Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Bacteriophage Isolation
2.3. Salmonella Challenge Tests
2.4. Morphology of LPSEYT
2.5. Phage Spot Test
2.6. Determination of Phage Adsorption Rate and One-Step Growth
2.7. pH and Temperature Tolerance
2.8. Phage Sequencing, Genome Annotation and Comparison
2.9. Infection Effect of Phage LPSEYT against Salmonella in Food
2.10. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Lytic Activity of the LPSEYT
3.2. Morphology and Biological Characteristics of LPSEYT
3.3. Investigation of the Infection Effect of Phage LPSEYT against Salmonella in Food
3.4. Sequencing and Bioinformatics Analysis
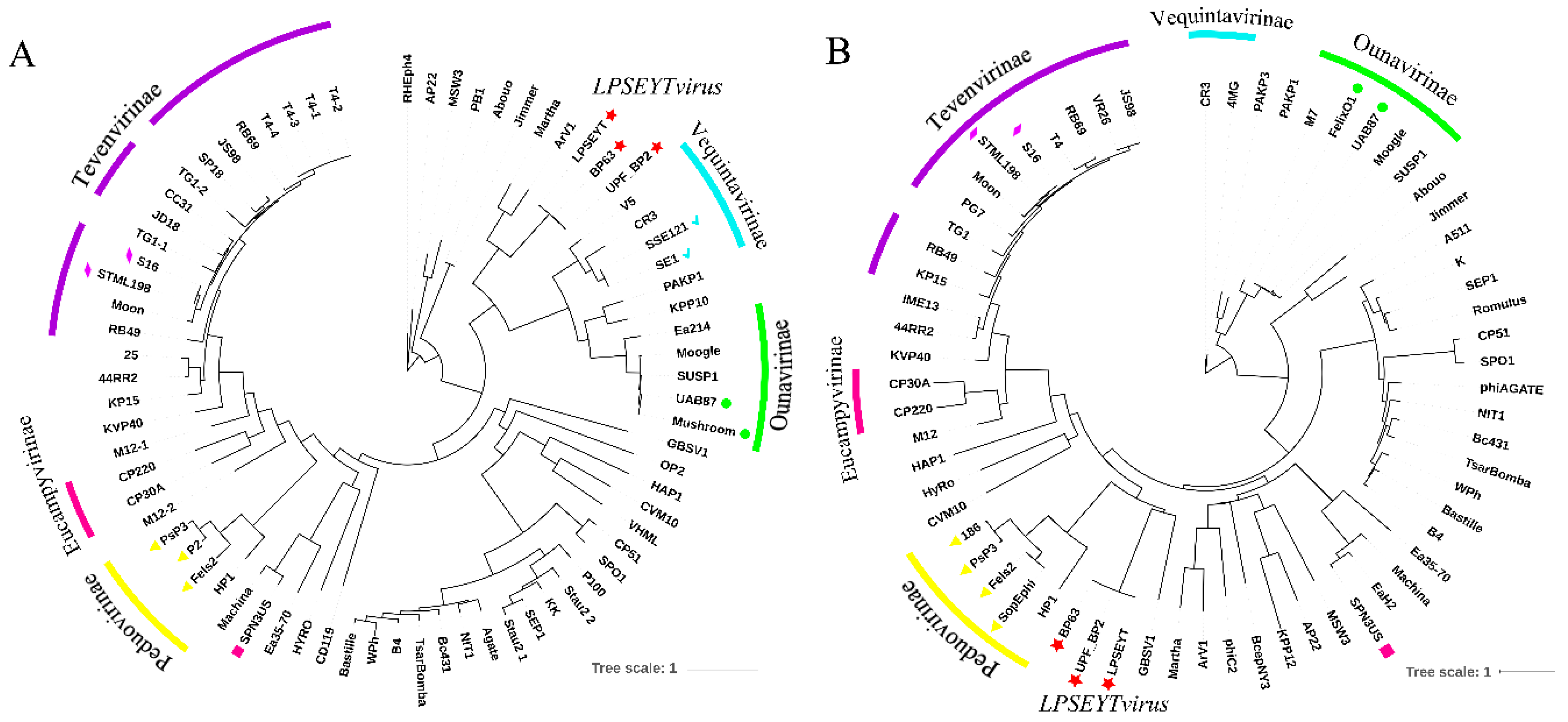
3.5. Classification of Lytic Phage LPSEYT and a New Genus LPSEYTvirus Can Be Proposed
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO. WHO Estimates of the Global Burden of Diseases-Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Lynch, M.F.; Tauxe, R.V.; Hedberg, C.W. The growing burden of foodborne outbreaks due to contaminated fresh produce: Risks and opportunities. Epidemiol. Infect. 2009, 137, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Duan, H.; Zhang, W.; Li, J.W. Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. Fems Immunol. Med. Microbiol. 2007, 51, 8–13. [Google Scholar] [CrossRef]
- Dewey-Mattia, D.; Manikonda, K.; Hall, A.J.; Wise, M.E.; Crowe, S.J. Surveillance for foodborne disease outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018, 67, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Finstad, S.; O′Bryan, C.A.; Marcy, J.A.; Crandall, P.G.; Ricke, S.C. Salmonella and broiler processing in the United States: Relationship to foodborne salmonellosis. Food Res. Int. 2012, 45, 789–794. [Google Scholar] [CrossRef]
- Braoudaki, M.; Hilton, A.C. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-Resistance to antimicrobial agents. J. Clin. Microbiol. 2004, 42, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Braoudaki, M.; Hilton, A.C. Low level of cross-Resistance between triclosan and antibiotics in Escherichia coli K-12 and E. coli O55 compared to E. coli O157. FEMS Microbiol. Lett. 2004, 235, 305–309. [Google Scholar] [CrossRef]
- Ren, D.; Chen, P.; Wang, Y.; Wang, J.; Liu, H.; Liu, H. Phenotypes and antimicrobial resistance genes in Salmonella isolated from retail chicken and pork in Changchun, China. J. Food Saf. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Bruttin, A.; Brussow, H. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob. Agents Chemother. 2005, 49, 2874–2878. [Google Scholar] [CrossRef]
- Meaden, S.; Koskella, B. Exploring the risks of phage application in the environment. Front. Microbiol. 2013, 4, 1–8. [Google Scholar] [CrossRef]
- Sarker, S.A.; McCallin, S.; Barretto, C.; Berger, B.; Pittet, A.-C.; Sultana, S.; Krause, L.; Huq, S.; Bibiloni, R.; Bruttin, A.; et al. Oral T4-Like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 2012, 434, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Intralytix: Food Safety. Available online: http://www.intralytix.com/index.php?page=food (accessed on 28 January 2020).
