Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria
Abstract
:1. Introduction
2. Microbial Resistance Mechanisms
3. Microbial Quorum Sensing System and Its Regulation Mechanism
4. Quorum Sensing and Biological Competition
5. Regulation of Microbial Resistance by Quorum Sensing
5.1. Regulation of Bacterial Efflux Pump by QS
5.2. Regulation of Bacterial Biofilm Formation by QS
5.3. Regulation of Bacterial Secretion System by QS
6. New Strategy for Preventive Treatment of Microbial Resistance
6.1. Inhibition of Signal Molecule Production
6.2. Degradation of Signal Molecule
6.3. Inhibition of Signal Molecule Conduction or Binding to Receptors
7. Future Outlook
8. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chioro, A.; Coll-Seck, A.M.; Hoie, B.; Moeloek, N.; Motsoaledi, A.; Rajatanavin, R.; Touraine, M. Antimicrobial resistance: A priority for global health action. Bull. World Health Organ. 2015, 93, 439. [Google Scholar] [CrossRef]
- Abdula, N.; Macharia, J.; Motsoaledi, A.; Swaminathan, S.; Vijay Raghavan, K. National action for global gains in antimicrobial resistance. Lancet 2016, 387, E3–E5. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Levy Hara, G.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef] [Green Version]
- Liao, X.; Ma, Y.; Daliri, E.B.-M.; Koseki, S.; Wei, S.; Liu, D.; Ye, X.; Chen, S.; Ding, T. Interplay of antibiotic resistance and food-associated stress tolerance in foodborne pathogens. Trends Food Sci. Technol. 2019. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug Resistance in Bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [Green Version]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Cassini, A.; Hogberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.K.; Johnson, N.; Cizmas, L.; McDonald, T.J.; Kim, H. A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 2016, 150, 702–714. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Isothermal Amplification Technologies for the Detection of Foodborne Pathogens. Food Anal. Meth. 2018. [Google Scholar] [CrossRef]
- Ma, Y.; Lan, G.; Li, C.; Cambaza, E.M.; Liu, D.; Ye, X.; Chen, S.; Ding, T. Stress tolerance of Staphylococcus aureus with different antibiotic resistance profiles. Microb. Pathog. 2019, 133. [Google Scholar] [CrossRef]
- Li, J.; Liu, D.H.; Tian, X.J.; Koseki, S.; Chen, S.G.; Ye, X.Q.; Ding, T. Novel antibacterial modalities against methicillin resistant Staphylococcus aureus derived from plants. Crit. Rev. Food Sci. Nutr. 2019, 59, S153–S161. [Google Scholar] [CrossRef]
- Munguia, J.; Nizet, V. Pharmacological Targeting of the Host-Pathogen Interaction: Alternatives to Classical Antibiotics to Combat Drug-Resistant Superbugs. Trends Pharmacol. Sci. 2017, 38, 473–488. [Google Scholar] [CrossRef] [Green Version]
- O’Niel, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; The Review on Antimicrobial Resistance; Welcome Trust: London, UK, 2016; Available online: https://amr-review.org/ (accessed on 15 March 2020).
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Haque, S.; Ahmad, F.; Dar, S.A.; Jawed, A.; Mandal, R.K.; Wahid, M.; Lohani, M.; Khan, S.; Singh, V.; Akhter, N. Developments in strategies for Quorum Sensing virulence factor inhibition to combat bacterial drug resistance. Microb. Pathog. 2018, 121, 293–302. [Google Scholar] [CrossRef]
- Jiang, Q.; Chen, J.; Yang, C.; Yin, Y.; Yao, K. Quorum Sensing: A Prospective Therapeutic Target for Bacterial Diseases. BioMed Res. Int. 2019. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.H.; Zhong, J.L.; Wei, C.J.; Lin, C.W.; Ding, T. Current Perspectives on Viable but Non-culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 24. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.H.; Zhao, F.H.; Zhong, N.J. Production of diacylglycerols through glycerolysis with SBA-15 supported Thermomyces lanuginosus lipase as catalyst. J. Sci. Food Agric. 2020, 100, 1426–1435. [Google Scholar] [CrossRef]
