Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Site and Microbe Isolation
2.2. Solubilization of Insoluble Phosphate
2.3. Screening for Indole-3-Acetic Acid (IAA) and Gibberellins (GAs) in Cell-Free Cultures
2.4. Screening of Endophytes on GA-Deficient Waito-C Dwarf Rice
2.5. Screening of Bacteria for Heavy Metal Stress and Germination of Seeds
2.6. Plant–Microbe Interaction under Heavy Metal Stress
2.7. DNA Extraction, Genome Sequencing, and Genome Assembly
2.8. Genome Annotation
2.9. Comparative Genome Analysis
3. Results and Discussion
3.1. Isolation of PGPR from Soil
3.2. GA Production and Phosphate Solubilization Potential of P. psychrotolerans CS51
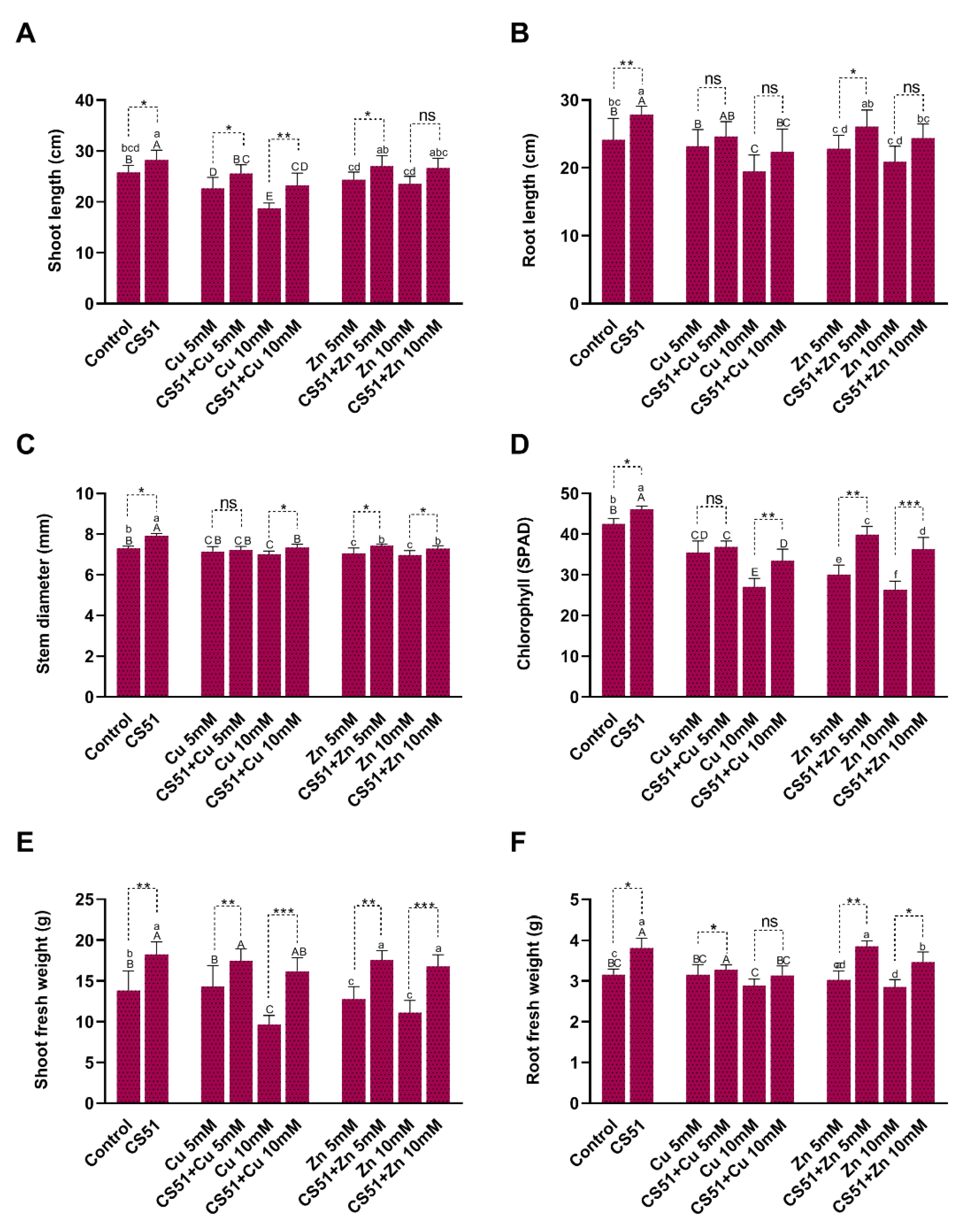
3.3. Promotion of Cucumber Plant Growth by P. Psychrotolerans CS51 under Heavy Metal Stress
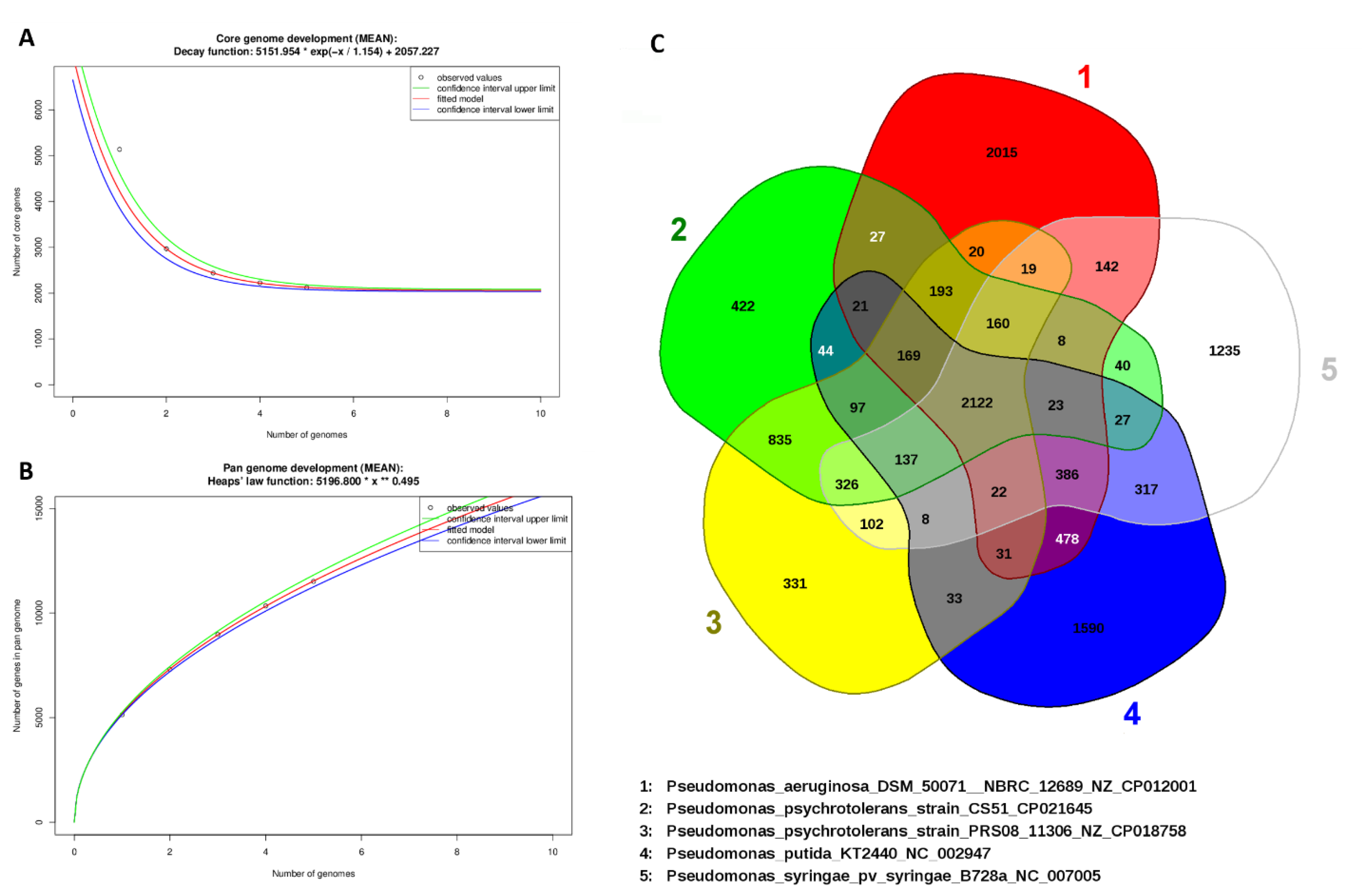
3.4. General Genomic Features of P. psychrotolerans CS51
3.5. Plant Growth-Promoting Potential of P. psychrotolerans CS51
3.6. Phytoremediation Strategies and Resistance to Heavy Metals
3.7. Comparison with Related Species
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Shahid, M.; Khalid, S.; Abbas, G.; Shahid, N.; Nadeem, M.; Sabir, M.; Aslam, M.; Dumat, C. Heavy metal stress and crop productivity. In Crop Production and Global Environmental Issues; Springer: Berlin, Germany, 2015; pp. 1–25. [Google Scholar]
- Förstner, U.; Wittmann, G.T. Metal Pollution in the Aquatic Environment; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Salla, V.; Hardaway, C.J.; Sneddon, J. Preliminary investigation of Spartina alterniflora for phytoextraction of selected heavy metals in soils from Southwest Louisiana. Microchem. J. 2011, 97, 207–212. [Google Scholar] [CrossRef]
- Xiong, T.; Leveque, T.; Shahid, M.; Foucault, Y.; Mombo, S.; Dumat, C. Lead and cadmium phytoavailability and human bioaccessibility for vegetables exposed to soil or atmospheric pollution by process ultrafine particles. J. Environ. Qual. 2014, 43, 1593–1600. [Google Scholar] [CrossRef] [PubMed]
- Moftah, A. Physiological responses of lead-polluted tomato and eggplant to the antioxidant ethylendiure. Minufiya J. Agric. Res. (Egypt) 2000, 25, 933–955. [Google Scholar]
- Chen, M.; Xu, P.; Zeng, G.; Yang, C.; Huang, D.; Zhang, J. Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs. Biotechnol. Adv. 2015, 33, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.; Sandhya, S.; Prasad, M.; Freitas, H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 2012, 30, 1562–1574. [Google Scholar] [CrossRef] [PubMed]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, T.U.; Bano, A.; Naz, I. Alleviation of heavy metals toxicity by the application of plant growth promoting rhizobacteria and effects on wheat grown in saline sodic field. Int. J. Phytoremediation 2017, 19, 522–529. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Ahmad, M.; Zahir, Z.A.; Javaid, A.; Ashraf, M. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 2014, 32, 429–448. [Google Scholar] [CrossRef]
- Dary, M.; Chamber-Pérez, M.; Palomares, A.; Pajuelo, E. “In Situ” phytostabilisation of heavy metal polluted soils using Lupinus luteus inoculated with metal resistant plant-growth promoting rhizobacteria. J. Hazard. Mater. 2010, 177, 323–330. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Kloepper, J.W.; Ryu, C.-M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Bharti, N.; Yadav, D.; Barnawal, D.; Maji, D.; Kalra, A. Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J. Microbiol. Biotechnol. 2013, 29, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.V.; Bogino, P.C.; Nocelli, N.; Cappellari Ldel, R.; Giordano, W.F.; Banchio, E. Analysis of Plant Growth-Promoting Effects of Fluorescent Pseudomonas Strains Isolated from Mentha piperita Rhizosphere and Effects of Their Volatile Organic Compounds on Essential Oil Composition. Front. Microbiol. 2016, 7, 1085. [Google Scholar] [CrossRef] [PubMed]