- Micreos: Food Safety. Available online: https://www.micreos.com/content/food-safety.aspx (accessed on 28 January 2020).
- Yuan, Y.; Peng, Q.; Yang, S.; Zhang, S.; Fu, Y.; Wu, Y.; Gao, M. Isolation of a novel Bacillus thuringiensis phage representing a new phage lineage and characterization of its endolysin. Viruses 2018, 10, 611. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Pirnay, J.-P.; Vos, D.D. Guidelines to compose an ideal bacteriophage cocktail. In Bacteriophage Therapy; Humana Press: New York, NY, USA, 2018; pp. 99–110. [Google Scholar]
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef] [PubMed]
- ICTV Master Species List 2018a v1. Available online: https://talk.ictvonline.org/files/master-species-lists/m/msl/7992 (accessed on 4 March 2019).
- Ackermann, H.W. Phage classification and characterization. Methods Mol. Biol. 2009, 501, 127–140. [Google Scholar]
- Calendar, R. The Bacteriophages; Oxford University Press: New York, NY, USA, 2006. [Google Scholar]
- Eriksson, H.; Maciejewska, B.; Latka, A.; Majkowska-Skrobek, G.; Hellstrand, M.; Melefors, Ö.; Wang, J.-T.; Kropinski, A.M.; Drulis-Kawa, Z.; Nilsson, A.S. A suggested new bacteriophage genus, “Kp34likevirus”, within the Autographivirinae subfamily of Podoviridae. Viruses 2015, 7, 1804–1822. [Google Scholar] [CrossRef]
- Lu, S.; Le, S.; Tan, Y.; Zhu, J.; Li, M.; Rao, X.; Zou, L.; Li, S.; Wang, J.; Jin, X.; et al. Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: Establishment of genus PaP1-Like phages. PLoS ONE 2013, 8, e62933. [Google Scholar] [CrossRef]
- Lavigne, R.; Darius, P.; Summer, E.J.; Seto, D.; Mahadevan, P.; Nilsson, A.S.; Ackermann, H.W.; Kropinski, A.M. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009, 9, 224. [Google Scholar] [CrossRef]
- Lopes, A.; Tavares, P.; Petit, M.-A.; Guérois, R.; Zinn-Justin, S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genom. 2014, 15, 1027. [Google Scholar] [CrossRef]
- Buttimer, C.; Lucid, A.; Neve, H.; Franz, C.M.; O’Mahony, J.; Turner, D.; Lavigne, R.; Coffey, A. Pectobacterium atrosepticum phage vB_PatP_CB5: A member of the proposed genus ‘Phimunavirus’. Viruses 2018, 10, 394. [Google Scholar] [CrossRef]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, W.S.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, characterization, and application of bacteriophage LPSE1 against Salmonella enterica in Ready to Eat (RTE) foods. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Islam, M.; Zhou, Y.; Liang, L.; Nime, I.; Liu, K.; Yan, T.; Wang, X.; Li, J. Application of a phage cocktail for control of Salmonella in foods and reducing biofilms. Viruses 2019, 11, 841. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Cunha, Â.; Delgadillo, I.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Bacteriophages with potential to inactivate Salmonella Typhimurium: Use of single phage suspensions and phage cocktails. Virus Res. 2016, 220, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Kwiatek, M.; Parasion, S.; Mizak, L.; Gryko, R.; Bartoszcze, M.; Kocik, J. Characterization of a bacteriophage, isolated from a cow with mastitis, that is lytic against Staphylococcus aureus strains. Arch. Virol. 2012, 157, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Lee, J.H.; Shin, H.; Kim, M.; Choi, J.; Kang, D.H.; Heu, S.; Ryu, S. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157: H7. Appl. Enviromental Microbiol. 2012, 78, 58–69. [Google Scholar] [CrossRef]
- Adams, M.H. Assay of phage by agar layer method. In Bacteriophages; Interscience Publishers Inc.: New York, NY, USA, 1959; pp. 450–454. [Google Scholar]
- López-Cuevas, O.; Campo, N.C.; León-Félix, J.; González-Robles, A.; Chaidez, C. Characterization of bacteriophages with a lytic effect on various Salmonella serotypes and Escherichia coli O157:H7. Can. J. Microbiol. 2011, 57, 1042–1051. [Google Scholar] [CrossRef] [PubMed]
- Tomat, D.; Casabonne, C.; Aquili, V.; Balagué, C.; Quiberoni, A. Evaluation of a novel cocktail of six lytic bacteriophages against Shiga toxin-producing Escherichia coli in broth, milk and meat. Food Microbiol. 2018, 76, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Spricigo, D.A.; Bardina, C.; Cortés, P.; Llagostera, M. Use of a bacteriophage cocktail to control Salmonella in food and the food industry. Int. J. Food Microbiol. 2013, 165, 169–174. [Google Scholar] [CrossRef]
- Carey-Smith, G.V.; Billington, C.; Cornelius, J.A.; Hudson, J.A.; Heinemann, J.A. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol. Lett. 2006, 258, 182–186. [Google Scholar] [CrossRef]
- Bao, H.; Zhang, P.; Zhang, H.; Zhou, Y.; Zhang, L.; Wang, R. Bio-Control of Salmonella Enteritidis in foods using bacteriophages. Viruses 2015, 7, 4836–4853. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Q.; Zhang, C.; Yang, J.; Lu, Z.; Lu, F.; Bie, X. Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res. 2017, 236, 14–23. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Prangishvili, D.; Lavigne, R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 2009, 11, 2775–2777. [Google Scholar] [CrossRef] [PubMed]
