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.G.; Cissell, R.; Liamthong, S. Antibiotic Resistance in Bacteria Associated with Food Animals: A United States Perspective of Livestock Production. Foodborne Pathog. Dis. 2007, 4, 115–133. [Google Scholar] [CrossRef] [Green Version]
- Thiele-Bruhn, S. Pharmaceutical antibiotic compounds in soils—A review. J. Plant Nutr. Soil Sci. 2003, 166, 145–167. [Google Scholar] [CrossRef]
- Karcı, A.; Balcıoğlu, I.A.; Karci, A.; Balcioglu, I.A. Investigation of the tetracycline, sulfonamide, and fluoroquinolone antimicrobial compounds in animal manure and agricultural soils in Turkey. Sci. Total. Environ. 2009, 407, 4652–4664. [Google Scholar] [CrossRef]
- Jensen, J. Veterinary Medicines and Soil Quality: The Danish Situation as an Example. ACS Symposium Series 2001, 791, 282–302. [Google Scholar]
- Tollefson, L.; Fedorka-Cray, P.J.; Angulo, F.J. Public health aspects of antibiotic resistance monitoring in the USA. Acta Veter- Scand. Suppl. 1999, 92, 67–75. [Google Scholar]
- Morris, A.K.; Masterton, R.G. Antibiotic resistance surveillance: action for international studies. J. Antimicrob. Chemother. 2002, 49, 7–10. [Google Scholar] [CrossRef] [Green Version]
- Hvistendahl, M. China Takes Aim at Rampant Antibiotic Resistance. Sci. 2012, 336, 795. [Google Scholar] [CrossRef]
- Su, H.-C.; Ying, G.-G.; Tao, R.; Zhang, R.-Q.; Fogarty, L.R.; Kolpin, D.W. Occurrence of antibiotic resistance and characterization of resistance genes and integrons in Enterobacteriaceae isolated from integrated fish farms in south China. J. Environ. Monit. 2011, 13, 3229. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.H.; Ma, W.; Dou, Z.; Ma, L.; Liu, X.L.; Xu, J.X.; Zhang, F.S. The estimation of the production amount of animal manure and its environmental effect in China [in Chinese]. Zhongguo Huanjing Kexue/China Environ. Sci. 2006, 26, 614–617. [Google Scholar]
- Alcock, R.; Sweetman, A.; Jones, K.C. Assessment of organic contanhnant fate in waste water treatment plants I: Selected compounds and physicochemical properties. Chemosphere 1999, 38, 2247–2262. [Google Scholar] [CrossRef]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.; Alm, E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nat. 2011, 480, 241–244. [Google Scholar] [CrossRef]
- Forsberg, K.; Reyes, A.; Wang, B.; Selleck, E.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Sci. 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Li, J.; Chen, P.; Ding, R.; Zhang, P.; Li, X. Occurrence of antibiotics and antibiotic resistances in soils from wastewater irrigation areas in Beijing and Tianjin, China. Environ. Pollut. 2014, 193, 94–101. [Google Scholar] [CrossRef]
- Sun, W.; Qian, X.; Gu, J.; Wang, X.-J.; Duan, M.-L. Mechanism and Effect of Temperature on Variations in Antibiotic Resistance Genes during Anaerobic Digestion of Dairy Manure. Sci. Rep. 2016, 6, 30237. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Carballo, E.; Barreiro, C.G.; Scharf, S.; Gans, O. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 2007, 148, 570–579. [Google Scholar] [CrossRef]
- Christian, T.; Schneider, R.J.; Färber, H.; Skutlarek, D.; Meyer, M.T.; Goldbach, H.E. Determination of Antibiotic Residues in Manure, Soil, and Surface Waters. Acta Hydrochim. et Hydrobiol. 2003, 31, 36–44. [Google Scholar] [CrossRef]
- Hamscher, G.; Pawelzick, H.T.; Höper, H.; Nau, H. Different behavior of tetracyclines and sulfonamides in sandy soils after repeated fertilization with liquid manure. Environ. Toxicol. Chem. 2005, 24, 861–868. [Google Scholar] [CrossRef]