- Anwar, N.; Abaydulla, G.; Zayadan, B.; Abdurahman, M.; Hamood, B.; Erkin, R.; Ismayil, N.; Rozahon, M.; Mamtimin, H.; Rahman, E. Pseudomonas populi sp. nov., an endophytic bacterium isolated from Populus euphratica. Int. J. Syst. Evol. Microbiol. 2016, 66, 1419–1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Pan, Y.; Wang, K.; Zhang, X.; Zhang, C.; Zhang, S.; Fu, X.; Jiang, J. Pseudomonas zhaodongensis sp. nov., isolated from saline and alkaline soils. Int. J. Syst. Evol. Microbiol. 2015, 65, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, E.; Ramírez-Bahena, M.H.; Fabryová, A.; Igual, J.M.; Benada, O.; Mateos, P.F.; Peix, A.; Kolařík, M.; García-Fraile, P. Pseudomonas coleopterorum sp. nov., a cellulase-producing bacterium isolated from the bark beetle Hylesinus fraxini. Int. J. Syst. Evol. Microbiol. 2015, 65, 2852–2858. [Google Scholar] [CrossRef]
- Pascual, J.; Lucena, T.; Ruvira, M.A.; Giordano, A.; Gambacorta, A.; Garay, E.; Arahal, D.R.; Pujalte, M.J.; Macián, M.C. Pseudomonas litoralis sp. nov., isolated from Mediterranean seawater. Int. J. Syst. Evol. Microbiol. 2012, 62, 438–444. [Google Scholar] [CrossRef]
- Zhong, Z.-P.; Liu, Y.; Hou, T.-T.; Liu, H.-C.; Zhou, Y.-G.; Wang, F.; Liu, Z.-P. Pseudomonas salina sp. nov., isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 2015, 65, 2846–2851. [Google Scholar] [CrossRef]
- Kohler, J.; Caravaca, F.; Carrasco, L.; Roldán, A. Contribution of Pseudomonas mendocina and Glomus intraradices to aggregate stabilization and promotion of biological fertility in rhizosphere soil of lettuce plants under field conditions. Soil Use Manag. 2006, 22, 298–304. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, Y.; Chen, P.; Lin, H.; Ye, G.; Wang, Z.; Ge, C.; Zhu, B.; Ren, D. Genomic and phenotypic analyses of Pseudomonas psychrotolerans PRS08-11306 reveal a turnerbactin biosynthesis gene cluster that contributes to nitrogen fixation. J. Biotechnol. 2017, 253, 10–13. [Google Scholar] [CrossRef]
- Roca, A.; Pizarro-Tobías, P.; Udaondo, Z.; Fernández, M.; Matilla, M.A.; Molina-Henares, M.A.; Molina, L.; Segura, A.; Duque, E.; Ramos, J.L. Analysis of the plant growth-promoting properties encoded by the genome of the rhizobacterium P seudomonas putida BIRD-1. Environ. Microbiol. 2013, 15, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Yanzhen, M.; Yang, L.; Xiangting, X.; Wei, H. Complete genome sequence of a bacterium Pseudomonas fragi P121, a strain with degradation of toxic compounds. J. Biotechnol. 2016, 224, 68–69. [Google Scholar] [CrossRef] [PubMed]
- Spiers, A.J.; Buckling, A.; Rainey, P.B. The causes of Pseudomonas diversity. Microbiology 2000, 146, 2345–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, E.; Kämpfer, P.; Busse, H.-J. Pseudomonas psychrotolerans sp. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1633–1637. [Google Scholar] [CrossRef] [Green Version]
- Santo, C.E.; Lin, Y.; Hao, X.; Wei, G.; Rensing, C.; Grass, G. Draft genome sequence of Pseudomonas psychrotolerans L19, isolated from copper alloy coins. Am. Soc. Microbiol. 2012. [Google Scholar] [CrossRef] [Green Version]
- Simmon, K.E.; Croft, A.C.; Petti, C.A. Application of SmartGene IDNS software to partial 16S rRNA gene sequences for a diverse group of bacteria in a clinical laboratory. J. Clin. Microbiol. 2006, 44, 4400–4406. [Google Scholar] [CrossRef] [Green Version]