- El-Dougdoug, N.K.; Cucic, S.; Abdelhamid, A.G.; Brovko, L.; Kropinski, A.M.; Griffiths, M.W.; Anany, H. Control of Salmonella Newport on cherry tomato using a cocktail of lytic bacteriophages. Int. J. Food Microbiol. 2019, 293, 60–71. [Google Scholar] [CrossRef]
- Lal, T.M.; Sano, M.; Ransangan, J. Isolation and characterization of large marine bacteriophage (Myoviridae), VhKM4 infecting Vibrio harveyi. J. Aquat. Anim. Health 2017, 29, 26–30. [Google Scholar] [CrossRef]
- Chang, H.C.; Chen, C.R.; Lin, J.W.; Shen, G.H.; Chang, K.M.; Tseng, Y.H.; Weng, S.F. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage φSMA5. Public Health Microbiol. 2005, 71, 1387–1393. [Google Scholar]
- Kęsik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dąbrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Łusiak-Szelachowska, M.; Żaczek, M.; Górski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Russell, N.; Elrod, B.; Dominguez, K. Stress-Induced prophage DNA replication in Salmonella enterica serovar Typhimurium. Infect. Genet. Evol. 2009, 9, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Berngruber, T.W.; Weissing, F.J.; Gandon, S. Inhibition of superinfection and the evolution of viral latency. J. Virol. 2010, 84, 10200–10208. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, M.; Lin, H.; Wang, J.; Jin, Y.; Han, F. Characterization of the novel T4-like Salmonella enterica bacteriophage STP4-A and its endolysin. Arch. Virol. 2016, 161, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771–10776. [Google Scholar] [CrossRef] [PubMed]
- Settle, L.L. Characterization of the Bacteriophage Felix O1 Endolysin and Potential Application for Salmonella Bioremediation. Ph.D. Thesis, the Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2012. [Google Scholar]
- Ramanculov, E.; Young, R. Genetic analysis of the T4 holin: Timing and topology. Gene 2001, 265, 25–36. [Google Scholar] [CrossRef]
- Lee, J.S.; Jang, H.B.; Kim, K.S.; Kim, T.H.; Im, I.S.; Kim, S.W.; Lazarte, J.M.S.; Kim, J.S.; Jung, T.S. Complete genomic and lysis-Cassette characterization of the novel phage, KBNP1315, which infects avian pathogenic Escherichia coli (APEC). PLoS ONE 2015, 10, e0142504. [Google Scholar] [CrossRef] [PubMed]