- Rajput, A.; Thakur, A.; Sharma, S.; Kumar, M. aBiofilm: A resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res. 2018, 46, D894–D900. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.Y.; Ye, C.X.; Soteyome, T.; Zhao, X.H.; Xia, J.; Xu, W.Y.; Mao, Y.Z.; Peng, R.X.; Chen, J.X.; Xu, Z.B.; et al. Inhibitory effects of two types of food additives on biofilm formation by foodborne pathogens. MicrobiologyOpen 2019, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Varela, M.F. Biochemistry of Bacterial Multidrug Efflux Pumps. Int. J. Mol. Sci. 2012, 13, 4484–4495. [Google Scholar] [CrossRef]
- Wasaznik, A.; Grinholc, M.; Bielawski, K.P. Active efflux as the multidrug resistance mechanism. Postepy Hig. I Med. Dosw. 2009, 63, 123–133. [Google Scholar]
- Sauvage, E.; Terrak, M. Glycosyltransferases and Transpeptidases/Penicillin-Binding Proteins: Valuable Targets for New Antibacterials. Antibiotics 2016, 5, 12. [Google Scholar] [CrossRef]
- Zhao, X.H.; Xia, J.; Liu, Y. Contrast of Real-Time Fluorescent PCR Methods for Detection of Escherichia coli O157:H7 and of Introducing an Internal Amplification Control. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Shriram, V.; Khare, T.; Bhagwat, R.; Shukla, R.; Kumar, V. Inhibiting Bacterial Drug Efflux Pumps via Phyto-Therapeutics to Combat Threatening Antimicrobial Resistance. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Poole, K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 2004, 10, 12–26. [Google Scholar] [CrossRef] [Green Version]
- De la Cruz, F.; Davies, J. Horizontal gene transfer and the origin of species: Lessons from bacteria. Trends Microbiol. 2000, 8, 128–133. [Google Scholar] [CrossRef]
- Luis Martinez, J. Ecology and Evolution of Chromosomal Gene Transfer between Environmental Microorganisms and Pathogens. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Mah, T.-F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.M.; Wu, G. Can Biofilm Be Reversed Through Quorum Sensing in Pseudomonas aeruginosa? Front. Microbiol. 2019, 10, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef]
- Balcazar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Hathroubi, S.; Mekni, M.A.; Domenico, P.; Dao, N.; Jacques, M. Biofilms: Microbial Shelters Against Antibiotics. Microb. Drug Resist. 2017, 23, 147–156. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Transcriptomic Analysis of Viable but Non-Culturable Escherichia coli O157:H7 Formation Induced by Low Temperature. Microorganisms 2019, 7, 634. [Google Scholar] [CrossRef] [Green Version]
- Bauerle, T.; Fischer, A.; Speck, T.; Bechinger, C. Self-organization of active particles by quorum sensing rules. Nat. Commun. 2018, 9, 8. [Google Scholar] [CrossRef]
- Turan, N.B.; Chormey, D.S.; Buyukpinar, C.; Engin, G.O.; Bakirdere, S. Quorum sensing: Little talks for an effective bacterial coordination. Trac-Trends Anal. Chem. 2017, 91, 1–11. [Google Scholar] [CrossRef]
- Wei, C.J.; Zhao, X.H. Induction of Viable but Nonculturable Escherichia coli O157:H7 by Low Temperature and Its Resuscitation. Front. Microbiol. 2018, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Monnet, V.; Gardan, R. Quorum-sensing regulators in Gram-positive bacteria: “cherchez le peptide”. Mol. Microbiol. 2015, 97, 181–184. [Google Scholar] [CrossRef]
