Optimization of Preservation Methods Allows Deeper Insights into Changes of Raw Milk Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Milk Sampling
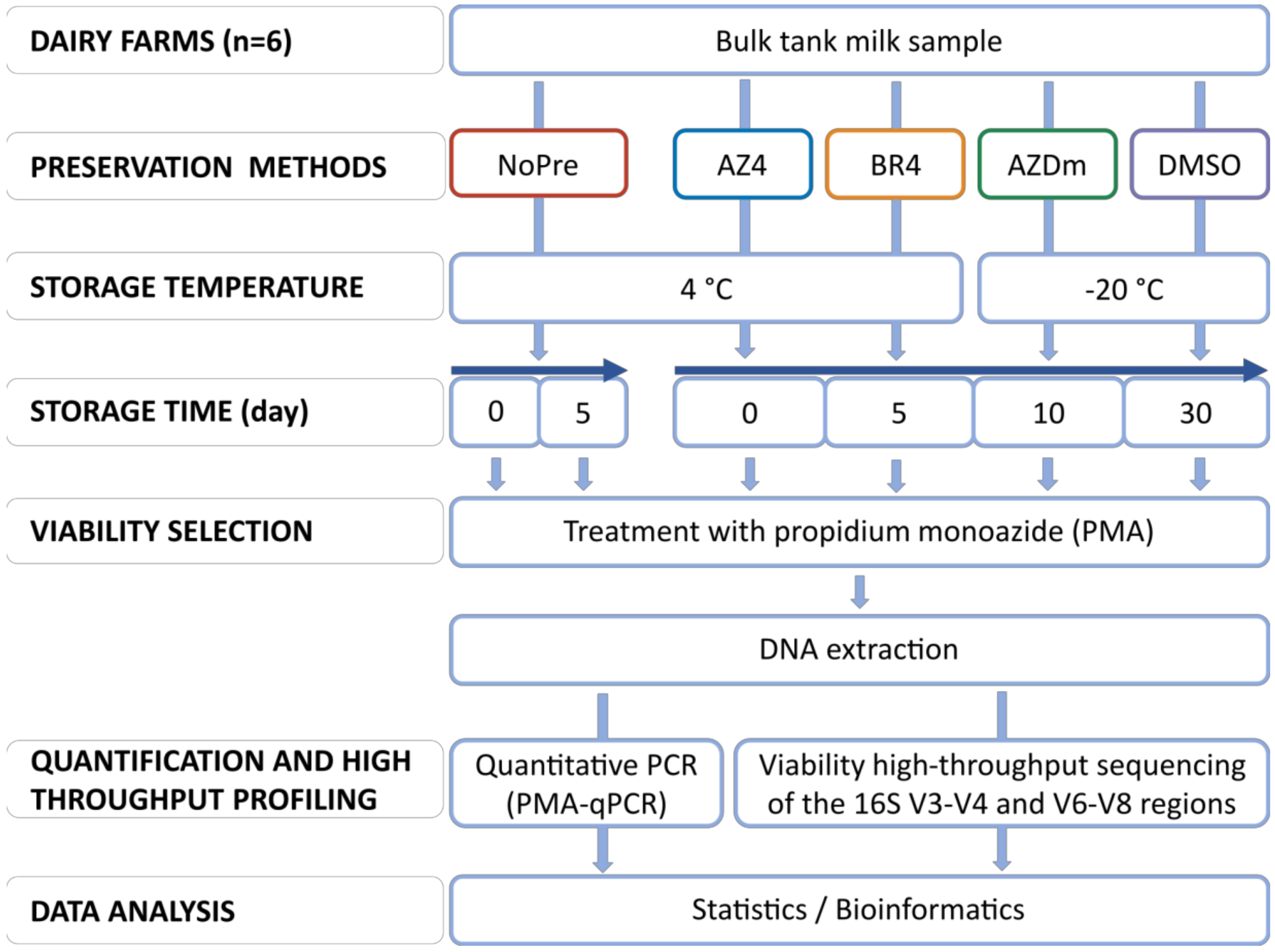
2.2. Preservation Treatments and Experimental Design
2.3. DNA Extraction and Sequencing of the 16S rRNA Gene
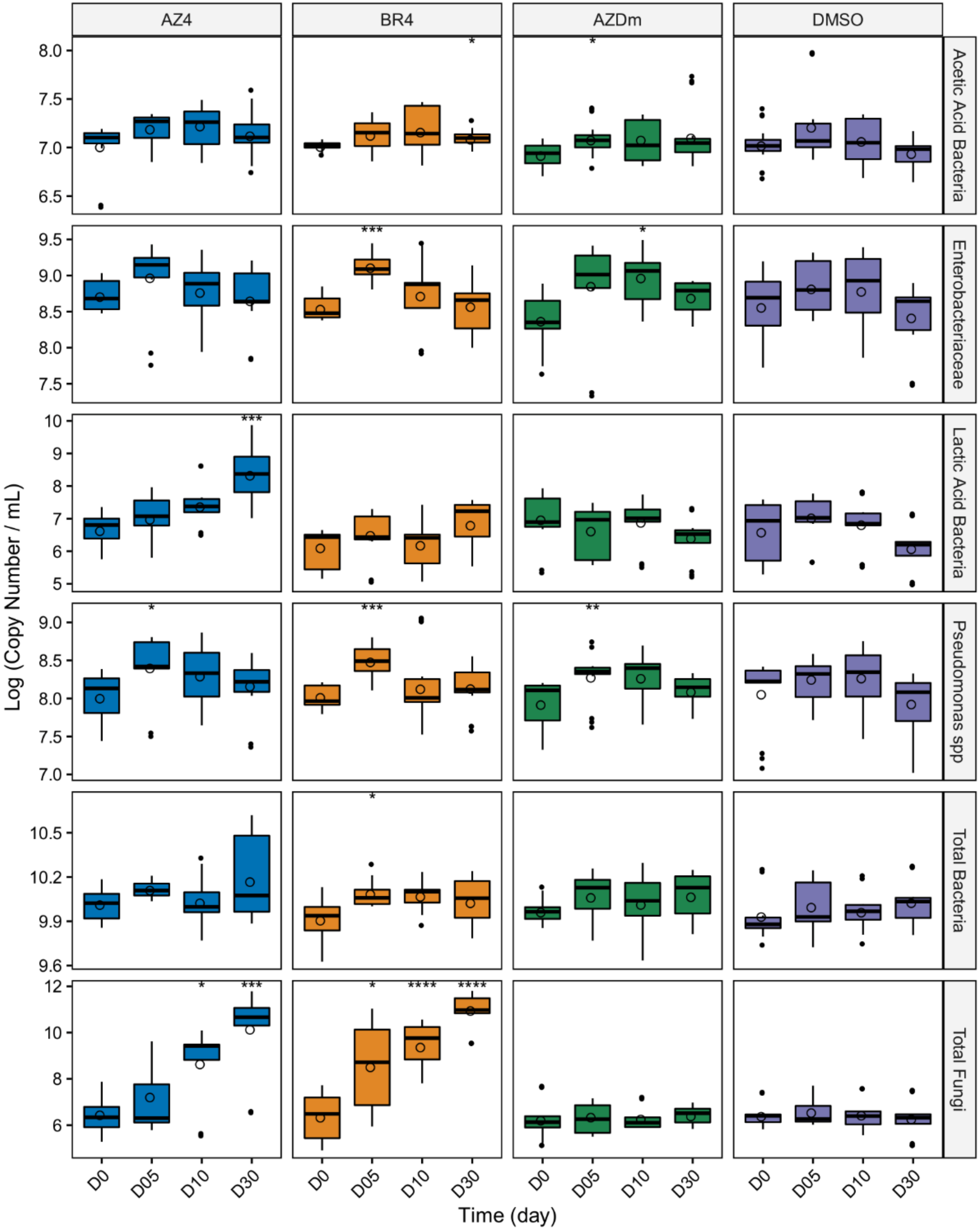
2.4. Quantitative PCR
2.5. 16S rRNA Gene Bioinformatics and Data Analysis
3. Results
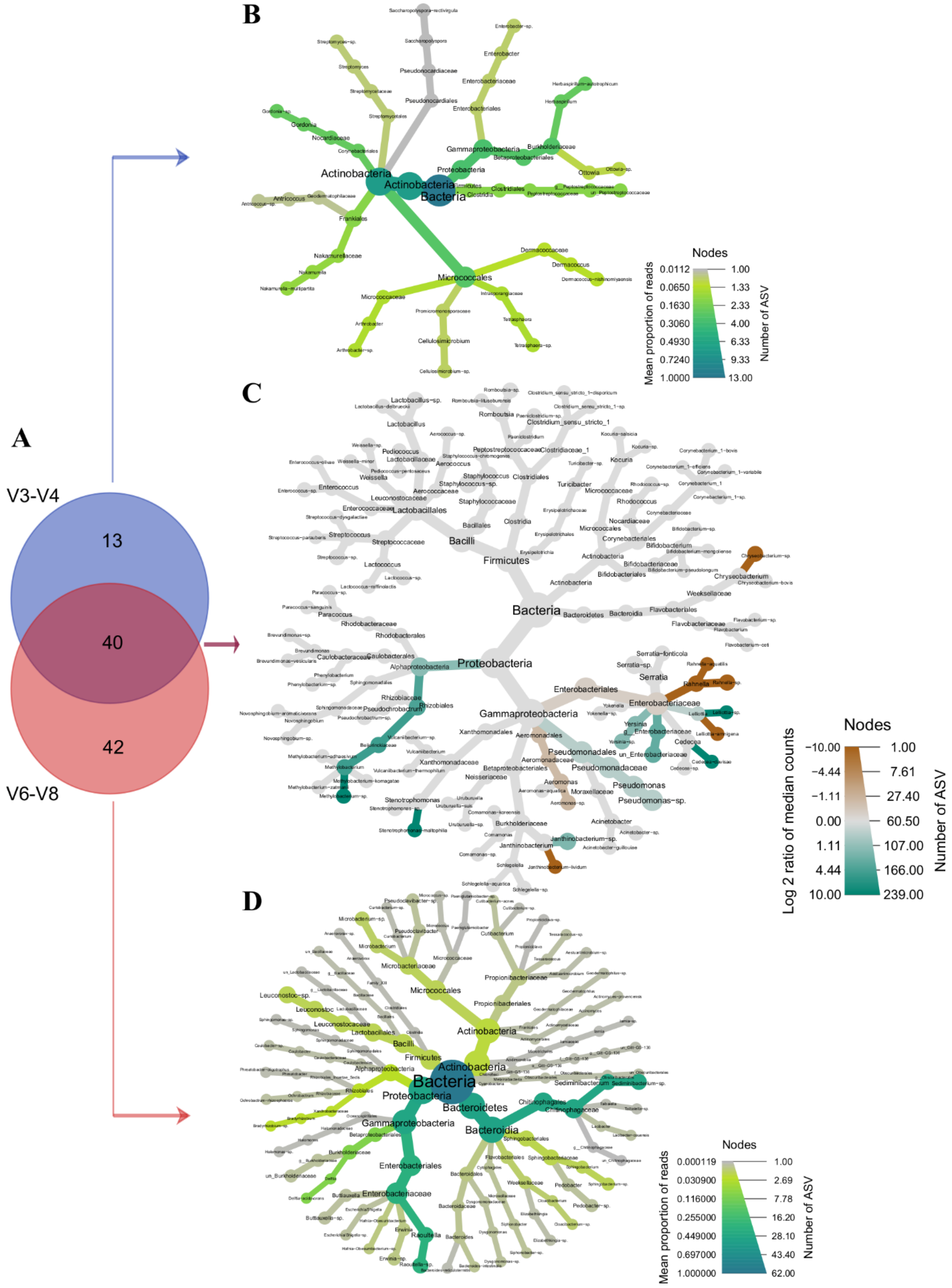
3.1. High-Throughput Sequencing of the 16S rRNA Hypervariable Regions V3-V4 and V6-V8 Reveals Different Taxonomic Profiles
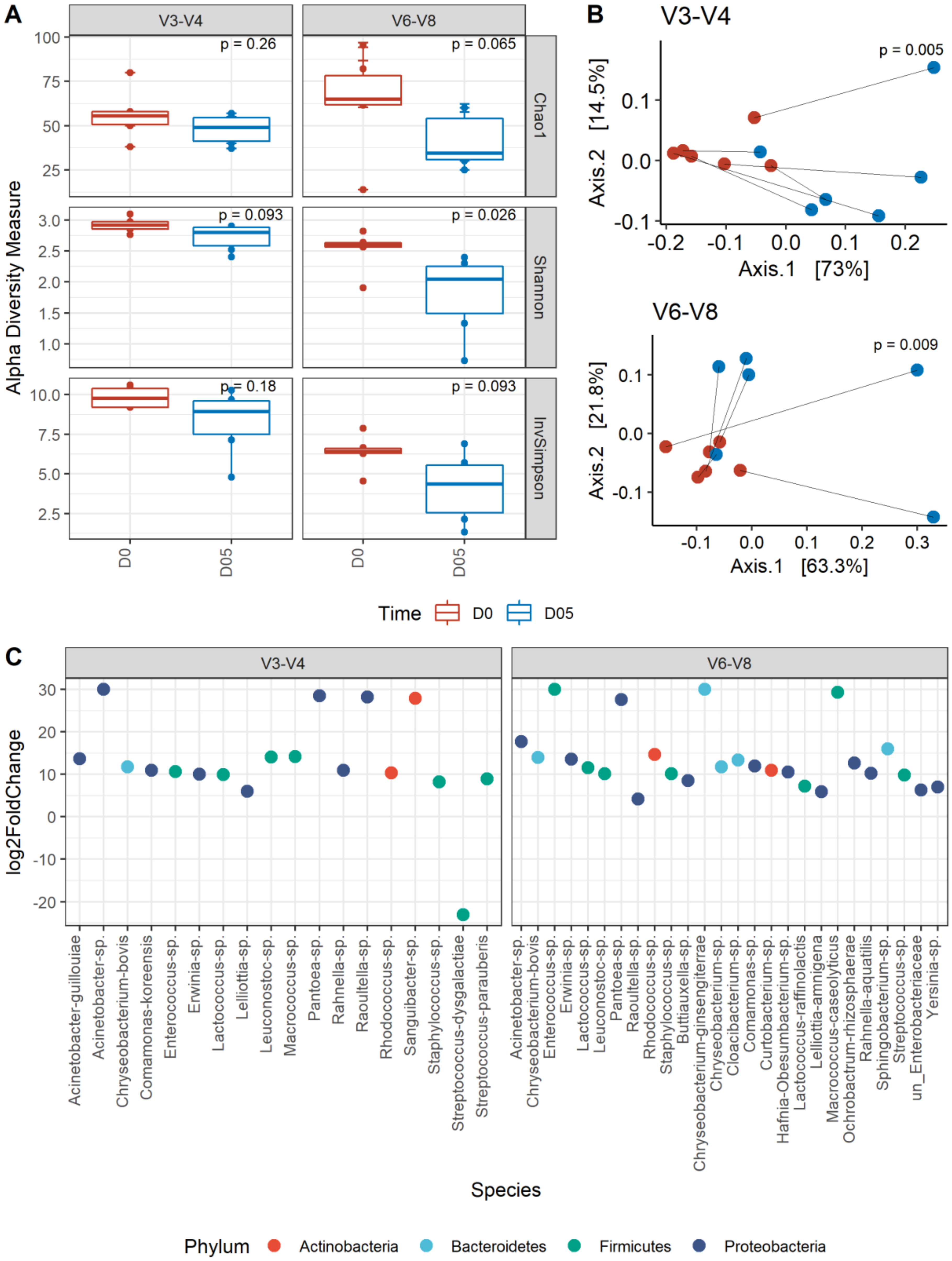
3.2. Short-Term Storage of Unpreserved Milk Resulted in Significant Alteration of the Microbial Community Structure
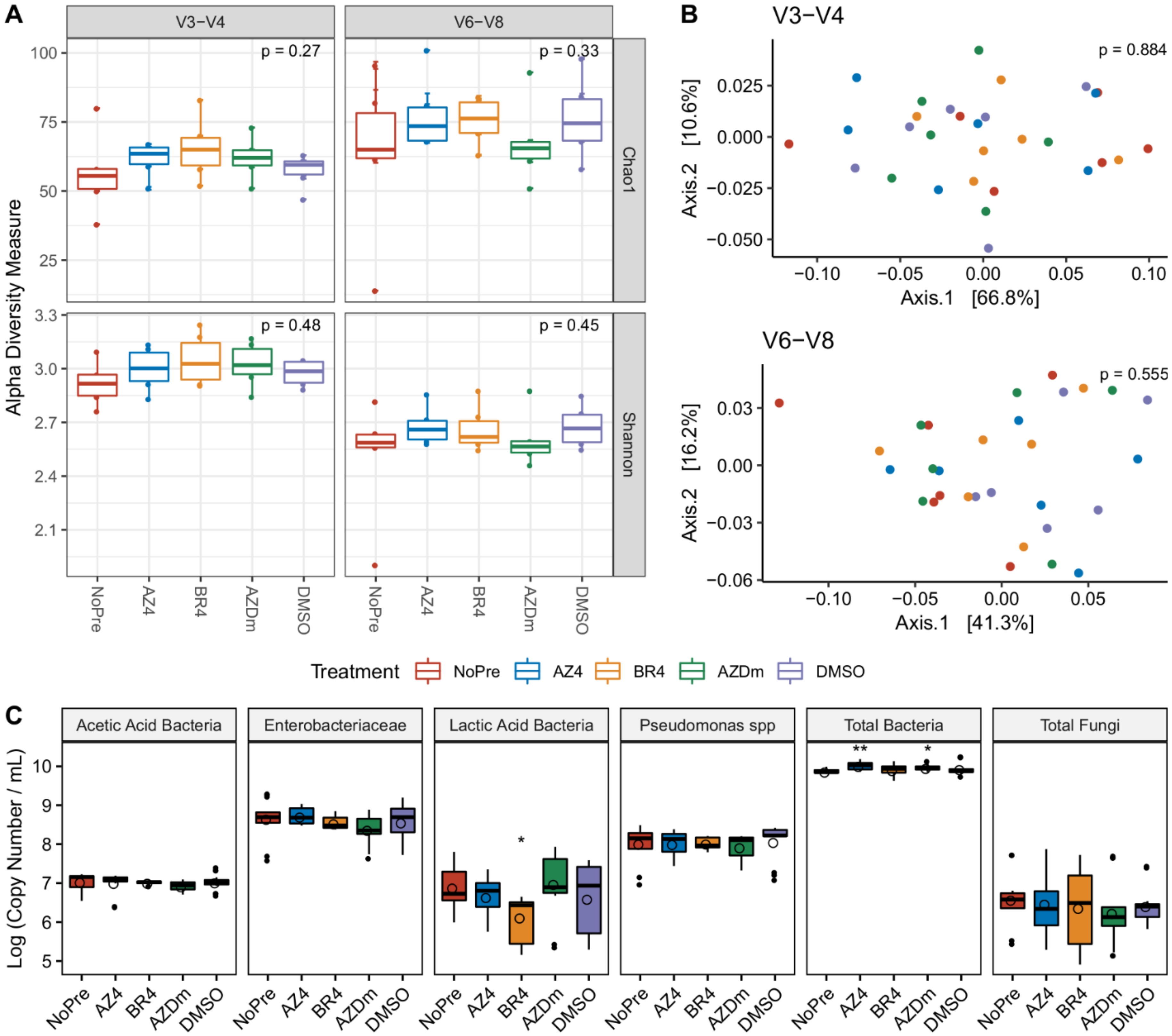
3.3. Impact of Preservative Chemicals on the Microbiota Once Mixed with Milk
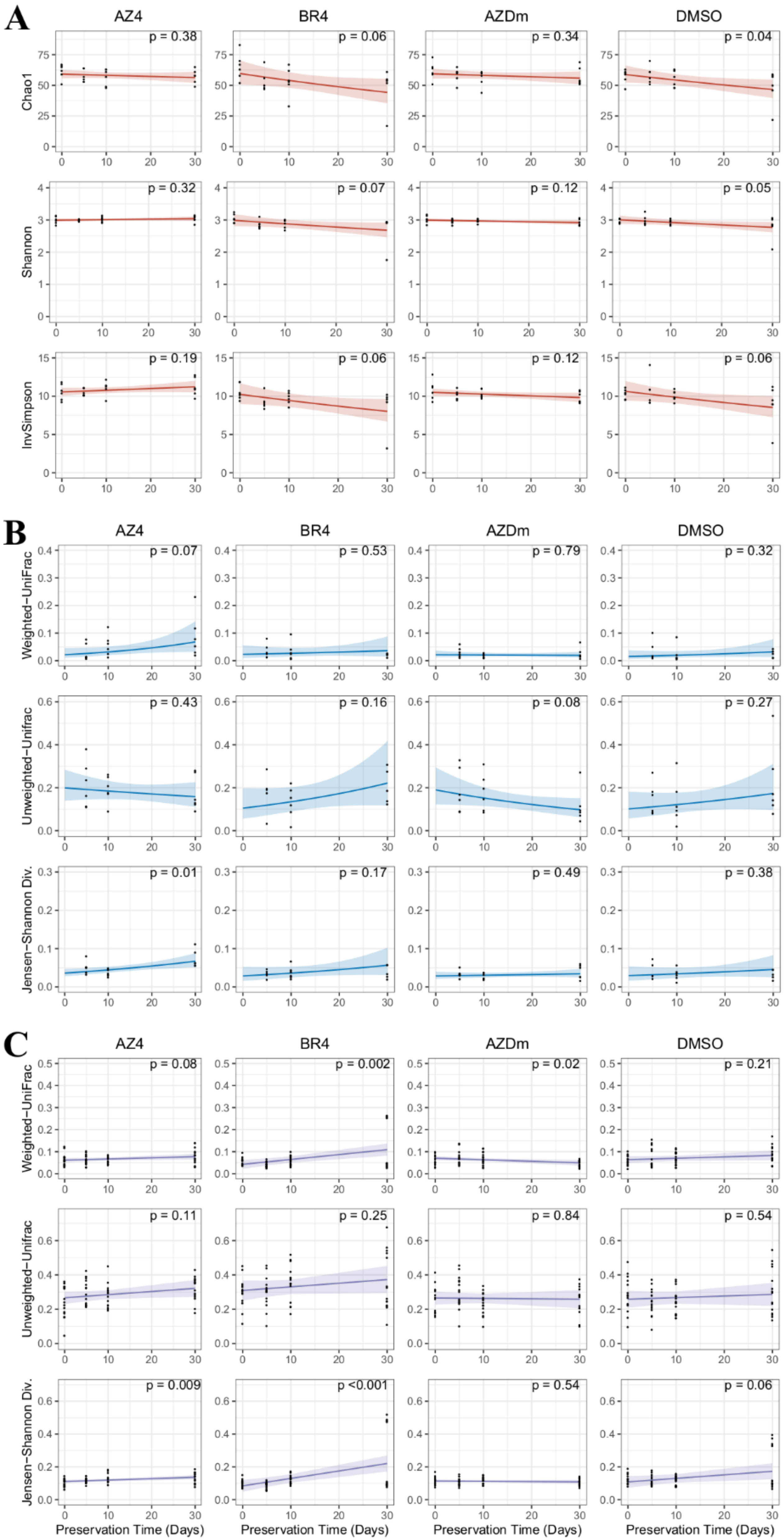
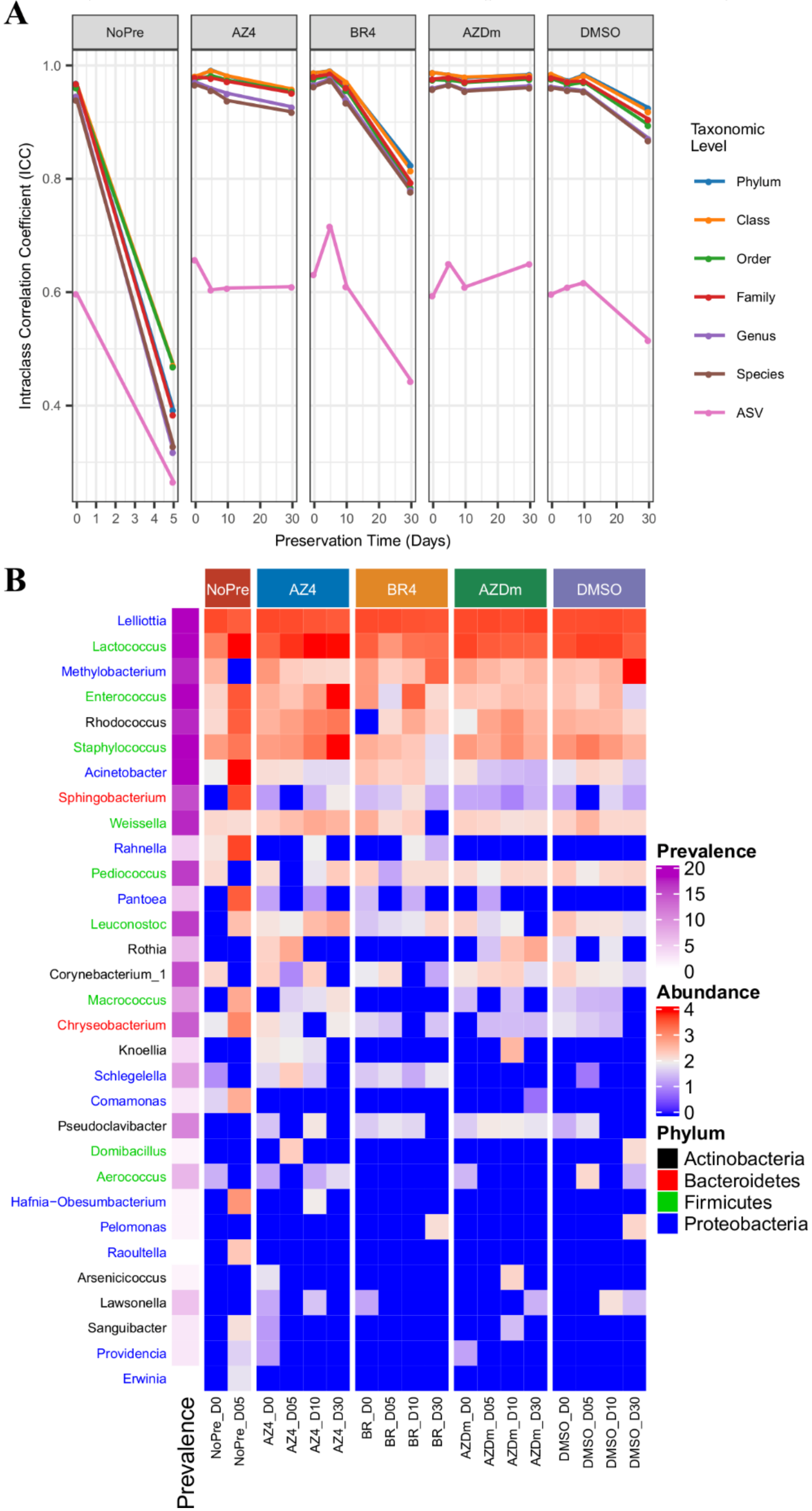
3.4. Impact of Preservation Methods on Microbial Community Stability over Time
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tilocca, B.; Costanzo, N.; Morittu, V.M.; Spina, A.A.; Soggiu, A.; Britti, D.; Roncada, P.; Piras, C. Milk microbiota: Characterization methods and role in cheese production. J. Proteom. 2020, 210, 103534. [Google Scholar] [CrossRef]
