Functions of Enyolreductase (ER) Domains of PKS Cluster in Lipid Synthesis and Enhancement of PUFAs Accumulation in Schizochytrium limacinum SR21 Using Triclosan as a Regulator of ER
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. Vector Construction, Transformation and Screening
2.3. Biomass Determination, Lipid Extraction and Fatty Acid Analysis
2.4. Quantitative Real-Time PCR Analysis (RT-qPCR)
2.5. Enzyme Activities Analysis
2.6. GC-MS Analysis
2.7. Statistical Analysis
3. Results and Discussion
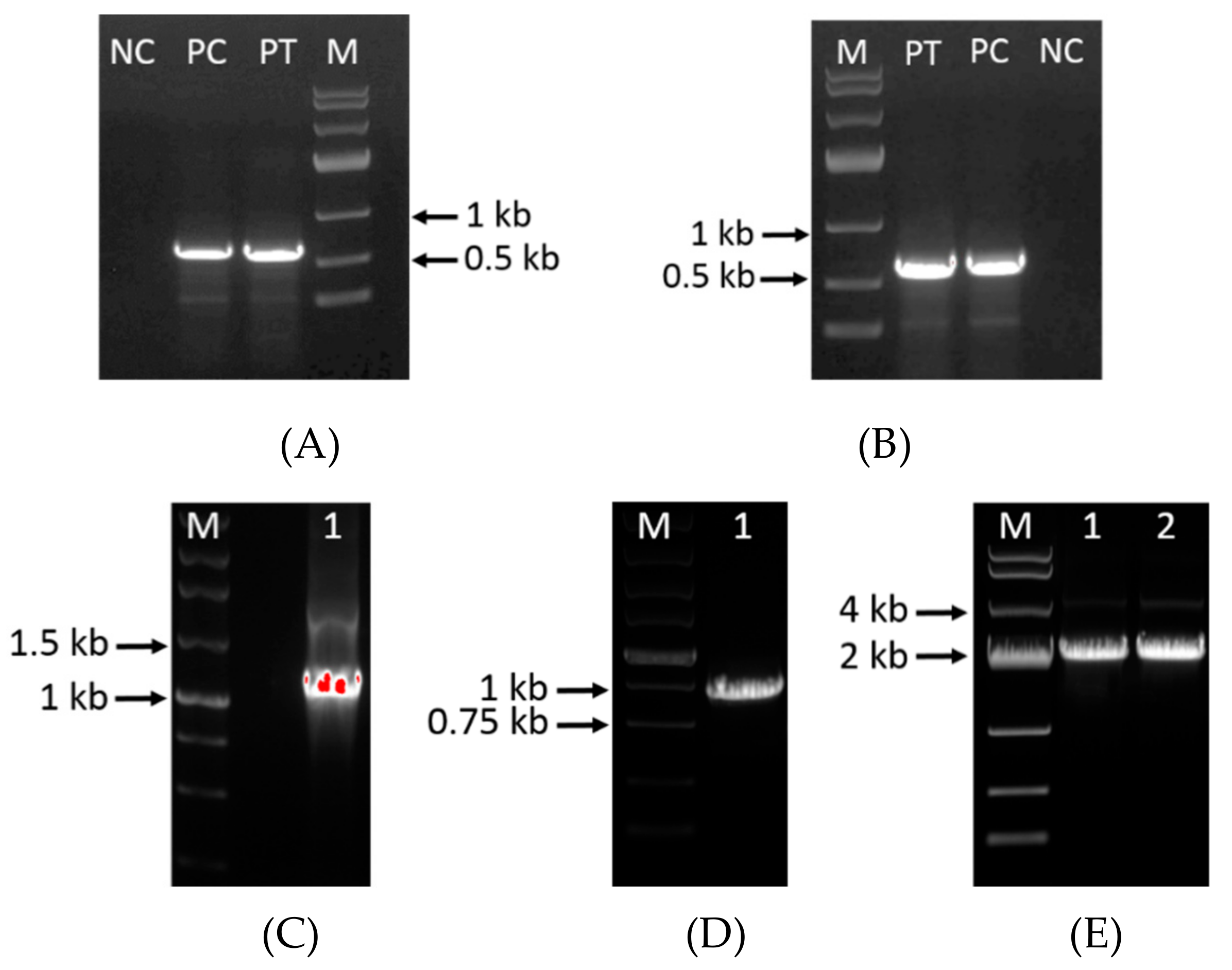
3.1. Construction of Deletion Mutants
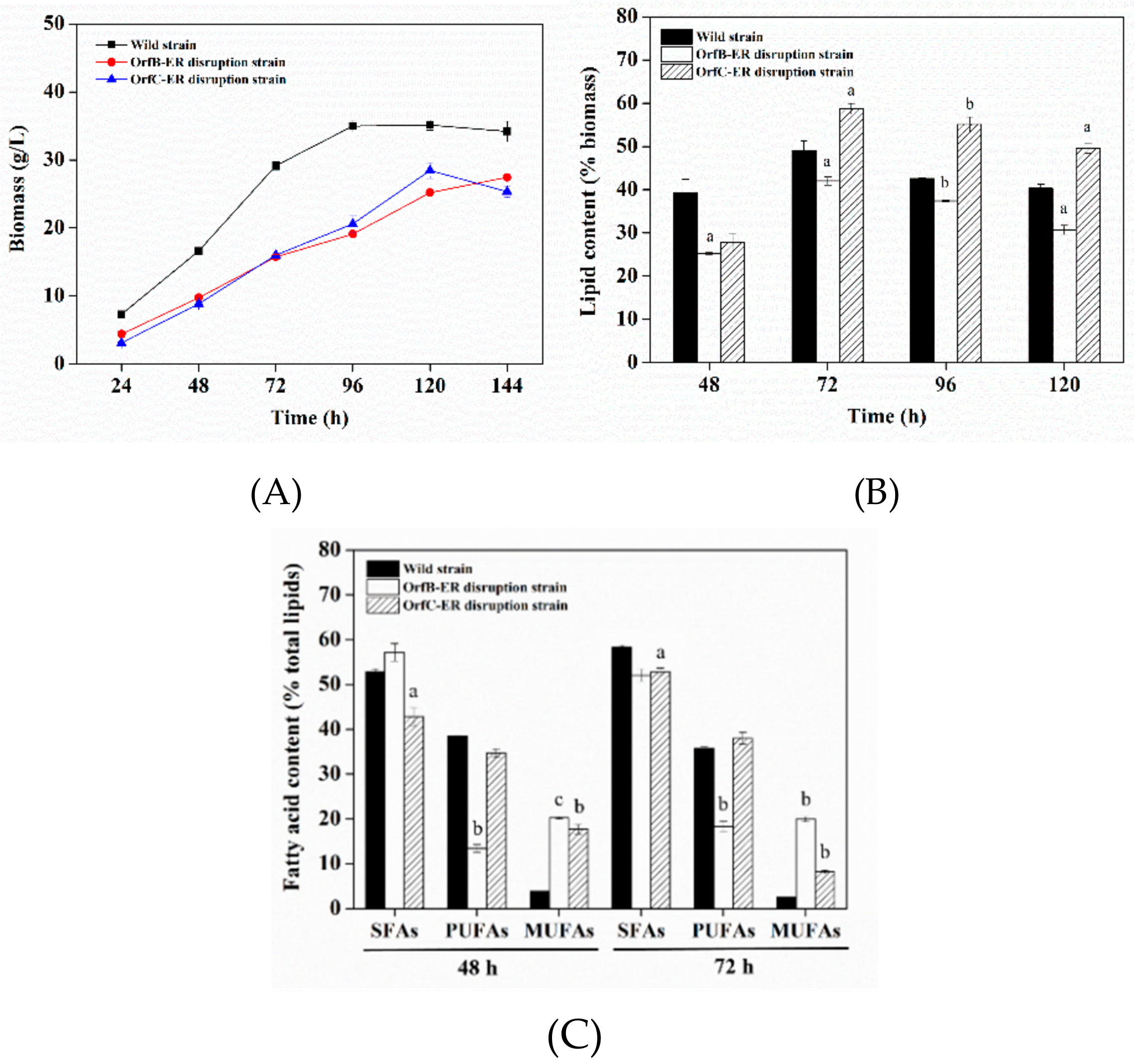
3.2. Biomass and Lipids Accumulation Analysis of Deletion Mutants
3.3. Lipid Profiles Analysis of Deletion Mutants
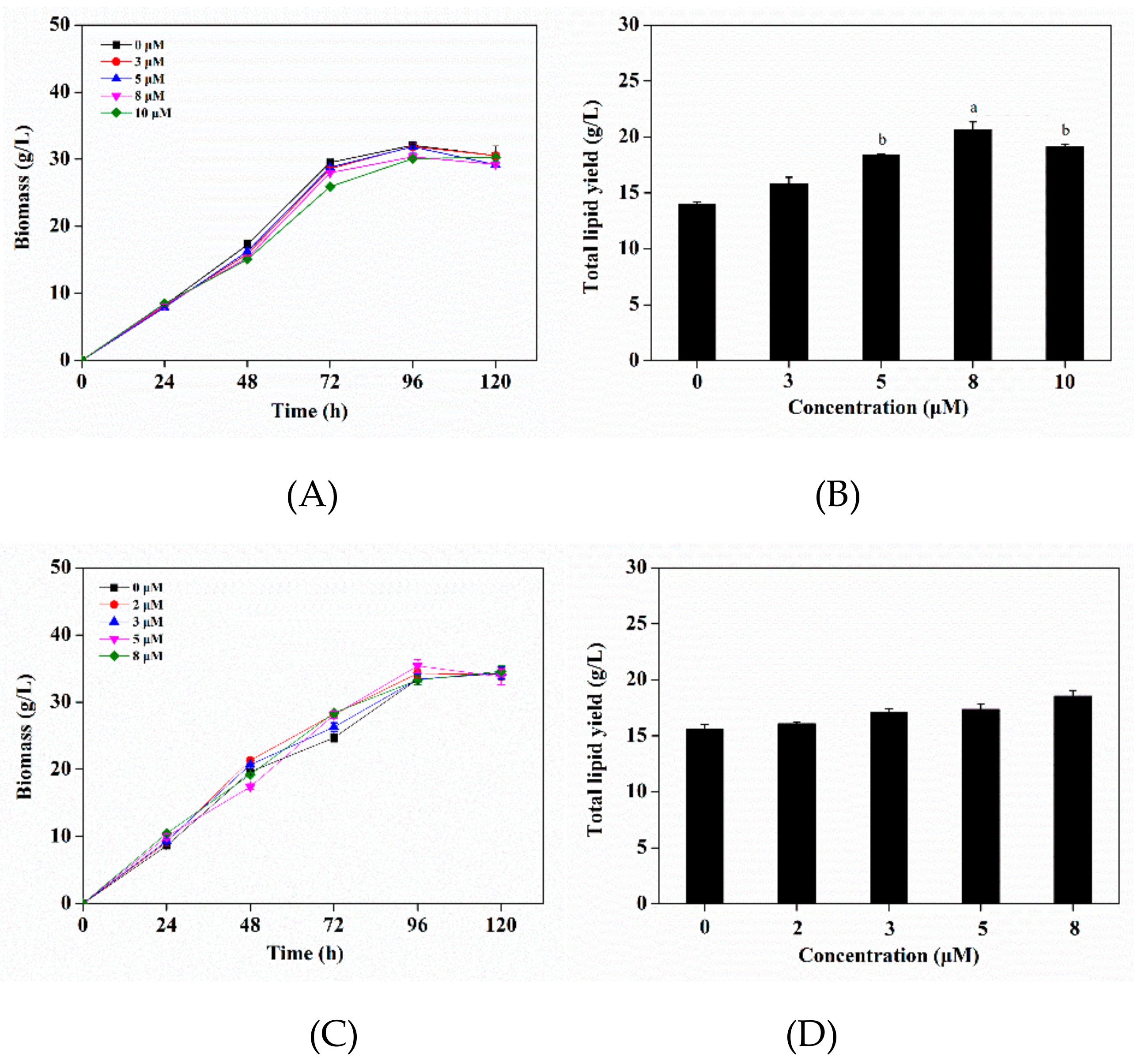
3.4. Effects of Triclosan as an Regulator of Enoylreductase (ER) on Growth and Lipid Production
3.5. Effects of Triclosan on Fatty Acid Composition
3.6. Effect of Triclosan on Expression Levels of Related Genes
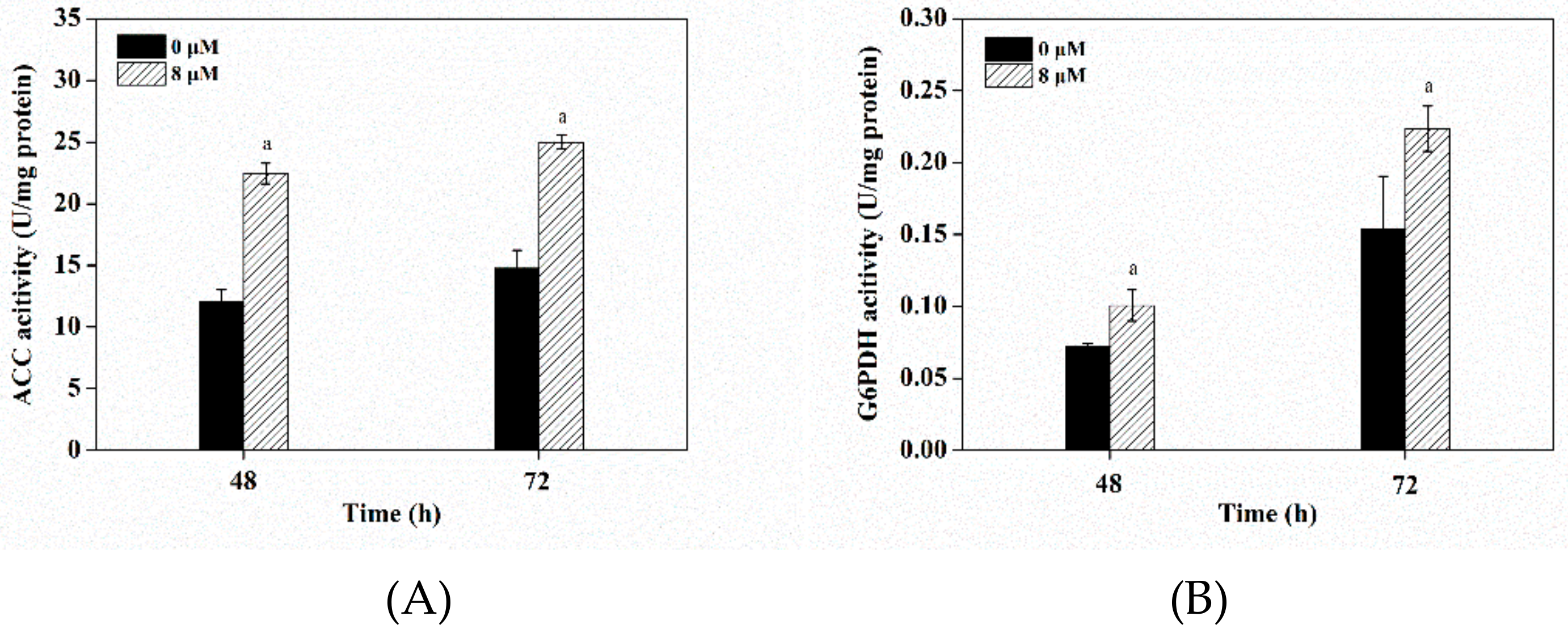
3.7. Effects of Triclosan on Key Enzyme Activities
3.8. Metabolite Analysis with Triclosan Treatment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Maki, K.C.; Palacios, O.M.; Bell, M.; Toth, P.P. Use of supplemental long-chain omega-3 fatty acids and risk for cardiac death: An updated meta-analysis and review of research gaps. J. Clin. Lipidol. 2017, 11, 1152–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canhada, S.; Castro, K.; Perry, I.S.; Luft, V.C. Omega-3 fatty acids’ supplementation in Alzheimer’s disease: A systematic review. Nutr. Neurosci. 2018, 21, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Tseng, P.T.; Chen, N.Y.; Lin, P.C.; Lin, P.Y.; Chang, J.P.C.; Kuo, F.Y.; Lin, J.; Wu, M.C.; Su, K.P. Safety and tolerability of prescription omega-3 fatty acids: A systematic review and meta-analysis of randomized controlled trials. Prostaglandins Leukot. Essent. Fat. Acids 2018, 129, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganuza, E.; Anderson, A.J.; Ratledge, C. High-cell-density cultivation of Schizochytrium sp. in an ammonium/pH-auxostat fed-batch system. Biotechnol. Lett. 2008, 30, 1559. [Google Scholar] [CrossRef]