- Zhang, W.W.; Li, C.H. Exploiting Quorum Sensing Interfering Strategies in Gram-Negative Bacteria for the Enhancement of Environmental Applications. Front. Microbiol. 2016, 6, 15. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Chen, Z.; Xiang, J. Advances in the research of LuxR family protein in quorum-sensing system of gram-negative bacteria. Chin. J. Burn. 2016, 32, 536–538. [Google Scholar] [CrossRef]
- Banerjee, G.; Ray, A.K. The talking language in some major Gram-negative bacteria. Arch. Microbiol. 2016, 198, 489–499. [Google Scholar] [CrossRef]
- Li, X.; Zhang, G.; Zhu, Y.; Bi, J.; Hao, H.; Hou, H. Effect of the luxI/R gene on AHL-signaling molecules and QS regulatory mechanism in Hafnia alvei H4. Amb Express 2019, 9, 197. [Google Scholar] [CrossRef] [Green Version]
- Ng, Y.K.; Grasso, M.; Wright, V.; Garcia, V.; Williams, P.; Atkinson, S. The Quorum Sensing System of Yersinia enterocolitica 8081 Regulates Swimming Motility, Host Cell Attachment, and Virulence Plasmid Maintenance. Genes 2018, 9, 14. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Quan, C.; Jin, L.; Chen, M. Production, detection and application perspectives of quorum sensing autoinducer-2 in bacteria. J. Biotechnol. 2018, 268, 53–60. [Google Scholar] [CrossRef]
- Park, H.; Lee, K.; Yeo, S.; Shin, H.; Holzapfel, W.H. Autoinducer-2 Quorum Sensing Influences Viability of Escherichia coli O157:H7 under Osmotic and In Vitro Gastrointestinal Stress Conditions. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-mediated signalling in bacteria. Fems Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef] [Green Version]
- Xavier, K.B. Bacterial interspecies quorum sensing in the mammalian gut microbiota. C. R. Biol. 2018, 341, 297–299. [Google Scholar] [CrossRef]
- Wang, Q.; He, Z.Y.; Hu, Y.J.; Jiang, Y.T.; Ma, R.; Tang, Z.S.; Liang, J.P.; Liu, Z.; Huang, Z.W. luxS Mutant Regulation: Quorum Sensing Impairment or Methylation Disorder? Sensors 2012, 12, 6176–6185. [Google Scholar] [CrossRef] [Green Version]
- Vendeville, A.; Winzer, K.; Heurlier, K.; Tang, C.M.; Hardie, K.R. Making ‘sense’ of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 2005, 3, 383–396. [Google Scholar] [CrossRef]
- Xue, T.; Yu, L.M.; Shang, F.; Li, W.C.; Zhang, M.; Ni, J.T.; Chen, X.L. Short communication: The role of autoinducer 2 (AI-2) on antibiotic resistance regulation in an Escherichia coli strain isolated from a dairy cow with mastitis. J. Dairy Sci. 2016, 99, 4693–4698. [Google Scholar] [CrossRef] [Green Version]
- Frezza, M.; Soulere, L.; Balestrino, D.; Gohar, M.; Deshayes, C.; Queneau, Y.; Forestier, C.; Doutheau, A. Ac2-DPD, the bis-(O)-acetylated derivative of 4,5-dihydroxy-2,3-pentanedione (DPD) is a convenient stable precursor of bacterial quorum sensing autoinducer AI-2. Bioorg. Med. Chem. Lett. 2007, 17, 1428–1431. [Google Scholar] [CrossRef]
- Stotani, S.; Gatta, V.; Medda, F.; Padmanaban, M.; Karawajczyk, A.; Tammela, P.; Giordanetto, F.; Tzalis, D.; Collina, S. A Versatile Strategy for the Synthesis of 4,5-Dihydroxy-2,3-Pentanedione (DPD) and Related Compounds as Potential Modulators of Bacterial Quorum Sensing. Molecules 2018, 23, 22. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef]
- Miller, S.T.; Xavier, K.B.; Campagna, S.R.; Taga, M.E.; Semmelhack, M.F.; Bassler, B.L.; Hughson, F.M. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 2004, 15, 677–687. [Google Scholar] [CrossRef]
- Cornforth, D.M.; Foster, K.R. Competition sensing: The social side of bacterial stress responses. Nat. Rev. Microbiol. 2013, 11, 285–293. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; Straight, P.D. Multifaceted interfaces of bacterial competition. J. Bacteriol. 2016, 198, 2145–2155. [Google Scholar] [CrossRef] [Green Version]
- Stubbendieck, R.M.; Vargas-Bautista, C.; Straight, P.D. Bacterial communities: Interactions to scale. Front. Microbiol. 2016, 7, 1234. [Google Scholar] [CrossRef] [Green Version]