- Feligini, M.; Panelli, S.; Sacchi, R.; Ghitti, M.; Capelli, E. Tracing the origin of raw milk from farm by using automated ribosomal intergenic spacer analysis (ARISA) fingerprinting of microbiota. Food Control. 2015, 50, 51–56. [Google Scholar] [CrossRef]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. Impacts of seasonal housing and teat preparation on raw milk microbiota: A high-throughput sequencing study. Appl. Environ. Microbiol. 2017, 83, e02694-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Sadiq, F.A.; Burmolle, M.; Wang, N.; He, G. Insights into psychrotrophic bacteria in rawmilk: A review. J. Food Prot. 2019, 82, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Bernier Gosselin, V.; Dufour, S.; Adkins, P.R.F.; Middleton, J.R. Persistence of coagulase negative staphylococcal intramammary infections in dairy goats. J. Dairy Res. 2019, 86, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Moineau-Jean, A.; Champagne, C.P.; Roy, D.; Raymond, Y.; LaPointe, G. Effect of Greek-style yoghurt manufacturing processes on starter and probiotic bacteria populations during storage. Int. Dairy J. 2019, 93, 35–44. [Google Scholar] [CrossRef]
- Colombo, M.; Castilho, N.P.A.; Todorov, S.D.; Nero, L.A. Beneficial properties of lactic acid bacteria naturally present in dairy production. BMC Microbiol. 2018, 18, 219. [Google Scholar] [CrossRef]
- Rasolofo, E.A.; St-Gelais, D.; LaPointe, G.; Roy, D. Molecular analysis of bacterial population structure and dynamics during cold storage of untreated and treated milk. Int. J. Food Microbiol. 2010, 138, 108–118. [Google Scholar] [CrossRef]