- Catalão, M.J.; Gil, F.; Moniz-Pereira, J.; Pimentel, M. Functional analysis of the holin-Like proteins of mycobacteriophage Ms6. J. Bacteriol. 2011, 193, 2793–2803. [Google Scholar] [CrossRef]
- Lim, J.A.; Shin, H.; Heu, S.; Ryu, S. Exogenous lytic activity of SPN9CC endolysin against gram-negative bacteria. J. Microbiol. Biotechnol. 2014, 24, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Born, Y.; Fieseler, L.; Marazzi, J.; Lurz, R.; Duffy, B.; Loessner, M.J. Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl. Enviromental Microbiol. 2011, 77, 5945–5954. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; Guinane, C.M.; Johnston, C.; Neve, H.; Coffey, A.; Ross, R.P.; McAuliffe, O.; O′Mahony, J. Genome analysis of Cronobacter phage vB_CsaP_Ss1 reveals an endolysin with potential for biocontrol of gram-negative bacterial pathogens. J. Gen. Virol. 2014, 96, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Lingohr, E.J.; Ackermann, H.W. The genome sequence of enterobacterial phage 7-11, which possesses an unusually elongated head. Arch. Virol. 2011, 156, 149–151. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.B.; Sun, E.C.; Yang, L.; Tong, Y.G.; Song, J.Y.; Wu, B. Characterization and complete genome sequence analysis of Shiga toxin-producing Escherichia bacteriophage vB_EcoM_PHB05. Chin. Vet. Sci. 2018, 48, 428–437. [Google Scholar]
- Dreiseikelmann, B.; Bunk, B.; Spröer, C.; Rohde, M.; Nimtz, M.; Wittmann, J. Characterization and genome comparisons of three Achromobacter phages of the family Siphoviridae. Arch. Virol. 2017, 162, 2191–2201. [Google Scholar] [CrossRef]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genom. 2012, 13, 223. [Google Scholar] [CrossRef]
- Kwan, T.; Liu, J.; DuBow, M.; Gros, P.; Pelletier, J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 2006, 188, 1184–1187. [Google Scholar] [CrossRef]
- Sepúlveda-Robles, O.; Kameyama, L.; Guarneros, G. High diversity and novel species of Pseudomonas aeruginosa bacteriophages. Appl. Environ. Microbiol. 2012, 78, 4510–4515. [Google Scholar] [CrossRef] [PubMed]
- Christiane, E.; Libera, L.; Cédric, M.; Yann, B.; Guillaume, L.; Nguetta, S.P.A.; Serge, L.; Arsher, C.; Kouassi, A.K.; Gilles, V.; et al. Investigation of a large collection of Pseudomonas aeruginosa bacteriophages collected from a single environmental source in Abidjan, Côte d’Ivoire. PLoS ONE 2015, 10, e0130548. [Google Scholar]