- Nadell, C.D.; Bassler, B.L. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc. Natl. Acad. Sci. USA 2011, 108, 14181–14185. [Google Scholar] [CrossRef] [Green Version]
- Khare, A.; Tavazoie, S. Multifactorial competition and resistance in a two-species bacterial system. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Li, Y.H.; Tian, X.L. Quorum sensing and bacterial social interactions in biofilms: Bacterial cooperation and competition. Stress Environ. Regul. Gene Expr. Adapt. Bact. 2016, 1195–1205. [Google Scholar]
- Evans, K.C.; Benomar, S.; Camuy-Vélez, L.A.; Nasseri, E.B.; Wang, X.; Neuenswander, B.; Chandler, J.R. Quorum-sensing control of antibiotic resistance stabilizes cooperation in Chromobacterium violaceum. Isme J. 2018, 12, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef] [Green Version]
- Doekes, H.M.; De Boer, R.J.; Hermsen, R. Toxin production spontaneously becomes regulated by local cell density in evolving bacterial populations. Plos Comput. Biol. 2019, 15, e1007333. [Google Scholar] [CrossRef] [Green Version]
- Subhadra, B.; Oh, M.H.; Choi, C.H. RND efflux pump systems in Acinetobacter, with special emphasis on their role in quorum sensing. J. Bacteriol. Virol. 2019, 49, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, B.; Grenier, D.; Yi, L. Regulatory Mechanisms of the LuxS/AI-2 System and Bacterial Resistance. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Pumbwe, L.; Skilbeck, C.A.; Wexler, H.M. Presence of quorum-sensing systems associated with multidrug resistance and biofilm formation in Bacteroides fragilis. Microb. Ecol. 2008, 56, 412–419. [Google Scholar] [CrossRef]
- Maseda, H.; Sawada, I.; Saito, K.; Uchiyama, H.; Nakae, T.; Nomura, N. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 1320–1328. [Google Scholar] [CrossRef] [Green Version]
- Alcalde-Rico, M.; Olivares-Pacheco, J.; Alvarez-Ortega, C.; Camara, M.; Martinez, J.L. Role of the Multidrug Resistance Efflux Pump MexCD-OprJ in the Pseudomonas aeruginosa Quorum Sensing Response. Front. Microbiol. 2018, 9, 16. [Google Scholar] [CrossRef]
- Li, X.Z.; Plesiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [Green Version]
- Bagge, N.; Schuster, M.; Hentzer, M.; Ciofu, O.; Givskov, M.; Greenberg, E.P.; Hoiby, N. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob. Agents Chemother. 2004, 48, 1175–1187. [Google Scholar] [CrossRef] [Green Version]
- Dhabaan, G.N.; AbuBakar, S.; Cerqueira, G.M.; Al-Haroni, M.; Pang, S.P.; Hassan, H. Imipenem Treatment Induces Expression of Important Genes and Phenotypes in a Resistant Acinetobacter baumannii Isolate. Antimicrob. Agents Chemother. 2016, 60, 1370–1376. [Google Scholar] [CrossRef] [Green Version]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Williams, P.; Camara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- An, S.Q.; Murtagh, J.; Twomey, K.B.; Gupta, M.K.; O’Sullivan, T.P.; Ingram, R.; Valvano, M.A.; Tang, J.L. Modulation of antibiotic sensitivity and biofilm formation in Pseudomonas aeruginosa by interspecies signal analogues. Nat. Commun. 2019, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Li, S.R.; Jiang, B.; Hu, X.M.; Li, S. Therapeutic Targeting of the Staphylococcus aureus Accessory Gene Regulator (agr) System. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- France, M.T.; Cornea, A.; Kehlet-Delgado, H.; Forney, L.J. Spatial structure facilitates the accumulation and persistence of antibiotic-resistant mutants in biofilms. Evol. Appl. 2019, 12, 498–507. [Google Scholar] [CrossRef] [Green Version]
- Saxena, P.; Joshi, Y.; Rawat, K.; Bisht, R. Biofilms: Architecture, Resistance, Quorum Sensing and Control Mechanisms. Indian J. Microbiol. 2019, 59, 3–12. [Google Scholar] [CrossRef]