- Hahne, J.; Isele, D.; Berning, J.; Lipski, A. The contribution of fast growing, psychrotrophic microorganisms on biodiversity of refrigerated raw cow’s milk with high bacterial counts and their food spoilage potential. Food Microbiol. 2019, 79, 11–19. [Google Scholar] [CrossRef]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. MBio 2016, 7, e00836-16. [Google Scholar] [CrossRef] [Green Version]
- McEniry, J.; O’Kiely, P.; Clipson, N.J.W.; Forristal, P.D.; Doyle, E.M. Assessing the impact of various ensilage factors on the fermentation of grass silage using conventional culture and bacterial community analysis techniques. J. Appl. Microbiol. 2010, 108, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Blekhman, R.; Tang, K.; Archie, E.A.; Barreiro, L.B.; Johnson, Z.P.; Wilson, M.E.; Kohn, J.; Yuan, M.L.; Gesquiere, L.; Grieneisen, L.E.; et al. Common methods for fecal sample storage in field studies yield consistent signatures of individual identity in microbiome sequencing data. Sci. Rep. 2016, 6, 31519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.J.; Amir, A.; Metcalf, J.L.; Amato, K.R.; Xu, Z.Z.; Humphrey, G.; Knight, R. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 2016, 1, e00021-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandeputte, D.; Tito, R.Y.; Vanleeuwen, R.; Falony, G.; Raes, J. Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 2017, 41, S154–S167. [Google Scholar] [PubMed]