- Le, T.; Munekage, Y.; Kato, S. Antibiotic resistance in bacteria from shrimp farming in mangrove areas. Sci. Total. Environ. 2005, 349, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Tien, Y.-C.; Li, B.; Zhang, T.; Scott, A.; Murray, R.; Sabourin, L.; Marti, R.; Topp, E. Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci. Total. Environ. 2017, 581, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Shi, P.; Hu, Q.; Li, B.; Zhang, T.; Zhang, X.-X. Bacterial Community Shift Drives Antibiotic Resistance Promotion during Drinking Water Chlorination. Environ. Sci. Technol. 2015, 49, 12271–12279. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Li, L.; Yang, S.; Lan, J.; He, H.; McElmurry, S.; Zhao, Y. Sulfamethoxazole, tetracycline and oxytetracycline and related antibiotic resistance genes in a large-scale landfill, China. Sci. Total. Environ. 2016, 551, 9–15. [Google Scholar] [CrossRef]
- Nelson, K.L.; Brozel, V.S.; Gibson, S.A.; Thaler, R.; Clay, S.A. Influence of manure from pigs fed chlortetracycline as growth promotant on soil microbial community structure. World J. Microb Biot 2011, 27, 659–668. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Wegener, H.C. The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microbes Infect. 1999, 1, 639–644. [Google Scholar] [CrossRef]
- Liu, C.; Wang, C.; Yao, H. Comprehensive Resource Utilization of Waste Using the Black Soldier Fly (Hermetia illucens (L.)) (Diptera: Stratiomyidae). Animals 2019, 9, 349. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zheng, N.; Wang, J.; Yao, H.; Qiu, Q.; Chapman, S.J. High turnover rate of free phospholipids in soil confirms the classic hypothesis of PLFA methodology. Soil Boil. Biochem. 2019, 135, 323–330. [Google Scholar] [CrossRef]
- Qian, M.; Wu, H.; Wang, J.; Zhang, H.; Zhang, Z.; Zhang, Y.; Lin, H.; Ma, J. Occurrence of trace elements and antibiotics in manure-based fertilizers from the Zhejiang Province of China. Sci. Total. Environ. 2016, 559, 174–181. [Google Scholar] [CrossRef]
- Lin, H.; Zhang, J.; Chen, H.; Wang, J.; Sun, W.; Zhang, X.; Yang, Y.; Wang, Q.; Ma, J. Effect of temperature on sulfonamide antibiotics degradation, and on antibiotic resistance determinants and hosts in animal manures. Sci. Total. Environ. 2017, 607, 725–732. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLOS ONE 2011, 6, e27310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, N.; An, X.-L.; Chen, Q.-L.; Yang, X.-R.; Christie, P.; Ke, X.; Wu, L.; Zhu, Y.-G. Antibiotics Disturb the Microbiome and Increase the Incidence of Resistance Genes in the Gut of a Common Soil Collembolan. Environ. Sci. Technol. 2018, 52, 3081–3090. [Google Scholar] [CrossRef]
- Li, Y.; Liao, H.; Yao, H. Prevalence of Antibiotic Resistance Genes in Air-Conditioning Systems in Hospitals, Farms, and Residences. Int. J. Environ. Res. Public Heal. 2019, 16, 683. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.-L.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92, 1–10. [Google Scholar] [CrossRef]
- Long, X.-E.; Yao, H.; Huang, Y.; Wei, W.; Zhu, Y.-G. Phosphate levels influence the utilisation of rice rhizodeposition carbon and the phosphate-solubilising microbial community in a paddy soil. Soil Boil. Biochem. 2018, 118, 103–114. [Google Scholar] [CrossRef]
- Chen, Q.-L.; An, X.-L.; Zhu, Y.-G.; Su, J.-Q.; Gillings, M.; Ye, Z.-L.; Cui, L. Application of Struvite Alters the Antibiotic Resistome in Soil, Rhizosphere, and Phyllosphere. Environ. Sci. Technol. 2017, 51, 8149–8157. [Google Scholar] [CrossRef]
- Klappenbach, J.A. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001, 29, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Kembel, S.W.; Wu, M.; Eisen, J.A.; Green, J.L. Incorporating 16S Gene Copy Number Information Improves Estimates of Microbial Diversity and Abundance. PLoS Comput. Boil. 2012, 8, e1002743. [Google Scholar] [CrossRef] [PubMed]
- Fruchterman, T.M.J.; Reingold, E.M. Graph drawing by force-directed placement. Software Pr. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. ICWSM 2009, 8, 361–362. [Google Scholar]