- Chebotar, I.V.; Mayansky, А.N.; Konchakova, Е.D.; Lazareva, А.V.; Chistyakova, V.P. Antimicrobial Resistance of Bacteria in Biofilms. Klin. Mikrobiol. I Antimikrobn. Khimioterapiya 2012, 14, 51–58. [Google Scholar]
- Qu, Y.; Daley, A.J.; Istivan, T.S.; Rouch, D.A.; Deighton, M.A. Densely adherent growth mode, rather than extracellular polymer substance matrix build-up ability, contributes to high resistance of Staphylococcus epidermidis biofilms to antibiotics. J. Antimicrob. Chemother. 2010, 65, 1405–1411. [Google Scholar] [CrossRef]
- Seixas, R.; Machado, J.; Bernardo, F.; Vilela, C.; Oliveira, M. Biofilm Formation by Salmonella Enterica Serovar 1,4, 5,12:i:- Portuguese Isolates: A Phenotypic, Genotypic, and Socio-geographic Analysis. Curr. Microbiol. 2014, 68, 670–677. [Google Scholar] [CrossRef]
- Han, X.; Li, Q.; Shen, L.; Hu, D.; Qu, Y. Correlation between the biofilm-forming ability, biofilm-related genes and antimicrobial resistance of Acinetobacter baumannii. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2014, 26, 639–643. [Google Scholar] [CrossRef]
- Pozzi, C.; Waters, E.M.; Rudkin, J.K.; Schaeffer, C.R.; Lohan, A.J.; Tong, P.; Loftus, B.J.; Pier, G.B.; Fey, P.D.; Massey, R.C.; et al. Methicillin Resistance Alters the Biofilm Phenotype and Attenuates Virulence in Staphylococcus aureus Device-Associated Infections. Plos Pathog. 2012, 8, 15. [Google Scholar] [CrossRef]
- McCluskey, J.; Hinds, J.; Husain, S.; Witney, A.; Mitchell, T.J. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 2004, 51, 1661–1675. [Google Scholar] [CrossRef] [Green Version]
- Coelho, L.R.; Souza, R.R.; Ferreira, F.A.; Guimaraes, M.A.; Ferreira-Carvalho, B.T.; Sa Figueiredo, A.M. agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of Staphylococcus aureus. Microbiol. Sgm. 2008, 154, 3480–3490. [Google Scholar] [CrossRef] [Green Version]
- Tamber, S.; Cheung, A.L. SarZ Promotes the Expression of Virulence Factors and Represses Biofilm Formation by Modulating SarA and agr in Staphylococcus aureus. Infect. Immun. 2009, 77, 419–428. [Google Scholar] [CrossRef] [Green Version]
- Gimza, B.D.; Larias, M.I.; Budny, B.G.; Shaw, L.N. Mapping the Global Network of Extracellular Protease Regulation in Staphylococcus aureus. mSphere 2019, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Virulence Mech. Bact. Pathog. 2016, 213–239. [Google Scholar]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Rapisarda, C.; Fronzes, R. Secretion Systems Used by Bacteria to Subvert Host Functions. Curr. Issues Mol. Biol. 2018, 25, 1–42. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Holland, I.B.; Schmitt, L. The type 1 secretion pathway—The hemolysin system and beyond. Biochim. Et Biophys. Acta (Bba)-Mol. Cell Res. 2014, 1843, 1629–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. Embo J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Maura, D.; Hazan, R.; Kitao, T.; Ballok, A.E.; Rahme, L.G. Evidence for direct control of virulence and defense gene circuits by the Pseudomonas aeruginosa quorum sensing regulator, MvfR. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Teschler, J.K.; Zamorano-Sánchez, D.; Utada, A.S.; Warner, C.J.; Wong, G.C.; Linington, R.G.; Yildiz, F.H. Living in the matrix: Assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 2015, 13, 255–268. [Google Scholar] [CrossRef] [Green Version]
- Grohmann, E.; Christie, P.J.; Waksman, G.; Backert, S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 2018, 107, 455–471. [Google Scholar] [CrossRef]