- Upadhyay, N.; Goyal, A.; Kumar, A.; Ghai, D.L.; Singh, R. Preservation of milk and milk products for analytical purposes. Food Rev. Int. 2014, 30, 203–224. [Google Scholar] [CrossRef]
- Martins, M.E.P.; Nicolau, E.S.; Mesquita, A.J.; Neves, R.B.S.; Oliveira, J.P. Bronopol and azidiol chemicals: Time and temperature influence in the total bacterial count of raw milk. Ciênc. Anim. Bras. 2009, 10, 627–633. [Google Scholar]
- Amores, J.; de la Fe, C.; Gómez-Martín, Á.; Corrales, J.C.; Contreras, A.; Sánchez, A. Preserved goat milk as a valid sample for the PCR detection of Mycoplasma agalactiae. Small Rumin. Res. 2011, 99, 61–64. [Google Scholar] [CrossRef]
- Lackey, K.A.; Williams, J.E.; Price, W.J.; Carrothers, J.M.; Brooker, S.L.; Shafii, B.; McGuire, M.A.; McGuire, M.K. Comparison of commercially-available preservatives for maintaining the integrity of bacterial DNA in human milk. J. Microbiol. Methods 2017, 141, 73–81. [Google Scholar] [CrossRef]
- Sharma, M.; An, D.; Liu, T.; Pinnock, T.; Cheng, F.; Voordouw, G. Biocide-mediated corrosion of coiled tubing. PLoS ONE 2017, 12, e0181934. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Wu, Y.; Li, X.; Zeng, J.; Lin, X. Continuing impacts of selective inhibition on bacterial and fungal communities in an agricultural soil. Microb. Ecol. 2019, 78, 927–935. [Google Scholar] [CrossRef]