- Li, Z.P.; Meng, T.; Ling, X.P.; Li, J.; Zheng, C.Q.; Shi, Y.Y.; Chen, Z.; Li, Z.Q.; Li, Q.B.; Lu, Y.H. Overexpression of Malonyl-CoA: ACP Transacylase in Schizochytrium sp. to Improve Polyunsaturated Fatty Acid Production. J. Agric. Food Chem. 2018, 66, 5382–5391. [Google Scholar] [CrossRef]
- Sun, X.M.; Ren, L.J.; Zhao, Q.Y.; Ji, X.J.; Huang, H. Microalgae for the production of lipid and carotenoids: A review with focus on stress regulation and adaptation. Biotechnol. Biofuels 2018, 11, 272. [Google Scholar] [CrossRef] [Green Version]
- Mamaeva, A.; Namsaraev, Z.; Maltsev, Y.; Gusev, E.; Kulikovskiy, M.; Petrushkina, M.; Filimonova, A.; Sorokin, B.; Zotko, N.; Vinokurov, V. Simultaneous increase in cellular content and volumetric concentration of lipids in Bracteacoccus bullatus cultivated at reduced nitrogen and phosphorus concentrations. J. Appl. Phycol. 2018, 30, 2237–2246. [Google Scholar] [CrossRef]
- Kumar, B.R.; Deviram, G.; Mathimani, T.; Duc, P.A.; Pugazhendhi, A. Microalgae as rich source of polyunsaturated fatty acids. Biocatal. Agric. Biotechnol. 2019, 17, 583–588. [Google Scholar] [CrossRef]
- Ghiffary, M.R.; Kim, H.U.; Chang, Y.K. Metabolic Engineering Strategies for the Enhanced Microalgal Production of Long-Chain Polyunsaturated Fatty Acids (LC-PUFAs). Biotechnol. J. 2019, 14, 1900043. [Google Scholar] [CrossRef]
- Li, X.P.; Liu, J.J.; Chen, G.Y.; Zhang, J.G.; Wang, C.B.; Liu, B. Extraction and purification of eicosapentaenoic acid and docosahexaenoic acid from microalgae: A critical review. Algal Res. 2019, 43, 101619. [Google Scholar] [CrossRef]
- Martins, D.; Custódio, L.; Barreira, L.; Pereira, H.; Ben-Hamadou, R.; Varela, J.; Abu-Salah, K. Alternative Sources of n-3 Long-Chain Polyunsaturated Fatty Acids in Marine Microalgae. Mar. Drugs 2013, 11, 2259–2281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metz, J.G.; Roessler, P.; Facciotti, D.; Levering, C.; Dittrich, F.; Lassner, M.; Valentine, R.; Lardizabal, K.; Domergue, F.; Yamada, A. Production of Polyunsaturated Fatty Acids by Polyketide Synthases in Both Prokaryotes and Eukaryotes. Science 2001, 293, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C. Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie 2004, 86, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Tan, Y.Z.; Liu, Y.J.; Zhang, J.T.; Liu, G.L.; Feng, Y.G.; Cui, Q. Different Impacts of Short-Chain Fatty Acids on Saturated and Polyunsaturated Fatty Acid Biosynthesis in Aurantiochytrium sp SD116. J. Agric. Food Chem. 2013, 61, 9876–9881. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Ueno, A.; Kawasaki, K.; Yumoto, I.; Ohgiya, S.; Hoshino, T.; Ishizaki, K.; Okuyama, H.; Morita, N. Isolation of clustered genes that are notably homologous to the eicosapentaenoic acid biosynthesis gene cluster from the docosahexaenoic acid-producing bacterium Vibrio marinus strain MP-1. Biotechnol. Lett. 1999, 21, 939–945. [Google Scholar] [CrossRef]
- Allen, E.E.; Facciotti, D.; Bartlett, D.H. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 1999, 65, 1710. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.J.; Chen, S.L.; Geng, L.J.; Ji, X.J.; Xu, X.; Song, P.; Gao, S.; Huang, H. Exploring the function of acyltransferase and domain replacement in order to change the polyunsaturated fatty acid profile of Schizochytrium sp. Algal Res. 2018, 29, 193–201. [Google Scholar] [CrossRef]
- Gong, Y.M.; Wan, X.; Jiang, M.L.; Hu, C.J.; Hu, H.H.; Huang, F.H. Metabolic engineering of microorganisms to produce omega-3 very long-chain polyunsaturated fatty acids. Prog. Lipid Res. 2014, 56, 19–35. [Google Scholar] [CrossRef]
- Xie, X.; Meesapyodsuk, D.; Qiu, X. Functional analysis of the dehydratase domains of a PUFA synthase from Thraustochytrium in Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 102, 847–856. [Google Scholar] [CrossRef]
- Song, X.J.; Zang, X.N.; Zhang, X.C. Production of high docosahexaenoic acid by Schizochytrium sp. using low-cost raw materials from food industry. J. Oleo Sci. 2015, 64, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.J.; Cao, Y.G.; Zhao, M.A. Biotechnological production of eicosapentaenoic acid: From a metabolic engineering point of view. Process Biochem. 2012, 47, 1320–1326. [Google Scholar] [CrossRef]
- Ursula, K.; Christian, H. Biosynthesis of polyunsaturated fatty acids by polyketide synthases. Angew. Chem. 2010, 41, 1866–1869. [Google Scholar] [CrossRef]
- Richard, J.; Heath, C.O.R. A triclosan-resistant bacterial enzyme. Nature 2000, 406, 13. [Google Scholar] [CrossRef]