- Peters, D.L.; Lynch, K.H.; Stothard, P.; Dennis, J.J. The isolation and characterization of two Stenotrophomonas maltophilia bacteriophages capable of cross-taxonomic order infectivity. BMC Genom. 2015, 16, 664. [Google Scholar] [CrossRef] [PubMed]
- Lal, T.M.; Ransangan, J. Complete genome sequence of VpKK5, a novel Vibrio parahaemolyticus lytic siphophage. Genome Announc. 2015, 3, 1–2. [Google Scholar] [CrossRef]
- Kanamaru, S.; Kondabagil, K.; Rossmann, M.G.; Rao, V.B. The functional domains of bacteriophage T4 terminase. J. Biol. Chem. 2004, 279, 40795–40801. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef]
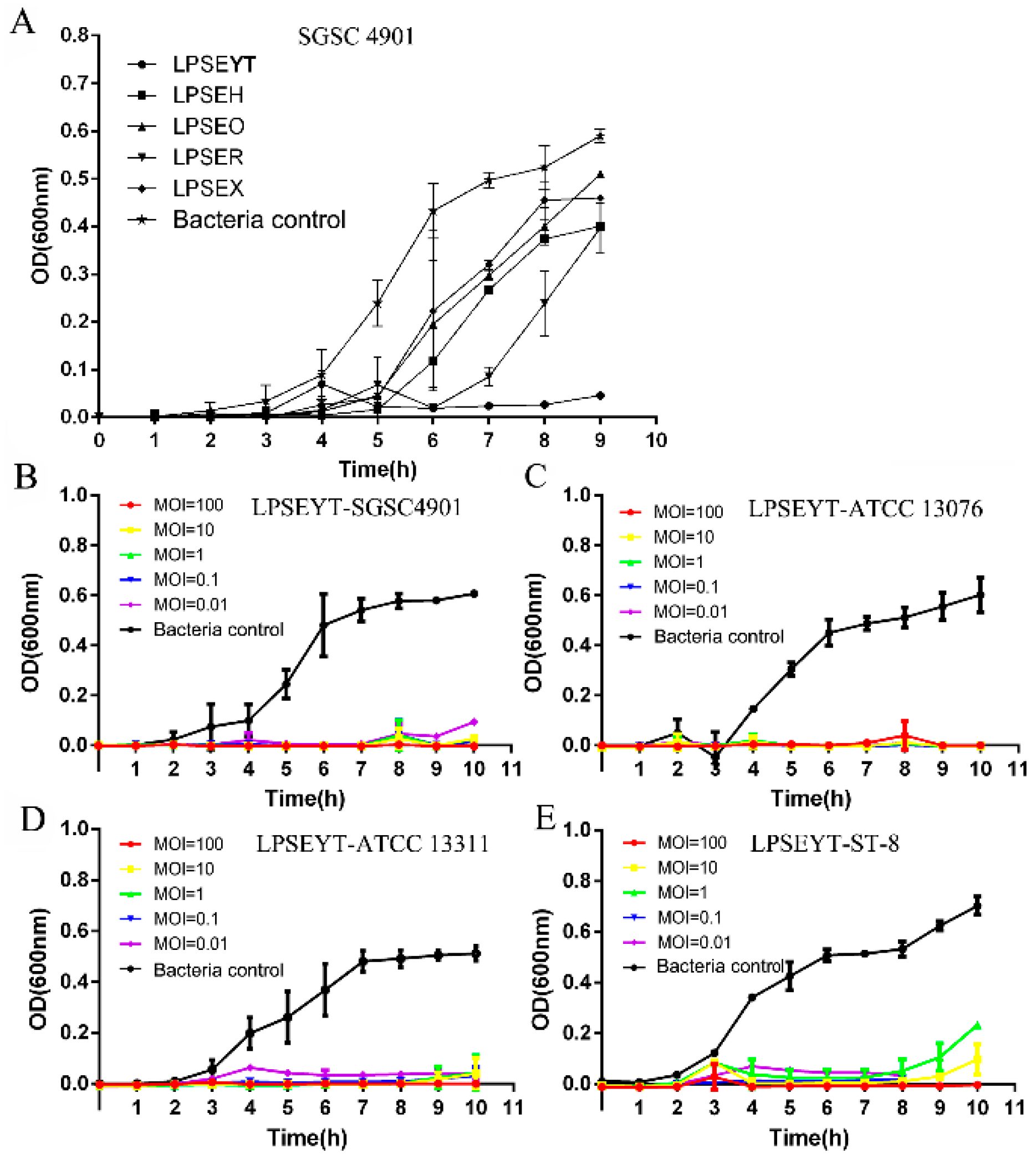
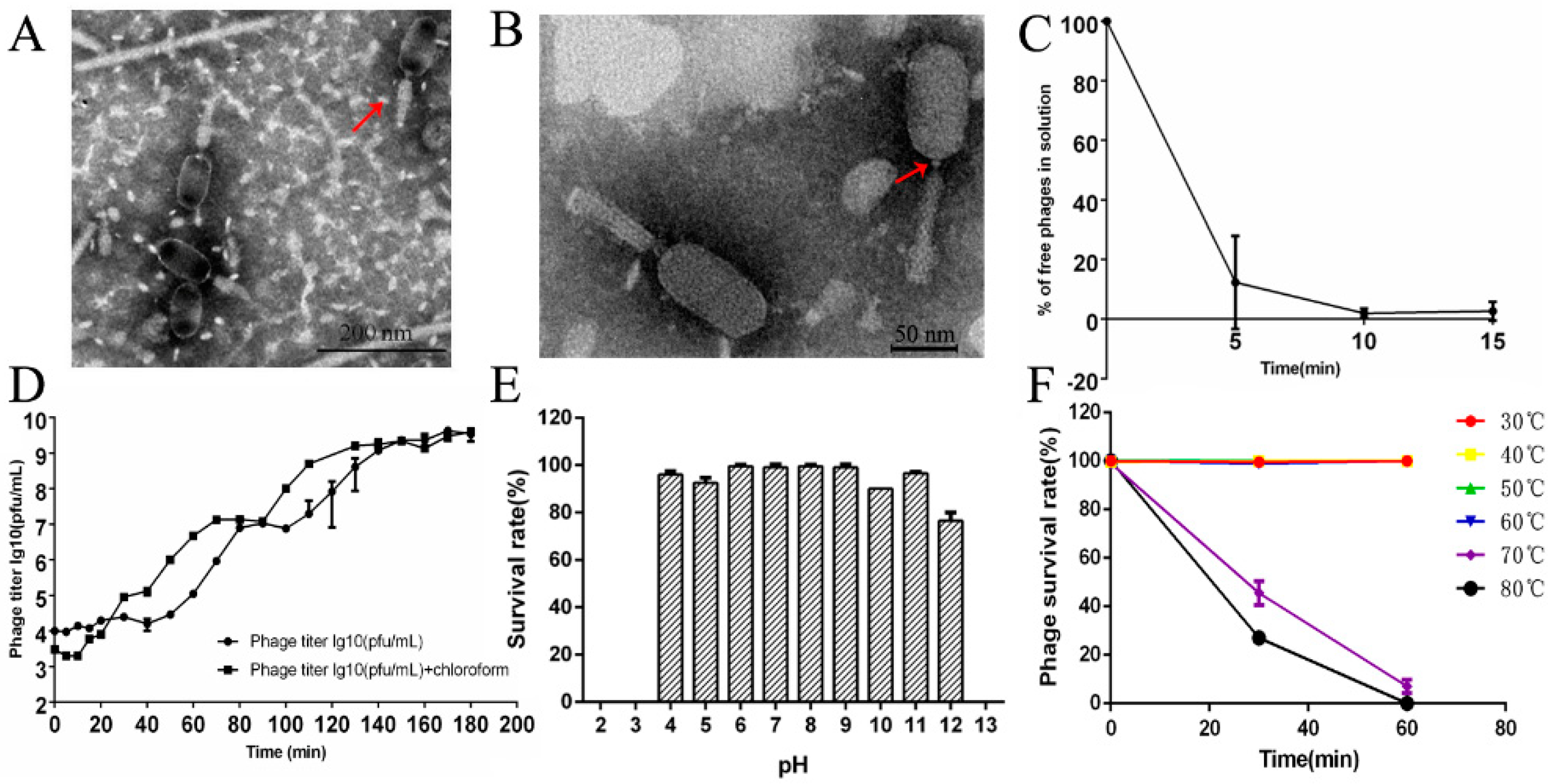
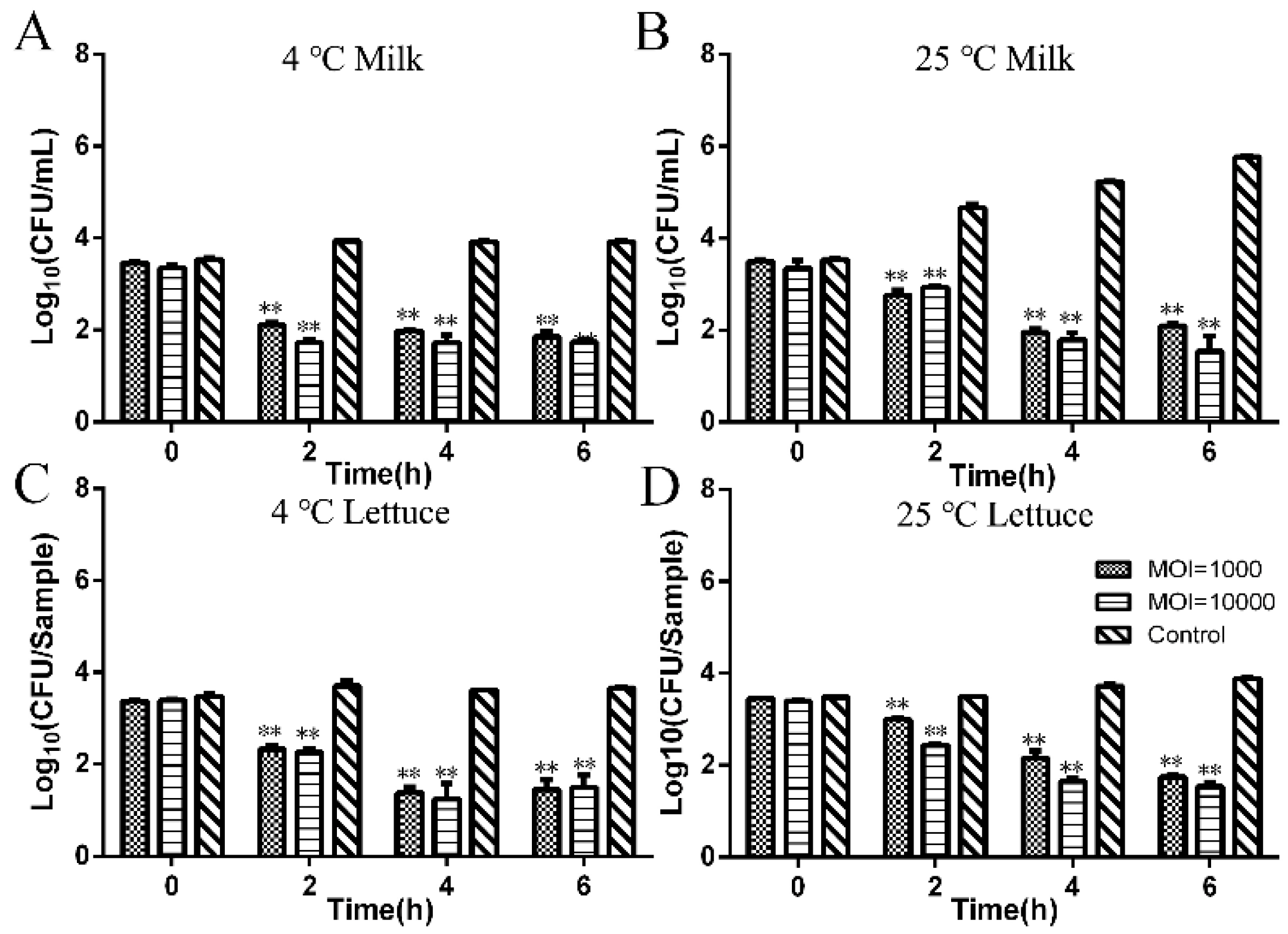
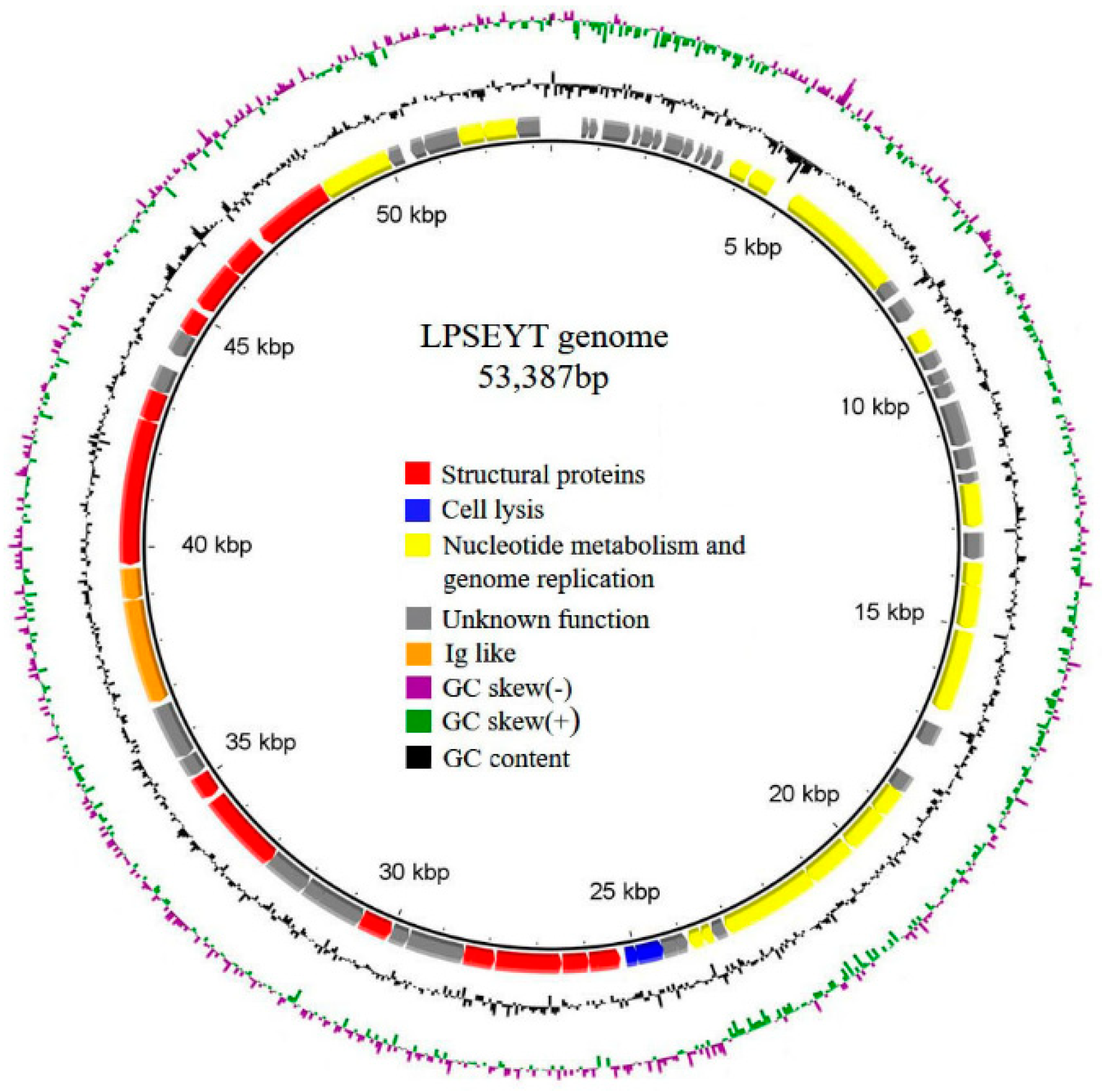
| Bacterial | Infectivity of Phage | ||||
|---|---|---|---|---|---|
| LPSEH | LPSEO | LPSER | LPSEX | LPSEYT | |
| S. Enteritidis ATCC 13076 | + | + | + | + | + |
| S. Enteritidis SJTUF 10978 | + | + | + | + | + |
| S. Enteritidis SJTUF 10984 | + | + | + | + | + |
| S. Enteritidis LSE4 (LK5) | + | + | + | + | + |
| S. Enteritidis SGSC 4901 | + | + | + | + | + |
| S. Typhimurium ATCC 14028 | + | + | + | + | + |
| S. Typhimurium ATCC 13311 | + | + | + | + | + |
| S. Typhimurium LST2 (ST-8) | + | + | + | + | + |
| S. Typhimurium LST4 (UK-1) | + | + | + | + | + |
| S. Typhimurium LST8 (SL1344) | + | + | + | + | + |
| S. Typhimurium SGSC 4903 | + | + | + | + | + |
| S. Typhi LSX1 (CT18) | + | + | – | + | + |
| S. Typhi LSX2 (Ty2) | + | + | – | + | + |
| S. pullorum CVCC 519 | + | + | + | + | + |
| S. pullorum CVCC 534 | + | – | + | – | + |
| Escherichia coli LEC4 (DH5α) | – | – | + | – | + |
| Escherichia coli LEC6 (BL21) | – | – | – | – | – |
| Escherichia coli LEC10 (D41) | – | – | – | – | + |
| Escherichia coli ATCC 10798 | – | – | – | – | + |
| Phage | Country | Accession | Genome Length (bp) | E Value | BLASTN Score | Query Cover | Identify | Ref. |
|---|---|---|---|---|---|---|---|---|
| Salmonella phage UPF_BP2 | Brazil | KX826077.1 | 54,894 | 0 | 9.0 × 104 | 96% | 97% | unpublished |
| Salmonella phage BP63 | Canada | KM366099.1 | 52,437 | 0 | 8.6 × 104 | 98% | 98% | unpublished |
| Erwinia phage vB_EamM-M7 | Switzerland | HQ728263.1 | 84,694 | 2 × 10−14 | 322 | 2% | 65% | [52] |
| Erwinia phage vB_EamM_Asesino | USA | KX397364.1 | 246,291 | 1 × 10−16 | 283 | 1% | 66% | unpublished |