- Li, P.; Tian, M.; Bao, Y.; Hu, H.; Liu, J.; Yin, Y.; Ding, C.; Wang, S.; Yu, S. Brucella rough mutant induce macrophage death via activating IRE1α pathway of endoplasmic reticulum stress by enhanced T4SS secretion. Front. Cell. Infect. Microbiol. 2017, 7, 422. [Google Scholar] [CrossRef] [Green Version]
- Saurav, K.; Bar-Shalom, R.; Haber, M.; Burgsdort, I.; Oliviero, G.; Costantino, V.; Morgenstern, D.; Steindler, L. In Search of Alternative Antibiotic Drugs: Quorum-Quenching Activity in Sponges and Their Bacterial Isolates. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Ciric, A.D.; Petrovic, J.D.; Glamoclija, J.M.; Smiljkovic, M.S.; Nikolic, M.M.; Stojkovic, D.S.; Sokovic, M.D. Natural products as biofilm formation antagonists and regulators of quorum sensing functions: A comprehensive review update and future trends. S. Afr. J. Bot. 2019, 120, 65–80. [Google Scholar] [CrossRef]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, L.H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.B.; Liu, J.H.; Liu, C.J.; Yang, A.D.; Qiao, J.J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 2009, 25. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.K.; Vinothkumar, K.; Rajpara, N. Bacterial quorum sensing inhibitors: Attractive alternatives for control of infectious pathogens showing multiple drug resistance. Recent Pat. Anti-Infect. Drug Discov. 2013, 8, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Lu, C. Biological activity and identification of a peptide inhibitor of LuxS from Streptococcus suis serotype 2. Fems Microbiol. Lett. 2009, 294, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schramm, V.L.; Gutierrez, J.A.; Cordovano, G.; Basu, I.; Guha, C.; Belbin, T.J.; Evans, G.B.; Tyler, P.C.; Furneaux, R.H. Transition state analogues in quorum sensing and SAM recycling. Nucleic acids Symp. Ser. 2008, 52, 75–76. [Google Scholar] [CrossRef]
- Storz, M.P.; Allegretta, G.; Kirsch, B.; Empting, M.; Hartmann, R.W. From in vitro to in cellulo: Structure-activity relationship of (2-nitrophenyl)methanol derivatives as inhibitors of PqsD in Pseudomonas aeruginosa. Org. Biomol. Chem. 2014, 12, 6094–6104. [Google Scholar] [CrossRef] [Green Version]
- Kalia, M.; Yadav, V.K.; Singh, P.K.; Dohare, S.; Sharma, D.; Narvi, S.S.; Agarwal, V. Designing quorum sensing inhibitors of Pseudomonas aeruginosa utilizing FabI: An enzymic drug target from fatty acid synthesis pathway. 3 Biotech 2019, 9, 10. [Google Scholar] [CrossRef]
- Zhang, J.W.; Xuan, C.G.; Lu, C.H.; Guo, S.; Yu, J.F.; Asif, M.; Jiang, W.J.; Zhou, Z.G.; Luo, Z.Q.; Zhang, L.Q. AidB, a Novel Thermostable N-Acylhomoserine Lactonase from the Bacterium Bosea sp. Appl. Environ. Microbiol. 2019, 85, 15. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Feng, T.; Du, R.; Tian, X.; Wang, Y.; Zhang, X.-H. Heterologous Expression of the Marine-Derived Quorum Quenching Enzyme MomL Can Expand the Antibacterial Spectrum of Bacillus brevis. Mar. Drugs 2019, 17, 128. [Google Scholar] [CrossRef] [Green Version]
- Cui, T.Q.; Bai, F.L.; Sun, M.T.; Lv, X.R.; Li, X.P.; Zhang, D.F.; Du, H. Lactobacillus crustorum ZHG 2-1 as novel quorum-quenching bacteria reducing virulence factors and biofilms formation of Pseudomonas aeruginosa. Lwt-Food Sci. Technol. 2020, 117, 8. [Google Scholar] [CrossRef]
- Yu, L.; Li, W.; Zhang, M.; Cui, Y.; Chen, X.; Ni, J.; Yu, L.; Shang, F.; Xue, T. Imidazole decreases the ampicillin resistance of an Escherichia coli strain isolated from a cow with mastitis by inhibiting the function of autoinducer 2. J. Dairy Sci. 2018, 101, 3356–3362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, V.; Fernandes, R.; Tsao, C.-Y.; Bentley, W.E. Cross species quorum quenching using a native AI-2 processing enzyme. Acs Chem. Biol. 2010, 5, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Bhasme, P.; Wang, Z.; Wang, L.; Wang, S.; Zeng, Y.; Wang, Y.; Ma, L.Z.; Li, Y. Chinese medicinal herb extract inhibits PQS-mediated quorum sensing system in Pseudomonas aeruginosa. J. Ethnopharmacol. 2020, 248, 112272. [Google Scholar] [CrossRef]