- Kolz, A.C.; Moorman, T.B.; Ong, S.K.; Scoggin, K.D.; Douglass, E.A. Degradation and metabolite production of tylosin in anaerobic and aerobic swine-manure lagoons. Water Environ. Res. 2005, 77, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Kolz, A.C.; Ong, S.K.; Moorman, T. Sorption of tylosin onto swine manure. Chemosphere 2005, 60, 284–289. [Google Scholar] [CrossRef]
- Loke, M.-L.; Tjørnelund, J.; Halling-Sørensen, B. Determination of the distribution coefficient (log Kd) of oxytetracycline, tylosin A, olaquindox and metronidazole in manure. Chemosphere 2002, 48, 351–361. [Google Scholar] [CrossRef]
- Tolls, J. Sorption of veterinary pharmaceuticals in soils: a review. Environ. Sci. Technol. 2001, 35, 3397–3406. [Google Scholar] [CrossRef]
- Boxall, A.B.; Blackwell, P.; Cavallo, R.; Kay, P.; Tolls, J. The sorption and transport of a sulphonamide antibiotic in soil systems. Toxicol. Lett. 2002, 131, 19–28. [Google Scholar] [CrossRef]
- Boxall, A.B. Prioritisation of veterinary medicines in the UK environment. Toxicol. Lett. 2003, 142, 207–218. [Google Scholar] [CrossRef]
- Ziglam, H.; Finch, R.G. Limitations of presently available glycopeptides in the treatment of Gram-positive infection. Clin. Microbiol. Infect. 2001, 7, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.M.; Ullman, J.L.; Bary, A.; Cogger, C.G.; Teel, A.L.; Watts, R.J. Antibiotic Degradation During Thermophilic Composting. Water, Air, Soil Pollut. 2015, 226, 13. [Google Scholar] [CrossRef]
- Bin Ho, Y.; Zakaria, M.P.; Latif, P.A.; Saari, N. Degradation of veterinary antibiotics and hormone during broiler manure composting. Bioresour. Technol. 2013, 131, 476–484. [Google Scholar]
- Ghosh, S.; LaPara, T.M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007, 1, 191–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sengeløv, G.; Halling-Sørensen, B.; Aarestrup, F.M. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet. Microbiol. 2003, 95, 91–101. [Google Scholar]
- Chen, Q.-L.; An, X.-L.; Li, H.; Zhu, Y.-G.; Su, J.-Q.; Cui, L. Do manure-borne or indigenous soil microorganisms influence the spread of antibiotic resistance genes in manured soil? Soil Boil. Biochem. 2017, 114, 229–237. [Google Scholar] [CrossRef]
- Wang, J.; Ben, W.; Zhang, Y.; Yang, M.; Qiang, Z. Effects of thermophilic composting on oxytetracycline, sulfamethazine, and their corresponding resistance genes in swine manure. Environ. Sci. Process. Impacts 2015, 17, 1654–1660. [Google Scholar] [CrossRef]
- Youngquist, C.P.; Mitchell, S.M.; Cogger, C.G. Fate of Antibiotics and Antibiotic Resistance during Digestion and Composting: A Review. J. Environ. Qual. 2016, 45, 537–545. [Google Scholar] [CrossRef]
- Chen, J.; Michel, F.; Sreevatsan, S.; Morrison, M.; Yu, Z. Occurrence and Persistence of Erythromycin Resistance Genes (erm) and Tetracycline Resistance Genes (tet) in Waste Treatment Systems on Swine Farms. Microb. Ecol. 2010, 60, 479–486. [Google Scholar] [CrossRef]
- Su, J.-Q.; Wei, B.; Ou-Yang, W.-Y.; Huang, F.-Y.; Zhao, Y.; Xu, H.-J.; Zhu, Y.-G. Antibiotic Resistome and Its Association with Bacterial Communities during Sewage Sludge Composting. Environ. Sci. Technol. 2015, 49, 7356–7363. [Google Scholar] [CrossRef]
- Yang, Q.; Ren, S.; Niu, T.; Guo, Y.; Qi, S.; Han, X.; Liu, D.; Pan, F. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ. Sci Pollut Res. Int 2014, 21, 1231–1241; [Google Scholar] [CrossRef]
- Tong, Z.; Xu-Xiang, Z.; Lin, Y.J. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. Plos One 2011, 6, e26041. [Google Scholar]
- Karkman, A.; Johnson, T.A.; Lyra, C.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Genet. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Costa, V.M.; Griffiths, E.; Wright, G.D. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 2007, 10, 481–489. [Google Scholar] [CrossRef]