- Adorada, D.L.; Stodart, B.J.; Tpoi, R.P.; Costa, S.S.; Ash, G.J. Bacteria associated with sheath browning and grain discoloration of rice in East Timor and implications for Australia’s biosecurity. Australas. Plant Dis. Notes 2013, 8, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Xie, G.-L.; Soad, A.; Swings, J.; Mew, T. Diversity of Gram negative bacteria antagonistic against major pathogens of rice from rice seed in the tropic environment. J. Zhejiang Univ. Sci. A 2003, 4, 463–468. [Google Scholar] [CrossRef]
- Karthik, M.; Pushpakanth, P.; Krishnamoorthy, R.; Senthilkumar, M. Endophytic bacteria associated with banana cultivars and their inoculation effect on plant growth. J. Hortic. Sci. Biotechnol. 2017, 92, 568–576. [Google Scholar] [CrossRef]
- Lee, K.E.; Kang, S.M.; Adhikari, A.; Lee, I.J. Effect of Silicate Solubilizing Bacteria Pseudomonas psychrotolerans CS51 Treatment on Soybean Crops at Paddy Soil. Proc. Korean Soc. Crop Sci. Conf. 2019, 2019, 46. [Google Scholar]
- Gaur, A. Phosphate Solubilizing Micro-Organisms as Biofertilizer; Omega Scientific Publishers: New Delhi, India, 1990. [Google Scholar]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asaf, S.; Khan, M.A.; Khan, A.L.; Waqas, M.; Shahzad, R.; Kim, A.-Y.; Kang, S.-M.; Lee, I.-J. Bacterial endophytes from arid land plants regulate endogenous hormone content and promote growth in crop plants: An example of Sphingomonas sp. and Serratia marcescens. J. Plant Interact. 2017, 12, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-M.; Asaf, S.; Kim, S.-J.; Yun, B.-W.; Lee, I.-J. Complete genome sequence of plant growth-promoting bacterium Leifsonia xyli SE134, a possible gibberellin and auxin producer. J. Biotechnol. 2016, 239, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Bilal, S.; Shahzad, R.; Khan, A.L.; Kang, S.-M.; Imran, Q.M.; Al-Harrasi, A.; Yun, B.-W.; Lee, I.-J. Endophytic microbial consortia of phytohormones-producing fungus Paecilomyces formosus LHL10 and bacteria Sphingomonas sp. LK11 to Glycine max L. regulates physio-hormonal changes to attenuate aluminum and zinc stresses. Front. Plant Sci. 2018, 9, 1273. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.G.; Yin, W.F.; Lim, Y.L. Complete Genome Sequence of Pseudomonas aeruginosa Strain YL84, a Quorum-Sensing Strain Isolated from Compost. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Markowitz, V.M.; Chen, I.M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, D115–D122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blom, J.; Albaum, S.P.; Doppmeier, D.; Pühler, A.; Vorhölter, F.-J.; Zakrzewski, M.; Goesmann, A. EDGAR: A software framework for the comparative analysis of prokaryotic genomes. BMC Bioinform. 2009, 10, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wu, J.; Yang, J.; Sun, S.; Xiao, J.; Yu, J. PGAP: Pan-genomes analysis pipeline. Bioinformatics 2012, 28, 416–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.L.; Waqas, M.; Kang, S.-M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.-Y. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef]
- Kang, S.-M.; Waqas, M.; Hamayun, M.; Asaf, S.; Khan, A.L.; Kim, A.-Y.; Park, Y.-G.; Lee, I.-J. Gibberellins and indole-3-acetic acid producing rhizospheric bacterium Leifsonia xyli SE134 mitigates the adverse effects of copper-mediated stress on tomato. J. Plant Interact. 2017, 12, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Tsukanova, K.; Meyer, J.; Bibikova, T. Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. S. Afr. J. Bot. 2017, 113, 91–102. [Google Scholar] [CrossRef]
- Kang, S.-M.; Waqas, M.; Khan, A.L.; Lee, I.-J. Plant-growth-promoting rhizobacteria: Potential candidates for gibberellins production and crop growth promotion. In Use of Microbes for the Alleviation of Soil Stresses; Springer: New York, NY, USA, 2014; Volume 1, pp. 1–19. [Google Scholar]