- Truchado, P.; Gimenez-Bastida, J.-A.; Larrosa, M.; Castro-Ibanez, I.; Carlos Espin, J.; Tomas-Barberan, F.A.; Teresa Garcia-Conesa, M.; Allende, A. Inhibition of Quorum Sensing (QS) in Yersinia enterocolitica by an Orange Extract Rich in Glycosylated Flavanones. J. Agric. Food Chem. 2012, 60, 8885–8894. [Google Scholar] [CrossRef]
- Ryu, E.-J.; Sim, J.; Sim, J.; Lee, J.; Choi, B.-K. D-Galactose as an autoinducer 2 inhibitor to control the biofilm formation of periodontopathogens. J. Microbiol. 2016, 54, 632–637. [Google Scholar] [CrossRef]
- Sun, B.; Zhang, M. Analysis of the antibacterial effect of an Edwardsiella tarda LuxS inhibitor. SpringerPlus 2016, 5, 92. [Google Scholar] [CrossRef] [Green Version]
- Halbedl, S.; Kratzer, M.C.; Rahm, K.; Crosta, N.; Masters, K.S.; Zippert, J.; Brase, S.; Gradl, D. Synthesis of novel inhibitors blocking Wnt signaling downstream of beta-Catenin. Febs Lett. 2013, 587, 522–527. [Google Scholar] [CrossRef]
- Takayama, H.; Jia, Z.J.; Kremer, L.; Bauer, J.O.; Strohmann, C.; Ziegler, S.; Antonchick, A.P.; Waldmann, H. Discovery of Inhibitors of the Wnt and Hedgehog Signaling Pathways through the Catalytic Enantioselective Synthesis of an Iridoid-Inspired Compound Collection. Angew. Chem. Int. Ed. 2013, 52, 12404–12408. [Google Scholar] [CrossRef]
- Blocher, R.; Ramirez, A.R.; Castro-Escarpulli, G.; Curiel-Quesada, E.; Reyes-Arellano, A. Design, Synthesis, and Evaluation of Alkyl-Quinoxalin-2(1H)-One Derivatives as Anti-Quorum Sensing Molecules, Inhibiting Biofilm Formation in Aeromonas caviae Sch3. Molecules 2018, 23, 11. [Google Scholar] [CrossRef] [Green Version]
- Bortolotti, D.; Trapella, C.; Bragonzi, A.; Marchetti, P.; Zanirato, V.; Alogna, A.; Gentili, V.; Cervellati, C.; Valacchi, G.; Sicolo, M.; et al. Conjugation of LasR Quorum-Sensing Inhibitors with Ciprofloxacin Decreases the Antibiotic Tolerance of P-aeruginosa Clinical Strains. J. Chem. 2019, 13. [Google Scholar] [CrossRef] [Green Version]
- Pena, R.T.; Blasco, L.; Ambroa, A.; Gonzalez-Pedrajo, B.; Fernandez-Garcia, L.; Lopez, M.; Bleriot, I.; Bou, G.; Garcia-Contreras, R.; Wood, T.K.; et al. Relationship Between Quorum Sensing and Secretion Systems. Front. Microbiol. 2019, 10, 14. [Google Scholar] [CrossRef] [Green Version]
- Grandclement, C.; Tannieres, M.; Morera, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. Fems Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Zhou, R.; Li, L.; Peters, B.M.; Li, B.; Lin, C.W.; Chuang, T.L.; Chen, D.Q.; Zhao, X.H.; Xiong, Z.Y.; et al. Viable but non-culturable state and toxin gene expression of enterohemorrhagic Escherichia coli 0157 under cryopreservation. Res. Microbiol. 2017, 168, 188–193. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in Rapid Detection Methods for Foodborne Pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [Green Version]
- Metz, B.; Jiskoot, W.; Hennink, W.E.; Crommelin, D.J.; Kersten, G.F. Physicochemical and immunochemical techniques predict the quality of diphtheria toxoid vaccines. Vaccine 2003, 22, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Meng, F.; Ma, X.; Song, X.; Liu, X.; Gao, W.; Dang, Y.; Meng, Y.; Cao, M.; Song, C. Regulation of bacteria population behaviors by AI-2 “consumer cells” and “supplier cells”. Bmc Microbiol. 2017, 17, 198. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Hixon, M.S.; Globisch, D.; Kaufmann, G.F.; Janda, K.D. Mechanistic insights into the LsrK kinase required for autoinducer-2 quorum sensing activation. J. Am. Chem. Soc. 2013, 135, 7827–7830. [Google Scholar] [CrossRef] [Green Version]