- Zouharova-Hubaakova, M.; Ryšánek, D. Effects of freezing milk samples on the recovery of alimentary pathogens and indicator microorganisms. Acta Vet. Brno 2007, 76, 301–307. [Google Scholar]
- Amores, J.; Sanchez, A.; Martin, A.G.; Corrales, J.C.; Contreras, A.; de la Fe, C. Viability of Mycoplasma agalactiae and Mycoplasma mycoides subsp. capri in goat milk samples stored under different conditions. Vet. Microbiol. 2010, 145, 347–350. [Google Scholar] [PubMed]
- Heylen, K.; Ettwig, K.; Hu, Z.; Jetten, M.; Kartal, B. Rapid and simple cryopreservation of anaerobic ammonium-oxidizing bacteria. Appl. Environ. Microbiol. 2012, 78, 3010–3013. [Google Scholar] [CrossRef] [Green Version]
- Nogva, H.K.; Drømtorp, S.M.; Nissen, H.; Rudi, K. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques 2003, 34, 804–813. [Google Scholar] [CrossRef]
- Nocker, A.; Cheung, C.-Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, M.; Nocker, A.; Codony, F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 2012, 91, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Elizaquível, P.; Aznar, R.; Sánchez, G. Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 2014, 116, 1–13. [Google Scholar] [CrossRef]
- Desfossés-Foucault, E.; Dussault-Lepage, V.; Le Boucher, C.; Savard, P.; Lapointe, G.; Roy, D. Assessment of probiotic viability during cheddar cheese manufacture and ripening using propidium monoazide-PCR quantification. Front. Microbiol. 