- Velly, H.; Bouix, M.; Passot, S.; Penicaud, C.; Beinsteiner, H.; Ghorbal, S.; Lieben, P.; Fonseca, F. Cyclopropanation of unsaturated fatty acids and membrane rigidification improve the freeze-drying resistance of Lactococcus lactis subsp. lactis TOMSC161. Appl. Microbiol. Biotechnol. 2015, 99, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.; White, S.; Rock, C. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 2002, 58, 695–703. [Google Scholar] [CrossRef]
- Lu, H.; Tonge, P.J. Inhibitors of FabI, an Enzyme Drug Target in the Bacterial Fatty Acid Biosynthesis Pathway. Acc. Chem. Res. 2008, 41, 11–20. [Google Scholar] [CrossRef]
- Liu, X.P.; Yu, H.Y.; Jiang, X.; Ai, G.M.; Yu, B.; Zhu, K. Biosynthesis of butenoic acid through fatty acid biosynthesis pathway in Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 99, 1795–1804. [Google Scholar] [CrossRef]
- Escalada, G.M. Triclosan inhibition of fatty acid synthesis and its effect on growth of Escherichia coli and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2005, 55, 879–882. [Google Scholar] [CrossRef] [Green Version]
- Cui, G.Z.; Ma, Z.G.; Liu, Y.J.; Feng, Y.G.; Sun, Z.J.; Cheng, Y.R.; Song, X.J.; Cui, Q.; Cui, G.Z.; Ma, Z.X. Overexpression of glucose-6-phosphate dehydrogenase enhanced the polyunsaturated fatty acid composition of Aurantiochytrium sp SD116. Algal Res. 2016, 19, 138–145. [Google Scholar] [CrossRef]
- Zhang, H.D.; Lu, D.; Li, X.; Feng, Y.G.; Cui, Q.; Song, X.J. Heavy ion mutagenesis combined with triclosan screening provides a new strategy for improving the arachidonic acid yield in Mortierella alpina. BMC Biotechnol. 2018, 18, 23. [Google Scholar] [CrossRef] [Green Version]
- Ling, X.P.; Guo, J.; Liu, X.T.; Zhang, X.; Wang, N.; Lu, Y.H.; Ng, I.S. Impact of carbon and nitrogen feeding strategy on high production of biomass and docosahexaenoic acid (DHA) by Schizochytrium sp. LU310. Bioresour. Technol. 