- Kim, M.-J.; Radhakrishnan, R.; Kang, S.-M.; You, Y.-H.; Jeong, E.-J.; Kim, J.-G.; Lee, I.-J. Plant growth promoting effect of Bacillus amyloliquefaciens H-2-5 on crop plants and influence on physiological changes in soybean under soil salinity. Physiol. Mol. Biol. Plants 2017, 23, 571–580. [Google Scholar] [CrossRef]
- Kang, S.-M.; Khan, A.L.; You, Y.-H.; Kim, J.-G.; Kamran, M.; Lee, I.-J. Gibberellin production by newly isolated strain Leifsonia soli SE134 and its potential to promote plant growth. J. Microbiol. Biotechnol. 2014, 24, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-M.; Khan, A.L.; Waqas, M.; Asaf, S.; Lee, K.-E.; Park, Y.-G.; Kim, A.-Y.; Khan, M.A.; You, Y.-H.; Lee, I.-J. Integrated phytohormone production by the plant growth-promoting rhizobacterium Bacillus tequilensis SSB07 induced thermotolerance in soybean. J. Plant Interact. 2019, 14, 416–423. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Jiang, W.; Cheng, Z.; Heikkila, J.J.; Glick, B.R. The Complete Genome Sequence of the Plant Growth-Promoting Bacterium Pseudomonas sp. UW4. PLoS ONE 2013, 8, e58640. [Google Scholar] [CrossRef] [Green Version]
- Otieno, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Şahin, F.; Çakmakçi, R.; Kantar, F. Sugar beet and barley yields in relation to inoculation with N 2-fixing and phosphate solubilizing bacteria. Plant Soil 2004, 265, 123–129. [Google Scholar] [CrossRef]
- Bar-Yosef, B.; Rogers, R.; Wolfram, J.; Richman, E. Pseudomonas cepacia–mediated rock phosphate solubilization in kaolinite and montmorillonite suspensions. Soil Sci. Soc. Am. J. 1999, 63, 1703–1708. [Google Scholar] [CrossRef]
- Puente, M.; Bashan, Y.; Li, C.; Lebsky, V. Microbial populations and activities in the rhizoplane of rock-weathering desert plants. I. Root colonization and weathering of igneous rocks. Plant Biol. 2004, 6, 629–642. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-M.; Shahzad, R.; Bilal, S.; Khan, A.L.; You, Y.-H.; Lee, W.-H.; Ryu, H.-L.; Lee, K.-E.; Lee, I.-J. Metabolism-mediated induction of zinc tolerance in Brassica rapa by Burkholderia cepacia CS2-1. J. Microbiol. 2017, 55, 955–965. [Google Scholar] [CrossRef]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Al-Hosni, K.; Kang, S.-M.; Seo, C.-W.; Lee, I.-J. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 2017, 68, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Rajkumar, M.; Freitas, H. Improvement of plant growth and nickel uptake by nickel resistant-plant-growth promoting bacteria. J. Hazard. Mater. 2009, 166, 1154–1161. [Google Scholar] [CrossRef] [Green Version]
- Belimov, A.; Hontzeas, N.; Safronova, V.; Demchinskaya, S.; Piluzza, G.; Bullitta, S.; Glick, B. Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 2005, 37, 241–250. [Google Scholar] [CrossRef]
- Hasnain, S.; Yasmin, S.; Yasmin, A. The effects of lead-resistant Pseudomonads on the growth of Triticum aestivum seedlings under lead stress. Environ. Pollut. 1993, 81, 179–184. [Google Scholar] [CrossRef]
- Rajkumar, M.; Nagendran, R.; Lee, K.J.; Lee, W.H.; Kim, S.Z. Influence of plant growth promoting bacteria and Cr6+ on the growth of Indian mustard. Chemosphere 2006, 62, 741–748. [Google Scholar] [CrossRef]
- Hemambika, B.; Balasubramanian, V.; Rajesh Kannan, V.; Arthur James, R. Screening of chromium-resistant bacteria for plant growth-promoting activities. Soil Sediment Contam. Int. J. 2013, 22, 717–736. [Google Scholar] [CrossRef]