2012, 3, 350. [Google Scholar] [CrossRef] [Green Version]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Paul Ross, R.; Fitzgerald, G.F.; Cotter, P.D. A comparison of methods used to extract bacterial DNA from raw milk and raw milk cheese. J. Appl. Microbiol. 2012, 113, 96–105. [Google Scholar] [CrossRef]
- Schmidt, R.J.; Hu, W.; Mills, J.A.; Kung, L. The development of lactic acid bacteria and Lactobacillus buchneri and their effects on the fermentation of alfalfa silage. J. Dairy Sci. 2009, 92, 5005–5010. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, D.M.; Muck, R.E.; Shinners, K.J.; Weimer, P.J. Use of real time PCR to determine population profiles of individual species of lactic acid bacteria in alfalfa silage and stored corn stover. Appl. Microbiol. Biotechnol. 2006, 71, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Andorrà, I.; Landi, S.; Mas, A.; Guillamón, J.M.; Esteve-Zarzoso, B. Effect of oenological practices on microbial populations using culture-independent techniques. Food Microbiol. 2008, 25, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Saito, R.; Miya, S.; Tanaka, Y.; Miyamura, N.; Kuda, T.; Kimura, B. Development of quantitative real-time PCR for detection and enumeration of Enterobacteriaceae. Int. J. Food Microbiol. 2017, 246, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Roosa, S.; Wauven, C.V.; Billon, G.; Matthijs, S.; Wattiez, R.; Gillan, D.C. The Pseudomonas community in metal-contaminated sediments as revealed by quantitative PCR: A link with metal bioavailability. Res. Microbiol. 2014, 165, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Wright, E.S. DECIPHER: Harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinform. 2015, 16, 322. [Google Scholar] [CrossRef] [Green Version]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, Z.S.L.; Sharpton, T.J.; Grünwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A. ggpubr: “ggplot2” Based Publication Ready Plots. R Package Version 0.2. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 12 June 2019).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2, 1st ed.; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D. R Core Team Nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-140. Available online: https://cran.r-project.org/package=nlme (accessed on 12 June 2019).
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shaw, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef] [Green Version]
- Lenth, R.; Singmann, H.; Love, J.; Buerkner, P.; Herve, M. Emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.4. 2018. Available online: https://cran.r-project.org/web/packages/emmeans/index.html (accessed on 7 February 2020).
- Gamer, M.; Lemon, J.; Singh, I. Irr: Various Coefficients of Interrater Reliability and Agreement. 2012. Available online: http://CRAN. R-project. org/package= irr] (accessed on 12 June 2019).