| Cronobacter phage vB_CsaP_GAP52 | Canada | JN882286.1 | 76,631 | 3 × 10−69 | 280 | 1% | 69% | unpublished |
| Acinetobacter phage SH-Ab 15497 | China | MG674163.1 | 43,420 | 1 × 10−30 | 276 | 1% | 66% | unpublished |
| Cronobacter phage vB_CsaP_Ss1 | Ireland | KM058087.1 | 42,205 | 1 × 10−17 | 267 | 1% | 67% | [53] |
| Salmonella phage SE131 | South Korea | MG873442.1 | 89,910 | 4 × 10−55 | 233 | 1% | 67% | unpublished |
| Salmonella phage 7-11 | Canada | HM997019.1 | 89,916 | 4 × 10−54 | 230 | 1% | 67% | [54] |
| Escherichia phage vB_EcoM_PHB05 | China | MF805809.1 | 147,659 | 5 × 10−15 | 211 | <1% | 72% | [55] |
| Enterobacteria phage ECGD1 | China | KU522583.1 | 146,647 | 3 × 10−12 | 204 | <1% | 71% | unpublished |
| Phage name | Phage Family | Genus | Genome Length (kb) | Accession | Homologous Genes with LPSEYT a | Homologous Proteins Rate | Reference |
|---|---|---|---|---|---|---|---|
| Salmonella phage BP63 | Caudovirales | unclassified | 52.437 | NC_031250 | 70 | 95.89% | unpublished |
| Salmonella phage UPF_BP2 | Caudovirales | unclassified | 54.894 | KX826077 | 47 | 64.38% | unpublished |
| Paracoccus phage vB_PmaS_IMEP1 | Siphoviridae | unclassified | 42.093 | NC_026608 | 7 | 9.59% | unpublished |
| Achromobacter phage JWX | Siphoviridae | Steinhofvirus | 49.714 | NC_028768 | 4 | 5.48% | [56] |
| Acinetobacter phage SH-Ab 15497 | unclassified | unclassified | 43.42 | MG674163 | 4 | 5.48% | unpublished |
| Burkholderia phage BcepGomr | Siphoviridae | unclassified | 52.414 | NC_009447 | 4 | 5.48% | unpublished |
| Burkholderia phage KL1 | Siphoviridae | Septimatrevirus | 42.832 | NC_018278 | 4 | 5.48% | [57] |
| Pantoea phage vB_PagS_Vid5 | Siphoviridae | Vidquintavirus | 61.437 | MG948468 | 4 | 5.48% | unpublished |
| Pseudomonas phage 73 | Siphoviridae | Septimatrevirus | 42.999 | NC_007806 | 4 | 5.48% | [58] |
| Pseudomonas phage JG054 | Siphoviridae | Nipunavirus | 57.839 | KX898400 | 4 | 5.48% | unpublished |
| Pseudomonas phage NP1 | Siphoviridae | Nipunavirus | 58.566 | NC_031058 | 4 | 5.48% | unpublished |
| Pseudomonas phage PaMx42 | Siphoviridae | Septimatrevirus | 43.225 | NC_028879 | 4 | 5.48% | [59] |
| Pseudomonas phage vB_PaeS_SCH_Ab26 | Siphoviridae | Septimatrevirus | 43.056 | HG962376 | 4 | 5.48% | [60] |
| Stenotrophomonas phage vB_SmaS-DLP_2 | Siphoviridae | Septimatrevirus | 42.593 | NC_029019 | 4 | 5.48% | [61] |
| Vibrio phage VpKK5 | Siphoviridae | unclassified | 56.637 | NC_026610 | 4 | 5.48% | [62] |