- Brown, S.D.; Utturkar, S.M.; Klingeman, D.M.; Johnson, C.M.; Martin, S.L.; Land, M.L.; Lu, T.Y.; Schadt, C.W.; Doktycz, M.J.; Pelletier, D.A. Twenty-one genome sequences from Pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of Populus deltoides. J. Bacteriol. 2012, 194, 5991–5993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Gopal, M.; Thomas, G.V.; Manikandan, V.; Gajewski, J.; Thomas, G.; Seshagiri, S.; Schuster, S.C.; Rajesh, P.; Gupta, R. Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS ONE 2014, 9, e104259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Shi, H.; Du, Z.; Wang, T.; Liu, X.; Chen, S. Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 2016, 6, 21329. [Google Scholar] [CrossRef]
- Smith, C.J.; Nedwell, D.B.; Dong, L.F.; Osborn, A.M. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl. Environ. Microbiol. 2007, 73, 3612–3622. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Deng, N.; Wu, D.; Hu, S. Quantitative response relationships between net nitrogen transformation rates and nitrogen functional genes during artificial vegetation restoration following agricultural abandonment. Sci. Rep. 2017, 7, 7752. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Kobayashi, Y.; Hulett, F.M. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J. Bacteriol. 1997, 179, 2534–2539. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, M.; Wasaki, J.; Tomizawa, Y.; Shinano, T.; Osaki, M. Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci. Plant Nutr. 2008, 54, 62–71. [Google Scholar] [CrossRef] [Green Version]
- Ram, B.; Fartyal, D.; Sheri, V.; Varakumar, P.; Borphukan, B.; James, D.; Yadav, R.; Bhatt, A.; Agrawal, P.K.; Achary, V.M.M. Characterization of phoA, a Bacterial Alkaline Phosphatase for Phi Use Efficiency in Rice Plant. Front. Plant Sci. 2019, 10, 37. [Google Scholar] [CrossRef] [Green Version]
- Mansilla, M.C.; de Mendoza, D. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology 2000, 146, 815–821. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Barajas, E.; Díaz-Pérez, C.; Ramírez-Díaz, M.I.; Riveros-Rosas, H.; Cervantes, C. Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 2011, 24, 687–707. [Google Scholar] [CrossRef] [PubMed]
- Kunito, T.; Kusano, T.; Oyaizu, H.; Senoo, K.; Kanazawa, S.; Matsumoto, S. Cloning and sequence analysis of czc genes in Alcaligenes sp. strain CT14. Biosci. Biotechnol. Biochem. 1996, 60, 699–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooksey, D.A. Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol. Rev. 1994, 14, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Voloudakis, A.E.; Bender, C.L.; Cooksey, D.A. Similarity between copper resistance genes from Xanthomonas campestris and Pseudomonas syringae. Appl. Environ. Microbiol. 1993, 59, 1627–1634. [Google Scholar] [CrossRef] [Green Version]
- Duncan, R.; Camakaris, J.; Lee, B.; Luke, R. Inducible plasmid-mediated copper resistance in Escherichia coli. Microbiology 1985, 131, 939–943. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Hendson, M.; Schroth, M. Cloning and characterization of copper-resistance genes from Xanthomonas campestris pv. juglandis. Phytopathology 1992, 82, 1125. [Google Scholar]
- Silver, S. Bacterial plasmid resistances to copper, cadmium, and zinc. Chem. Copp. Zinc Triads 1993, 39–53. [Google Scholar]
- Cha, J.-S.; Cooksey, D.A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 1991, 88, 8915–8919. [Google Scholar] [CrossRef] [Green Version]
- Cooksey, D.A.; Azad, H.R. Accumulation of copper and other metals by copper-resistant plant-pathogenic and saprophytic pseudomonads. Appl. Environ. Microbiol. 1992, 58, 274–278. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Olson, B.H. Occurrence of cop-like copper resistance genes among bacteria isolated from a water distribution system. Can. J. Microbiol. 1995, 41, 642–646. [Google Scholar] [CrossRef]