2015, 184, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.K.; Heo, S.Y.; Oh, B.R.; Kim, C.H.; Sohn, J.H.; Yang, J.W.; Kondo, A.; Seo, J.W. A transgene expression system for the marine microalgae Aurantiochytrium sp. KRS101 using a mutant allele of the gene encoding ribosomal protein L44 as a selectable transformation marker for cycloheximide resistance. Bioprocess Biosyst. Eng. 2013, 36, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.J.; Huang, H.; Xiao, A.H.; Lian, M.; Jin, L.J.; Ji, X.J. Enhanced docosahexaenoic acid production by reinforcing acetyl-CoA and NADPH supply in Schizochytrium sp. HX-308. Bioprocess Biosyst. Eng. 2009, 32, 837. [Google Scholar] [CrossRef] [PubMed]
- Langdon, R.G. Glucose 6-phosphate dehydrogenase from erythrocytes. Methods Enzymol. 1966, 9, 126–131. [Google Scholar] [CrossRef]
- Yu, X.J.; Sun, J.; Zheng, J.Y.; Sun, Y.Q.; Wang, Z. Metabolomics analysis reveals 6-benzylaminopurine as a stimulator for improving lipid and DHA accumulation of Aurantiochytrium sp. J. Chem. Technol. Biotechnol. 2016, 91, 1199–1207. [Google Scholar] [CrossRef] [Green Version]
- Strelkov, S.; Von Elstermann, M.; Schomburg, D. Comprehensive analysis of metabolites in Corynebacterium glutamicum by gas chromatography/mass spectrometry. Biol. Chem. 2004, 758, 81–861. [Google Scholar] [CrossRef]
- Chang, G.F.; Luo, Z.F.; Gu, S.T.; Wu, Q.H.; Ming, C.; Wang, X.G. Fatty acid shifts and metabolic activity changes of Schizochytrium sp. S31 cultured on glycerol. Bioresour. Technol. 2013, 142, 255–260. [Google Scholar] [CrossRef]
- Nishida, I.; Murata, N. Chilling sensitivity in plants and cyanobacteria: The Crucial Contribution of Membrane Lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 541–568. [Google Scholar] [CrossRef] [Green Version]
- Heath, R.J.; Rock, C.O. Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 1996, 271, 27795–27801. [Google Scholar] [CrossRef] [Green Version]
- Heath, R.J.; Rock, C.O. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J. Biol. Chem. 1995, 270, 26538–26542. [Google Scholar] [CrossRef] [Green Version]
- Durnoford, E.; Shahidi, F. Comparison of FA compositions of selected tissues of phocid seals of Eastern Canada using one-way and multivariate techniques. J. Am. Oil Chem. Soc. 2002, 79, 1095–1102. [Google Scholar] [CrossRef]