- Cheng, W.; Chen, H.; Su, C.; Yan, S. Abundance and persistence of antibiotic resistance genes in livestock farms: A comprehensive investigation in eastern China. Environ. Int. 2013, 61, 1–7. [Google Scholar] [CrossRef]
- Wang, F.-H.; Qiao, M.; Su, J.-Q.; Chen, Z.; Zhou, X.; Zhu, Y.-G. High Throughput Profiling of Antibiotic Resistance Genes in Urban Park Soils with Reclaimed Water Irrigation. Environ. Sci. Technol. 2014, 48, 9079–9085. [Google Scholar] [CrossRef]
- Stasinakis, A.S. Review on the fate of emerging contaminants during sludge anaerobic digestion. Bioresour. Technol. 2012, 121, 432–440. [Google Scholar] [CrossRef]
- An, J.; Chen, H.; Wei, S.; Gu, J. Antibiotic contamination in animal manure, soil, and sewage sludge in Shenyang, northeast China. Environ. Earth Sci. 2015, 74, 5077–5086. [Google Scholar] [CrossRef]
- Nilmini, B.; Masahiro, I.; Lateef, S.A.; Takaki, Y.; Ikko, I.; Kazutaka, U.J. The survival of multidrug-resistant bacteria in thermophilic and mesophilic anaerobic co-digestion of dairy manure and waste milk. Anim. Sci. J. 2013, 84, 426–433. [Google Scholar]
- Diehl, D.L.; LaPara, T.M. Effect of Temperature on the Fate of Genes Encoding Tetracycline Resistance and the Integrase of Class 1 Integrons within Anaerobic and Aerobic Digesters Treating Municipal Wastewater Solids. Environ. Sci. Technol. 2010, 44, 9128–9133. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Lu, X.; Rensing, C.; Friman, V.P.; Geisen, S.; Chen, Z.; Yu, Z.; Wei, Z.; Zhou, S.; Zhu, Y. Hyperthermophilic Composting Accelerates the Removal of Antibiotic Resistance Genes and Mobile Genetic Elements in Sewage Sludge. Environ. Sci Technol 2018, 52, 266–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Oda, Y.; Grewal, S.; Morrison, M.; Michel, F.C.; Yu, Z. Persistence of resistance to erythromycin and tetracycline in swine manure during simulated composting and lagoon treatments. Microb. Ecol. 2012, 63, 32–40; [Google Scholar] [CrossRef]
- Perreten, V.; Vorlet-Fawer, L.; Slickers, P.; Ehricht, R.; Kuhnert, P.; Frey, J. Microarray-Based Detection of 90 Antibiotic Resistance Genes of Gram-Positive Bacteria. J. Clin. Microbiol. 2005, 43, 2291–2302. [Google Scholar] [CrossRef] [Green Version]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. [Google Scholar]
- Stegmann, E.; Frasch, H.-J.; Kilian, R.; Pozzi, R. Self-resistance mechanisms of actinomycetes producing lipid II-targeting antibiotics. Int. J. Med Microbiol. 2015, 305, 190–195. [Google Scholar] [CrossRef]
- Forsberg, K.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Knight, R.; Fierer, N.; Dantas, G. Bacterial phylogeny structures soil resistomes across habitats. Nature 2014, 509, 612–616. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Yao, H. Effects of Composting Different Types of Organic Fertilizer on the Microbial Community Structure and Antibiotic Resistance Genes. Microorganisms 2020, 8, 268. https://doi.org/10.3390/microorganisms8020268
Zhou Z, Yao H. Effects of Composting Different Types of Organic Fertilizer on the Microbial Community Structure and Antibiotic Resistance Genes. Microorganisms. 2020; 8(2):268. https://doi.org/10.3390/microorganisms8020268
Chicago/Turabian StyleZhou, Zeming, and Huaiying Yao. 2020. "Effects of Composting Different Types of Organic Fertilizer on the Microbial Community Structure and Antibiotic Resistance Genes" Microorganisms 8, no. 2: 268. https://doi.org/10.3390/microorganisms8020268
APA StyleZhou, Z., & Yao, H. (2020). Effects of Composting Different Types of Organic Fertilizer on the Microbial Community Structure and Antibiotic Resistance Genes. Microorganisms, 8(2), 268. https://doi.org/10.3390/microorganisms8020268