- Cooksey, D.A. Copper uptake and resistance in bacteria. Mol. Microbiol. 1993, 7. [Google Scholar] [CrossRef] [PubMed]
- Hmiel, S.; Snavely, M.; Florer, J.; Maguire, M.; Miller, C. Magnesium transport in Salmonella typhimurium: Genetic characterization and cloning of three magnesium transport loci. J. Bacteriol. 1989, 171, 4742–4751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hmiel, S.; Snavely, M.; Miller, C.; Maguire, M. Magnesium transport in Salmonella typhimurium: Characterization of magnesium influx and cloning of a transport gene. J. Bacteriol. 1986, 168, 1444–1450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groisman, E.A.; Hollands, K.; Kriner, M.A.; Lee, E.-J.; Park, S.-Y.; Pontes, M.H. Bacterial Mg2+ homeostasis, transport, and virulence. Annu. Rev. Genet. 2013, 47, 625–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eitinger, T.; Suhr, J.; Moore, L.; Smith, J.A.C. Secondary transporters for nickel and cobalt ions: Theme and variations. Biometals 2005, 18, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.L.; Grass, G.; Rensing, C. Copper toxicity and the origin of bacterial resistance—New insights and applications. Metallomics 2011, 3, 1109–1118. [Google Scholar] [CrossRef]
- Magnani, D.; Solioz, M. How bacteria handle copper. In Molecular Microbiology of Heavy Metals; Springer: Berlin/Heidelberg, Germany, 2007; pp. 259–285. [Google Scholar]
- Wei, G.; Fan, L.; Zhu, W.; Fu, Y.; Yu, J.; Tang, M. Isolation and characterization of the heavy metal resistant bacteria CCNWRS33-2 isolated from root nodule of Lespedeza cuneata in gold mine tailings in China. J. Hazard. Mater. 2009, 162, 50–56. [Google Scholar] [CrossRef]
- Mellano, M.A.; Cooksey, D.A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 1988, 170, 2879–2883. [Google Scholar] [CrossRef] [Green Version]
- Lejon, D.P.; Nowak, V.; Bouko, S.; Pascault, N.; Mougel, C.; Martins, J.M.; Ranjard, L. Fingerprinting and diversity of bacterial copA genes in response to soil types, soil organic status and copper contamination. FEMS Microbiol. Ecol. 2007, 61, 424–437. [Google Scholar] [CrossRef] [Green Version]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef] [Green Version]
- Konstantinidis, K.T.; Ramette, A.; Tiedje, J.M. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Z.; Halachev, M.R.; Loman, N.J.; Constantinidou, C.; Pallen, M.J. Defining bacterial species in the genomic era: Insights from the genus Acinetobacter. BMC Microbiol. 2012, 12, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Ke, R.; Hughes, D.; Nilsson, M.; Andersson, D.I. Genome-wide detection of spontaneous chromosomal rearrangements in bacteria. PLoS ONE 2012, 7, e42639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leekitcharoenphon, P.; Lukjancenko, O.; Friis, C.; Aarestrup, F.M.; Ussery, D.W. Genomic variation in Salmonella enterica core genes for epidemiological typing. BMC Genom. 2012, 13, 88. [Google Scholar] [CrossRef] [Green Version]
- Adékambi, T.; Butler, R.W.; Hanrahan, F.; Delcher, A.L.; Drancourt, M.; Shinnick, T.M. Core Gene Set As the Basis of Multilocus Sequence Analysis of the Subclass Actinobacteridae. PLoS ONE 2011, 6, e14792. [Google Scholar] [CrossRef] [Green Version]
- Urwin, R.; Maiden, M.C.J. Multi-locus sequence typing: A tool for global epidemiology. Trends Microbiol. 2003, 11, 479–487. [Google Scholar] [CrossRef]




| Annotation Statistics | |
|---|---|
| Genome size (bp) | 5,364,174 |
| GC (%) | 64.71 |
| Total number of genes | 4859 |
| Number of CDSs | 4771 |
| Coding genes | 4683 |