- Lippmeier, J.C.; Crawford, K.S.; Owen, C.B.; Rivas, A.A.; Metz, J.G.; Apt, K.E. Characterization of Both Polyunsaturated Fatty Acid Biosynthetic Pathways in Schizochytrium sp. Lipids 2009, 44, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Hauvermale, A.; Kuner, J.; Rosenzweig, B.; Guerra, D.; Diltz, S.; Metz, J.G. Fatty acid production in Schizochytrium sp.: Involvement of a polyunsaturated fatty acid synthase and a type I fatty acid synthase. Lipids 2006, 41, 739. [Google Scholar] [CrossRef]
- Hamid, A.A.; Mokhtar, N.F.; Taha, E.M.; Omar, O.; Wan, M.W.Y. The role of ATP citrate lyase, malic enzyme and fatty acid synthase in the regulation of lipid accumulation in Cunninghamella sp. 2A1. Ann. Microbiol. 2011, 61, 463–468. [Google Scholar] [CrossRef]
- Ashcraft, B.A.; Fillers, W.S.; Augustine, S.L.; Clarke, S.D. Polymer-protomer transition of acetyl-CoA carboxylase occurs in vivo and varies with nutritional conditions. J. Biol. Chem. 1980, 255, 10033–10035. [Google Scholar] [PubMed]
- Qiao, K.J.; Abidi, S.H.I.; Liu, H.J.; Zhang, H.R.; Chakraborty, S.; Watson, N.; Ajikumar, P.K.; Stephanopoulos, G. Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 2015, 29, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.H.; Kim, M.D.; Jin, Y.S.; Seo, J.H. Engineering of NADPH regenerators in Escherichia coli for enhanced biotransformation. Appl. Microbiol. Biotechnol. 2013, 97, 2761–2772. [Google Scholar] [CrossRef]
- Pfleger, B.F.; Gossing, M.; Nielsen, J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab. Eng. 2015, 29, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhou, Z.; Yi, C.F.; Wang, F.L.; Niu, Y.P.; Li, H. Intracellular metabolic changes in Saccharomyces cerevisiae and promotion of ethanol tolerance during the bioethanol fermentation process. RSC Adv. 2016, 6. [Google Scholar] [CrossRef]
- Rathinasabapathi, B. Metabolic Engineering for Stress Tolerance: Installing Osmoprotectant Synthesis Pathways. Ann. Bot. 2000, 86, 709–716. [Google Scholar] [CrossRef]
- Ruijter, G.J.; Bax, M.; Patel, H.; Flitter, S.J.; Pj, V.D.V.; de Vries, R.P.; Vankuyk, P.A.; Visser, J. Mannitol Is Required for Stress Tolerance in Aspergillus niger Conidiospores. Eukaryot. Cell 2003, 2, 690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B.; Jensen, R.G.; Bohnert, H.J. Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol. 1997, 113, 1177–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B.; Jensen, R.G.; Bohnert, H.J. Mannitol Protects against Oxidation by Hydroxyl Radicals. Plant Physiol. 1997, 115, 527–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siniossoglou, S. Phospholipid metabolism and nuclear function: Roles of the lipin family of phosphatidic acid phosphatases. Biochim. Biophys. Acta 2013, 1831, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Nikawa, J. Phosphatidylserine synthase from yeast. Biochim. Biophys. Acta 1997, 1348, 228. [Google Scholar] [CrossRef]
- Li, J.; Zhou, H.; Pan, X.S.; Li, Z.P.; Lu, Y.H.; He, N.; Meng, T.; Yao, C.Y.; Chen, C.X.; Ling, X.P. The role of fluconazole in the regulation of fatty acid and unsaponifiable matter biosynthesis in Schizochytrium sp. MYA 1381. BMC Microbiol. 2019, 19, 256. [Google Scholar] [CrossRef]
- Sun, L.; Ren, L.J.; Zhuang, X.Y.; Ji, X.J.; Yan, J.C.; Huang, H. Differential effects of nutrient limitations on biochemical constituents and docosahexaenoic acid production of Schizochytrium sp. Bioresour. Technol. 2014, 159, 199–206. [Google Scholar] [CrossRef]
- Cui, M.Y.; Lin, Y.C.; Zu, Y.G.; Efferth, T.; Li, D.W.; Tang, Z.H. Ethylene increases accumulation of compatible solutes and decreases oxidative stress to improve plant tolerance to water stress in Arabidopsis. J. Plant Biol. 2015, 58, 193–201. [Google Scholar] [CrossRef]
- Yang, D.; Sheng, J.P.; Li, S.Y.; Ying, N.; Zhao, J.H.; Zhen, Z.; Wang, Z.D.; Tang, X.M. The role of gibberellins in the mitigation of chilling injury in cherry tomato (Solanum lycopersicum L.) fruit. Postharvest Biol. Technol. 2015, 101, 88–95. [Google Scholar] [CrossRef]
- Kumar, S.; Punekar, N.S. The metabolism of 4-aminobutyrate (GABA) in fungi. Mycol. Res. 1997, 101, 403–409. [Google Scholar] [CrossRef]


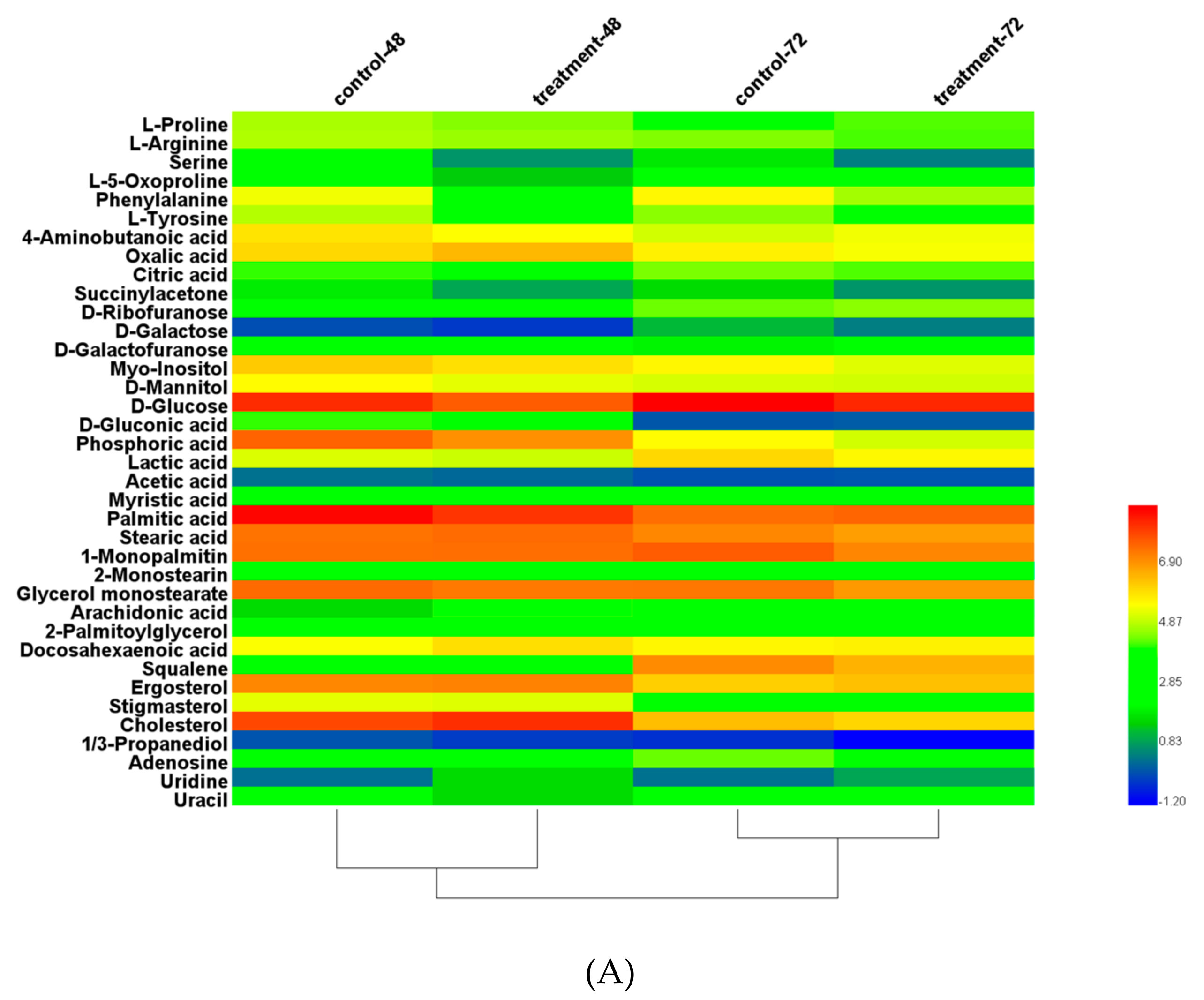
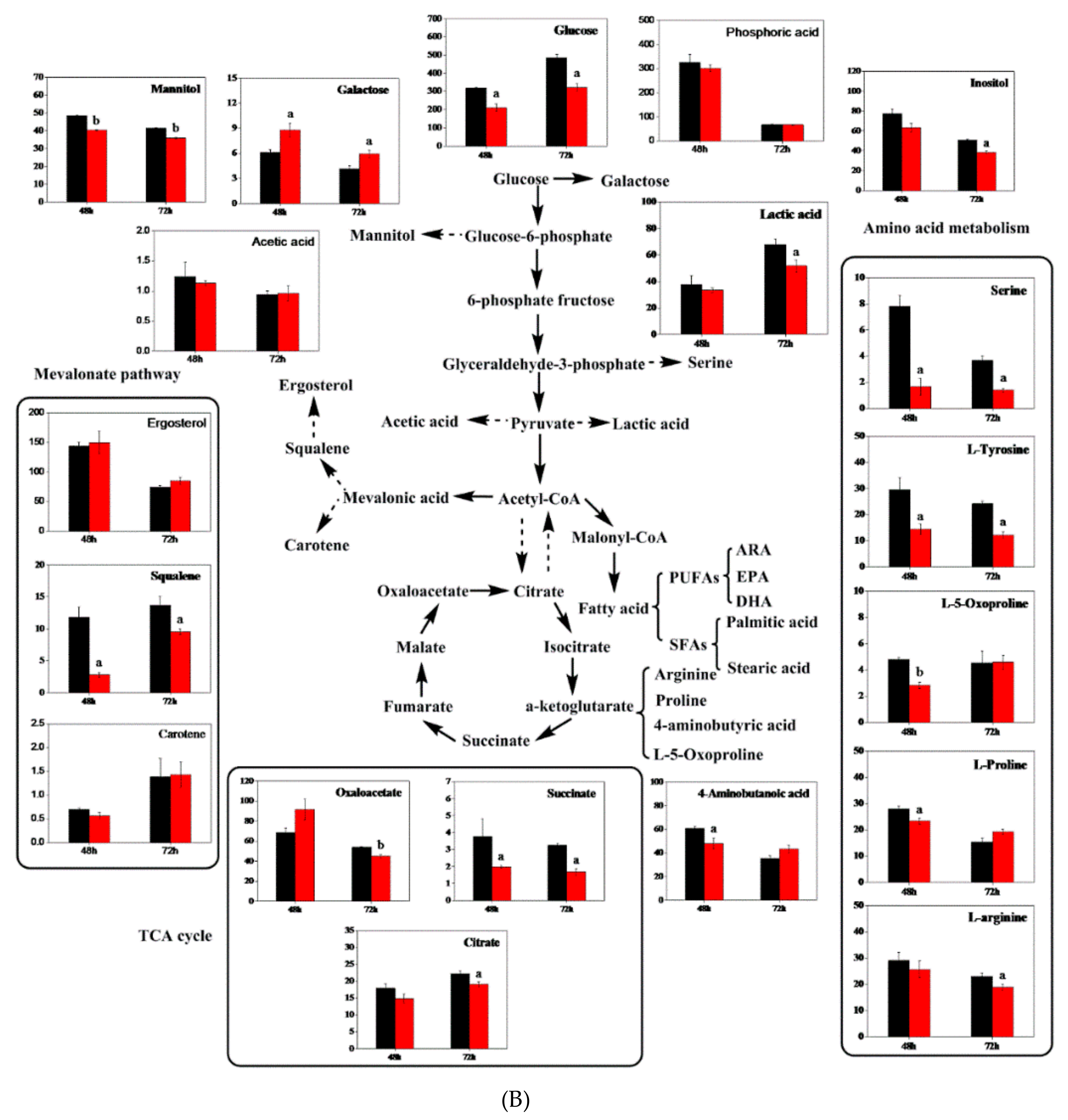

| Lipid/PUFAs Yield | No Treatment | With Triclosan | Increase Ratio |
|---|---|---|---|
| Total lipids (g/L) | 13.96 ± 0.26 | 20.61 ± 0.74a | 47.6% |
| Total PUFAs (g/L) | 4.89 ± 0.19 | 7.42 ± 0.06b | 51.7% |
| EPA (mg/L) | 53.75 ± 0.70 | 82.44 ± 1.09a | 53.4% |
| DHA (mg/L) | 4019.78 ± 154.26 | 6002.66 ± 9.27a | 49.3% |
| Fatty Acid Composition | 48 h | 72 h | ||
|---|---|---|---|---|