- Bender, J.M.; Li, F.; Adisetiyo, H.; Lee, D.; Zabih, S.; Hung, L.; Wilkinson, T.A.; Pannaraj, P.S.; She, R.C.; Bard, J.D.; et al. Quantification of variation and the impact of biomass in targeted 16S rRNA gene sequencing studies. Microbiome 2018, 6, 155. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivière, A.; Gagnon, M.; Weckx, S.; Roy, D.; De Vuyst, L. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl. Environ. Microbiol. 2015, 81, 7767–7781. [Google Scholar] [PubMed] [Green Version]
- Gagnon, M.; Savard, P.; Rivière, A.; LaPointe, G.; Roy, D. Bioaccessible antioxidants in milk fermented by Bifidobacterium longum subsp. longum strains. Biomed. Res. Int. 2015, 2015, 169381. [Google Scholar] [PubMed] [Green Version]
- Joseph, L.-M.; Denis, R.; Luc, L. Viability of probiotic bacteria in a maple sap beverage during refrigerated storage. LWT Food Sci. Technol. 2016, 74, 160–167. [Google Scholar]
- Roy, D.; Savard, P.; Guertin, N.; Martoni, C.J.; Jones, M.L.; Champagne, C.P. Viability of Lactobacillus reuteri ncimb 30242 during storage in fruit juice and soy beverage. J. Microbiol. Biotechnol. Food Sci. 2016, 05, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Hui, P.C.; Hui, M.; Yeoh, Y.K.; Wong, P.Y.; Chan, M.C.W.; Wong, M.C.S.; Ng, S.C.; Chan, F.K.L.; Chan, P.K.S. Impact of preservation method and 16S rRNA hypervariable region on gut microbiota profiling. mSystems 2019, 4, e00271-18. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.W.; Martin, J.C.; Scott, P.; Parkhill, J.; Flint, H.J.; Scott, K.P. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Teng, F.; Darveekaran Nair, S.S.; Zhu, P.; Li, S.; Huang, S.; Li, X.; Xu, J.; Yang, F. Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Sci. Rep. 2018, 8, 16321. [Google Scholar] [CrossRef]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.S.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.L.; Tringe, S.G. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef] [Green Version]
- Sierra, D.; Sánchez, A.; Contreras, A.; Luengo, C.; Corrales, J.C.; de la Fe, C.; Guirao, I.; Morales, C.T.; Gonzalo, C. Short communication: Effect of storage and preservation on total bacterial counts determined by automated flow cytometry in bulk tank goat milk. J. Dairy Sci. 2009, 92, 4841–4845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Boeren, S.; van Hooijdonk, A.C.M.; Vervoort, J.M.; Hettinga, K.A. A proteomic perspective on the changes in milk proteins due to high somatic cell count. J. Dairy Sci. 2015, 98, 5339–5351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thongboonkerd, V.; Saetun, P. Bacterial overgrowth affects urinary proteome analysis: Recommendation for centrifugation, temperature, duration, and the use of preservatives during sample collection. J. Proteome Res. 2007, 6, 4173–4181. [Google Scholar] [CrossRef] [PubMed]
- Soggiu, A.; Piras, C.; Mortera, S.L.; Alloggio, I.; Urbani, A.; Bonizzi, L.; Roncada, P. Unravelling the effect of clostridia spores and lysozyme on microbiota dynamics in Grana Padano cheese: A metaproteomics approach. J. Proteom. 2016, 147, 21–27. [Google Scholar] [CrossRef]
| Microorganisms | Primer Name | Primer Sequence (5′-3′) | Gene | Amplicon Size | Reference |
|---|---|---|---|---|---|
| Lactobacillus buchneri | LBF2 LBR1 | GAAACAGGTGCTAATACCGTATAACAACCA CGCCTTGGTAGGCCGTTACCTTACCAACA | 16S rRNA | 129 | [30] |
| Lactobacillus plantarum | LacPlan1F LacPlan1R | AGGCGCGGCTGATGTCA CGCGATTGTCTTGGTTTTGTT | recA | 68 | [31] |
| Lactic acid bacteria | WLAB1 WLAB2 | TCCGGATTTATTGGGCGTAAAGCGA TCGAATTAAACCACATGCTCCA | 16S rRNA | 407 | [32] |
| Acetic acid bacteria | AQ1F AQ2R | TCAAGTCCTCATGGCCCTTATG CGCCATTGTAGCACGTGTGTA | 16S rRNA | 55 | |
| Enterobacteriaceae | rplP 1F rplP 185R | ATGTTACAACCAAAGCGTACA TTACCYTGACGCTTAACTGC | rplP | 185 | [33] |
| Pseudomonas spp. | Pse435F Pse686R | ACTTTAAGTTGGGAGGAAGGG ACACAGGAAATTCCACCACCC | 16S rRNA | 251 | [34] |
| Total bacteria | Uni334F Uni514R | ACTCCTACGGGAGGCAGCAGT ATTACCGCGGCTGCTGGC | 16S rRNA | 180 | [35] |
| Total fungi | ITS1f 5.8S | TCCGTAGGTGAACCTGCGG CGCTGCGTTCTTCATCG | rRNA | 300 | [36] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kennang Ouamba, A.J.; LaPointe, G.; Dufour, S.; Roy, D. Optimization of Preservation Methods Allows Deeper Insights into Changes of Raw Milk Microbiota. Microorganisms 2020, 8, 368. https://doi.org/10.3390/microorganisms8030368
Kennang Ouamba AJ, LaPointe G, Dufour S, Roy D. Optimization of Preservation Methods Allows Deeper Insights into Changes of Raw Milk Microbiota. Microorganisms. 2020; 8(3):368. https://doi.org/10.3390/microorganisms8030368
Chicago/Turabian StyleKennang Ouamba, Alexandre J., Gisèle LaPointe, Simon Dufour, and Denis Roy. 2020. "Optimization of Preservation Methods Allows Deeper Insights into Changes of Raw Milk Microbiota" Microorganisms 8, no. 3: 368. https://doi.org/10.3390/microorganisms8030368