| Pseudogenes | 88 |
| RNA | 88 |
| rRNA (5 S, 16 S, 23 S) | 5, 5, 5 |
| rRNA genes | 15 |
| tRNA genes | 67 |
| ncRNA | 6 |
| Protein-coding genes with function prediction | 3950 |
| Protein-coding genes without function prediction | 733 |
| Protein-coding genes encoding enzymes | 1323 |
| Protein coding genes connected to KEGG pathways | 1577 |
| Protein coding genes connected to KEGG Orthology (KO) | 2726 |
| Protein coding genes connected to MetaCyc pathways | 1128 |
| Protein coding genes with COGs | 3683 |
| Activity Description | Genes in Growth Promoting Activities | Gene Annotation | Gene Names |
|---|---|---|---|
| Plant Hormones | Auxin biosynthesis | Tryptophan synthase alpha chain (Tsa) | (Tsa) |
| Auxin biosynthesis | Anthranilate phosphoribosyltransferase (APRT) | (APRT) | |
| Auxin biosynthesis | Tryptophan synthase beta chain (TSB) | (TSB) | |
| Auxin biosynthesis | Phosphoribosylanthranilate isomerase (PRAI) | (PRAI) | |
| Siderophores | Siderophore Enterobactin | Enterobactin esterase (Fes) | (Fes) |
| Siderophore Enterobactin | Enterobactin synthetase component F, serine activating enzyme (EC 2.7.7.) | EntF | |
| Siderophore Enterobactin | Enterobactin exporter EntS | EntS | |
| Siderophore Enterobactin | Ferric enterobactin transport system permease protein FepG (TC 3.A.1.14.2) | FepG | |
| Siderophore Enterobactin | Ferric enterobactin-binding periplasmic protein FepB (TC 3.A.1.14.2) | FepB | |
| Siderophore Enterobactin | Ferric enterobactin transport system permease protein FepD (TC 3.A.1.14.2) | FepD | |
| Siderophore Enterobactin | Ferric enterobactin transport ATP-binding protein FepC (TC 3.A.1.14.2) | FepC | |
| Nitrogen Metabolism | Cyanate hydrolysis | Cyanate transport protein CynX | CynX |
| Cyanate hydrolysis | Cyanate hydratase (EC 4.2.1.104) | CynS | |
| Nitrosative stress | Anaerobic nitric oxide reductase transcription regulator NorR | NorR | |
| Amidase clustered with urea and nitrile hydratase functions | Amidase clustered with urea ABC transporter and nitrile hydratase functions | ||
| Nitrate and nitrite ammonification | Nitrate ABC transporter, ATP-binding protein | ||
| Nitrate and nitrite ammonification | Response regulator NasT | NasT | |
| Ammonia assimilation | Glutamate synthase [NADPH] putative GlxC chain (EC 1.4.1.13) | GlxC | |
| Ammonia assimilation | Glutamine amidotransferase protein GlxB (EC 2.4.2.-) | GlxB | |
| Phosphorus Metabolism | Phosphate metabolism | Phosphate regulon transcriptional regulatory protein PhoB (SphR) | PhoB |
| Phosphate metabolism | Phosphate transport system permease protein PstC (TC 3.A.1.7.1) | PstC | |
| Phosphate metabolism | Phosphate transport ATP-binding protein PstB (TC 3.A.1.7.1) | PstB | |
| Phosphate metabolism | Phosphate transport system permease protein PstA (TC 3.A.1.7.1) | PstA | |
| Phosphate metabolism | Phosphate transport system regulatory protein PhoU | PhoU | |
| Phosphate metabolism | Predicted ATPase related to phosphate starvation-inducible protein PhoH | PhoH | |
| Phosphate metabolism | Phosphate ABC transporter, periplasmic phosphate-binding protein PstS (TC 3.A.1.7.1) | PstS | |
| Phosphate metabolism | Phosphate starvation-inducible protein PhoH, predicted ATPase | PhoH | |
| Phosphate metabolism | Phosphate regulon sensor protein PhoR (SphS) (EC 2.7.13.3) | PhoR | |
| Sulfur Metabolism | Inorganic Sulfur Assimilation | Putative sulfate permease | Sulp3 |
| Inorganic Sulfur Assimilation | Adenylylsulfate kinase (EC 2.7.1.25) | ASK | |
| Inorganic Sulfur Assimilation | Sulfate and thiosulfate binding protein CysP | CysP | |
| Inorganic Sulfur Assimilation | 3’(2’),5’-bisphosphate nucleotidase (EC 3.1.3.7) | CysQ | |