| No Treatment | With Triclosan | No Treatment | With Triclosan | |
| C14:0 | 3.68 ± 0.23 | 2.71 ± 0.17a | 4.46 ± 0.12 | 3.45 ± 0.28a |
| C14:1 | 2.13 ± 0.14 | 3.20 ± 0.09a | 1.42 ± 0.16 | 2.49 ± 0.06a |
| C16:0 | 49.19 ± 0.48 | 45.41 ± 0.38a | 51.48 ± 0.39 | 49.34 ± 0.37a |
| C18:0 | 2.21 ± 0.25 | 1.84 ± 0.13a | 1.73 ± 0.08 | 1.67 ± 0.07 |
| EPA | 0.26 ± 0.02 | 0.27 ± 0.05 | 0.21 ± 0.09 | 0.25 ± 0.02 |
| DPA | 5.86 ± 0.23 | 6.56 ± 0.36 | 5.16 ± 0.21 | 6.05 ± 0.23a |
| DHA | 30.79 ± 0.56 | 33.68 ± 0.21a | 30.26 ± 0.13 | 31.94 ± 0.48a |
| SFAs | 55.53 ± 0.26 | 49.96 ± 0.36a | 57.65 ± 0.13 | 55.33 ± 0.03a |
| PUFAs | 36.95 ± 0.27 | 39.98 ± 0.58a | 36.39 ± 0.14 | 38.08 ± 0.16a |
| PUFAs/SFAs | 64.52 ± 0.13 | 76.20 ± 0.21b | 60.30 ± 0.24 | 67.15 ± 0.18a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, X.; Zhou, H.; Yang, Q.; Yu, S.; Li, J.; Li, Z.; He, N.; Chen, C.; Lu, Y. Functions of Enyolreductase (ER) Domains of PKS Cluster in Lipid Synthesis and Enhancement of PUFAs Accumulation in Schizochytrium limacinum SR21 Using Triclosan as a Regulator of ER. Microorganisms 2020, 8, 300. https://doi.org/10.3390/microorganisms8020300
Ling X, Zhou H, Yang Q, Yu S, Li J, Li Z, He N, Chen C, Lu Y. Functions of Enyolreductase (ER) Domains of PKS Cluster in Lipid Synthesis and Enhancement of PUFAs Accumulation in Schizochytrium limacinum SR21 Using Triclosan as a Regulator of ER. Microorganisms. 2020; 8(2):300. https://doi.org/10.3390/microorganisms8020300
Chicago/Turabian StyleLing, Xueping, Hao Zhou, Qinghua Yang, Shengyang Yu, Jun Li, Zhipeng Li, Ning He, Cuixue Chen, and Yinghua Lu. 2020. "Functions of Enyolreductase (ER) Domains of PKS Cluster in Lipid Synthesis and Enhancement of PUFAs Accumulation in Schizochytrium limacinum SR21 Using Triclosan as a Regulator of ER" Microorganisms 8, no. 2: 300. https://doi.org/10.3390/microorganisms8020300
APA StyleLing, X., Zhou, H., Yang, Q., Yu, S., Li, J., Li, Z., He, N., Chen, C., & Lu, Y. (2020). Functions of Enyolreductase (ER) Domains of PKS Cluster in Lipid Synthesis and Enhancement of PUFAs Accumulation in Schizochytrium limacinum SR21 Using Triclosan as a Regulator of ER. Microorganisms, 8(2), 300. https://doi.org/10.3390/microorganisms8020300