| Xanthomonas phage XAJ2 | unclassified | unclassified | 49.241 | KU197014 | 4 | 5.48% | unpublished |
| Family | Subfamily | Genus | Phage | Accession | Country | Genome Length (kb) |
|---|---|---|---|---|---|---|
| Myoviridae | unknown | Spn3virus | Salmonella virus SPN3US | NC_027402 | South Korea | 240.413 |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus FelixO1 | AF320576 | USA | 86.155 |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus HB2014 | unknown | unknown | unknown |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus Mushroom | KP143762 | USA | 87.709 |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus UAB87 | JN225449 | Espanya | 87.603 |
| Myoviridae | Peduovirinae | P2virus | Salmonella virus Fels2 | NC_010463 | USA | 33.693 |
| Myoviridae | Peduovirinae | P2virus | Salmonella virus PsP3 | NC_005340 | USA | 30.636 |
| Myoviridae | Peduovirinae | P2virus | Salmonella virus SopEphi | unknown | unknown | unknown |
| Myoviridae | Tevenvirinae | S16virus | Salmonella virus S16 | NC_020416 | Switzerland | 160.221 |
| Myoviridae | Tevenvirinae | S16virus | Salmonella virus STML198 | NC_027344 | USA | 158.099 |
| Myoviridae | Vequintavirinae | Se1virus | Salmonella virus SE1 | GU070616 | Portugal | 145.964 |
| Myoviridae | Vequintavirinae | Se1virus | Salmonella virus SSE121 | NC_027351 | USA | 147.745 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, T.; Liang, L.; Yin, P.; Zhou, Y.; Mahdy Sharoba, A.; Lu, Q.; Dong, X.; Liu, K.; Connerton, I.F.; Li, J. Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods. Microorganisms 2020, 8, 400. https://doi.org/10.3390/microorganisms8030400
Yan T, Liang L, Yin P, Zhou Y, Mahdy Sharoba A, Lu Q, Dong X, Liu K, Connerton IF, Li J. Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods. Microorganisms. 2020; 8(3):400. https://doi.org/10.3390/microorganisms8030400
Chicago/Turabian StyleYan, Ting, Lu Liang, Ping Yin, Yang Zhou, Ashraf Mahdy Sharoba, Qun Lu, Xingxing Dong, Kun Liu, Ian F. Connerton, and Jinquan Li. 2020. "Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods" Microorganisms 8, no. 3: 400. https://doi.org/10.3390/microorganisms8030400
APA StyleYan, T., Liang, L., Yin, P., Zhou, Y., Mahdy Sharoba, A., Lu, Q., Dong, X., Liu, K., Connerton, I. F., & Li, J. (2020). Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods. Microorganisms, 8(3), 400. https://doi.org/10.3390/microorganisms8030400