| Inorganic Sulfur Assimilation | Sulfate transport system permease protein CysW | CysW | |
| Inorganic Sulfur Assimilation | Sulfate transport system permease protein CysT | CysT | |
| Inorganic Sulfur Assimilation | Sulfate adenylyltransferase subunit 2 (EC 2.7.7.4) | SAT2 | |
| Inorganic Sulfur Assimilation | Sulfate adenylyltransferase subunit 1 (EC 2.7.7.4) | SAT1 | |
| Inorganic Sulfur Assimilation | Sulfate and thiosulfate import ATP-binding protein CysA (EC 3.6.3.25) | CysA | |
| Inorganic Sulfur Assimilation | Ferredoxin | FDX | |
| Protein Metabolism | GroEL GroES | Heat shock protein GrpE | GrpE |
| GroEL GroES | Heat shock protein 60 family co-chaperone GroES | GroES | |
| GroEL GroES | Heat shock protein 60 family chaperone GroEL | GroEL | |
| Protein chaperones | Heat shock protein GrpE | GrpE |
| Subsystem | Role | Gene Names |
|---|---|---|
| Cobalt-zinc-cadmium resistance | Cobalt-zinc-cadmium resistance protein CzcD | CzcD |
| Copper homeostasis: copper tolerance | Magnesium and cobalt efflux protein CorC | CorC |
| Magnesium transport | Magnesium and cobalt transport protein CorA | CorA |
| Magnesium transport | Magnesium and cobalt efflux protein CorC | CorC |
| Transport of Nickel and Cobalt | Predicted cobalt transporter CbtC | CbtC |
| Transport of Nickel and Cobalt | Predicted cobalt transporter CbtA | CbtA |
| G3E family of P-loop GTPases (metallocenter biosynthesis) | GTPase involved in cobalt insertion for B12 biosynthesis | CobW |
| Copper homeostasis | Multicopper oxidase | |
| Copper homeostasis | Copper resistance protein CopC | CopC |
| Copper homeostasis | Multidrug resistance transporter, Bcr/CflA family | |
| Copper homeostasis | Cu(I)-responsive transcriptional regulator | |
| Copper homeostasis | Cytochrome c heme lyase subunit CcmF | CcmF |
| Copper homeostasis | Copper resistance protein CopD | CopD |
| Copper homeostasis | Copper chaperone | |
| Copper homeostasis | Copper-translocating P-type ATPase (EC 3.6.3.4) | |
| Copper homeostasis | Copper resistance protein B | copB |
| Copper homeostasis: copper tolerance | Copper homeostasis protein CutE | CutE |
| Copper homeostasis: copper tolerance | Magnesium and cobalt efflux protein CorC | CorC |
| Copper uptake system CopCD | Copper resistance protein CopD | CopD |
| Copper uptake system CopCD | Copper resistance protein CopC | CopC |
| Copper transport system | Copper-translocating P-type ATPase (EC 3.6.3.4) | |
| Copper transport system | Copper resistance protein CopC | CopC |
| Copper transport and blue copper proteins | Azurin | |
| Copper homeostasis | Copper resistance protein A | CopA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, S.-M.; Asaf, S.; Khan, A.L.; Lubna; Khan, A.; Mun, B.-G.; Khan, M.A.; Gul, H.; Lee, I.-J. Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions. Microorganisms 2020, 8, 382. https://doi.org/10.3390/microorganisms8030382
Kang S-M, Asaf S, Khan AL, Lubna, Khan A, Mun B-G, Khan MA, Gul H, Lee I-J. Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions. Microorganisms. 2020; 8(3):382. https://doi.org/10.3390/microorganisms8030382
Chicago/Turabian StyleKang, Sang-Mo, Sajjad Asaf, Abdul Latif Khan, Lubna, Adil Khan, Bong-Gyu Mun, Muhammad Aaqil Khan, Humaira Gul, and In-Jung Lee. 2020. "Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions" Microorganisms 8, no. 3: 382. https://doi.org/10.3390/microorganisms8030382
APA StyleKang, S.-M., Asaf, S., Khan, A. L., Lubna, Khan, A., Mun, B.-G., Khan, M. A., Gul, H., & Lee, I.-J. (2020). Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions. Microorganisms, 8(3), 382. https://doi.org/10.3390/microorganisms8030382