- Xavier, K.B.; Miller, S.T.; Lu, W.; Kim, J.H.; Rabinowitz, J.; Pelczer, I.; Semmelhack, M.F.; Bassler, B.L. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. Acs Chem. Biol. 2007, 2, 128–136. [Google Scholar] [CrossRef]
- Stotani, S.; Gatta, V.; Medarametla, P.; Padmanaban, M.; Karawajczyk, A.; Giordanetto, F.; Tammela, P.i.; Laitinen, T.; Poso, A.; Tzalis, D. DPD-inspired discovery of novel LsrK kinase inhibitors: An opportunity to fight antimicrobial resistance. J. Med. Chem. 2019, 62, 2720–2737. [Google Scholar] [CrossRef] [Green Version]
- Neiditch, M.B.; Federle, M.J.; Pompeani, A.J.; Kelly, R.C.; Swem, D.L.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 2006, 126, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.; Shang, W.; Yang, Z.; Sun, Z.; Li, Y.; Guo, J.; Wang, X.; Zou, D.; Wang, S.; Lei, H. LuxS-dependent AI-2 regulates versatile functions in Enterococcus faecalis V583. J. Proteome Res. 2012, 11, 4465–4475. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yaohari, H.; Zhou, Z.; Sze, C.C.; Stuckey, D.C. Autoinducer-2-mediated quorum sensing partially regulates the toxic shock response of anaerobic digestion. Water Res. 2019, 158, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.M.; Queneau, Y.; Soulere, L.; von Bodman, S.; Doutheau, A. Mechanisms and Synthetic Modulators of AHL-Dependent Gene Regulation. Chem. Rev. 2011, 111, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Bu, Y.; Suga, H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 2003, 10, 563–571. [Google Scholar] [CrossRef] [Green Version]
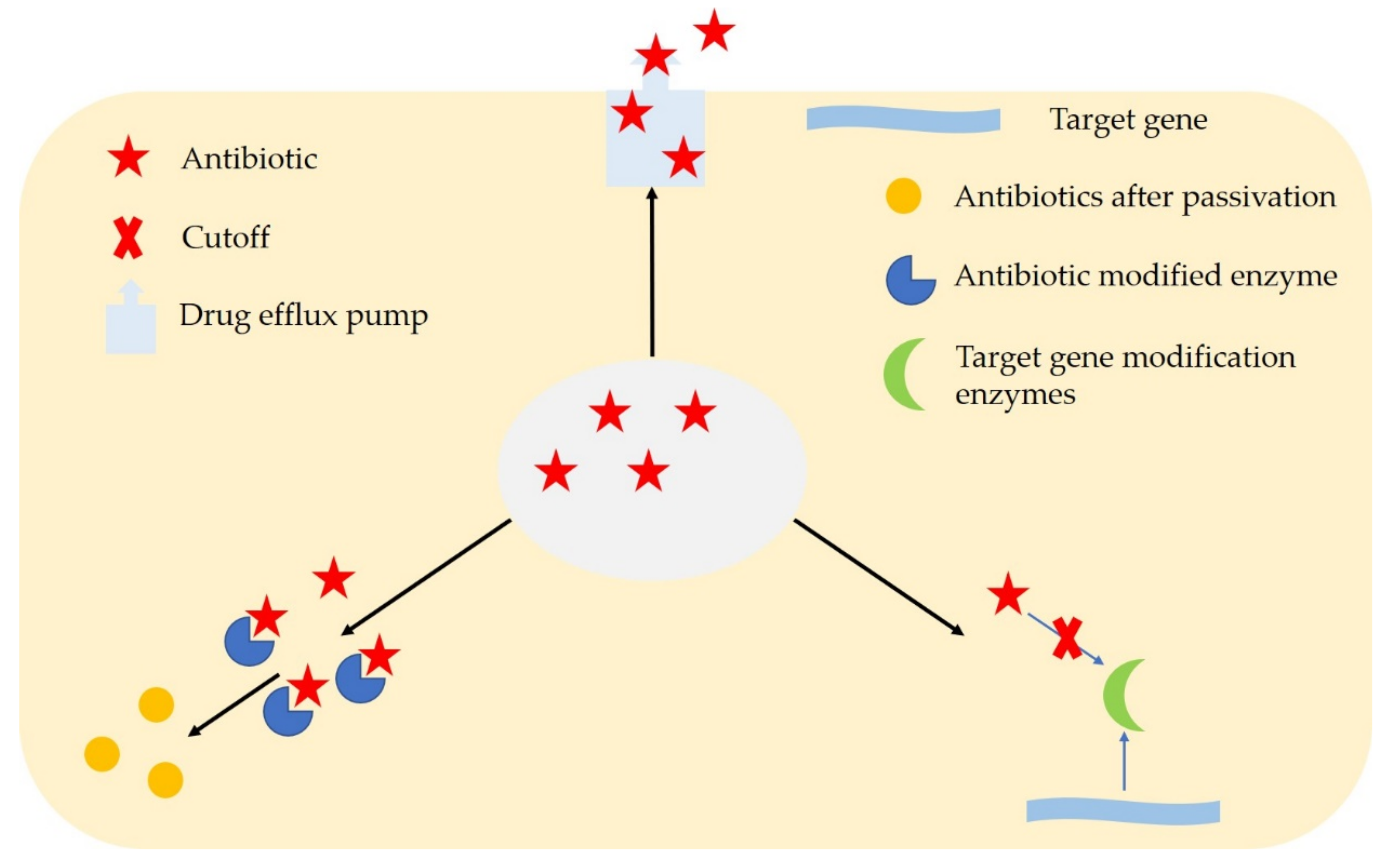
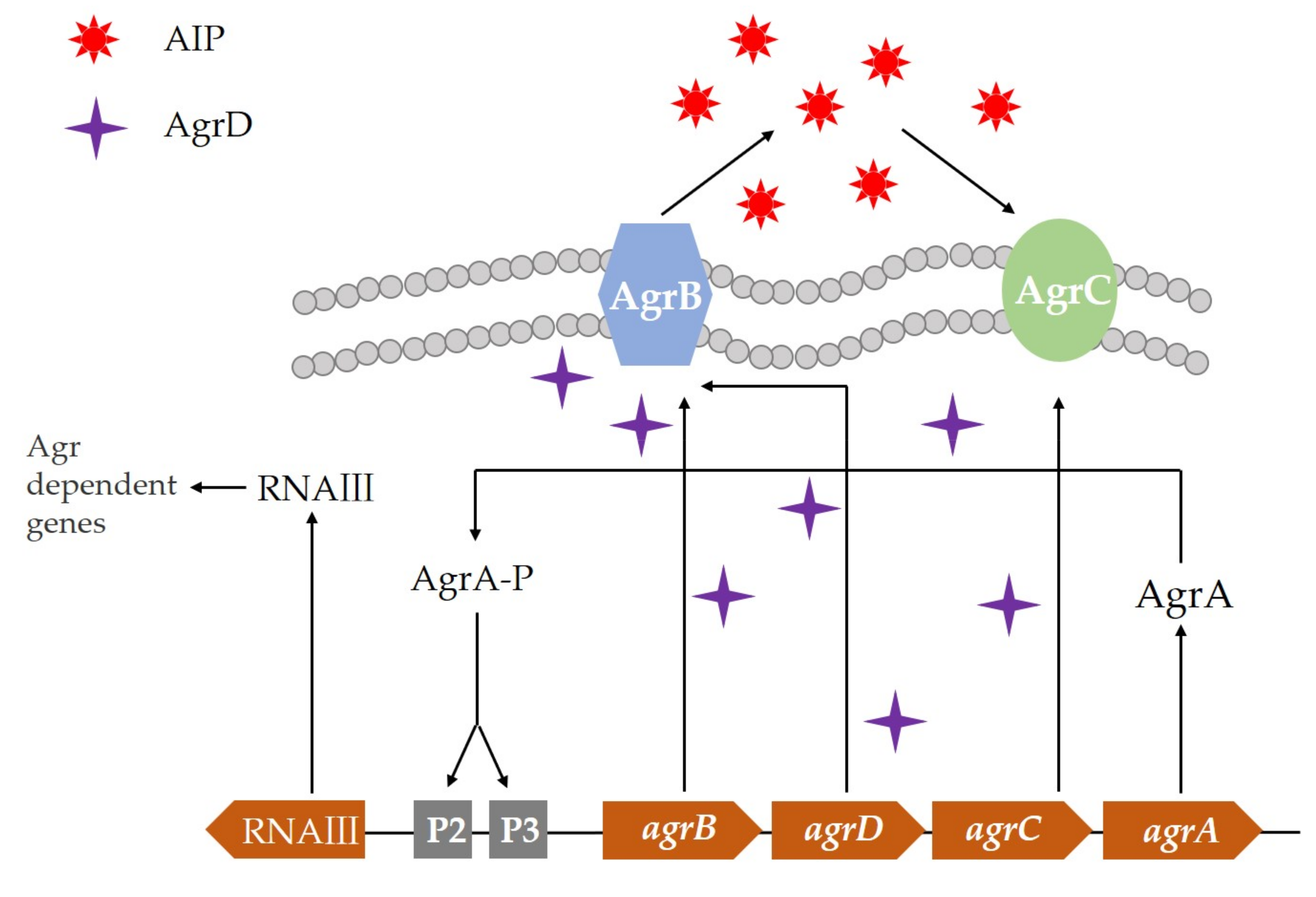
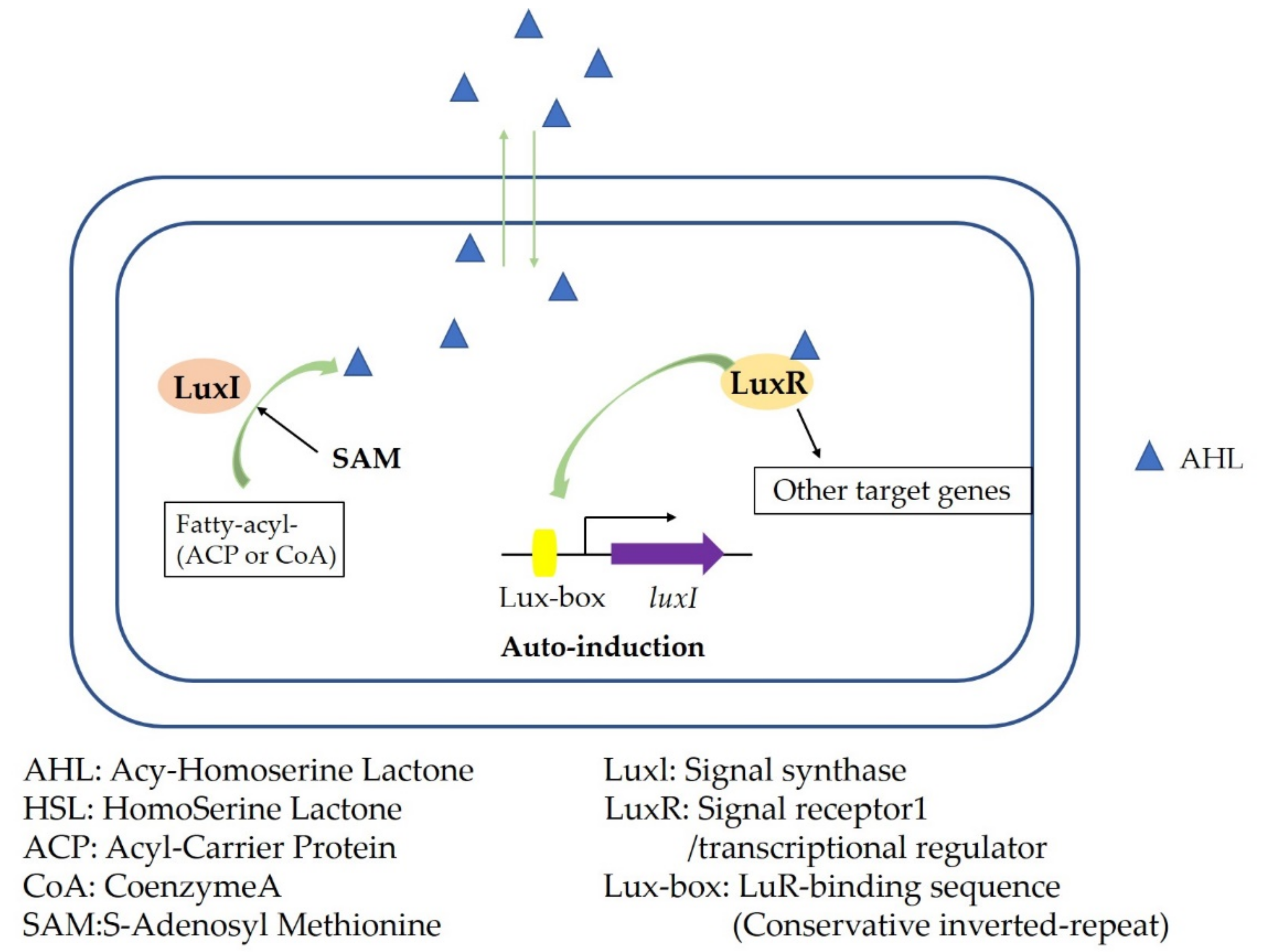


| Resistance Mechanism | Action Mechanism |
|---|---|
| Chemical modification | Change the chemical structure of antibiotic drugs |
| Efflux pump system | Discharge intracellular antibiotic drugs |
| Modification of drug-targeting genes | Change drug-targeting genes |
| Global cellular adaptation | Adapt and cope with stress |
| Biofilm itself | Reduce the permeability of antibiotics |
| Special internal environment in biofilm | Intracellular thallus produce heterogeneity |
| Extreme environment outside the biofilm | Intracellular thallus produce resistance |
| Regulation Type | Regulation Method | Biological Effect | Reference |
|---|---|---|---|
| Bacterial active efflux pumps | |||
| Autoinducer C6-HSL and C8-HSL | QS regulate the expression of efflux pump genes | Upregulate the expression of bmeB efflux pump | [70] |
| Quorum-sensing autoinducer C4-HSL | QS regulate the expression of efflux pump genes | Upregulate resistance pump MexAB-OprM | [71] |
| 4-hydroxy-2-heptylquinoline excretion | QS affected by the expression level of efflux pump | Shut down the QS response | [72] |
| RND efflux pump | Self-inducible molecules excretion | Exacerbate bacterial infections | [73] |
| Biofilm | |||
| Polysaccharides | Form molecular barrier and charge barrier | Prevent or delay the penetration of antibiotics | [76] |
| Permeation limitation | Lack of nutrients | Less sensitive to antibiotics | [36] |
| lasl/lasR and rhlI/rhlI signaling system | Activate related transcription regulators | Form biofilms | [77] |
| Interspecies signal analogues | Regulate or inhibit enzyme activity | Altered biofilm formation | [78] |
| WalK/WalR two-component system | Directly regulates biofilm formation | [79] | |
| Stimulating factors | Promote QS system adjustment | Regulate biofilm formation | [80] |
| Quenching Way | Function Method | Biological Effect | Reference |
|---|---|---|---|
| Inhibition of signal molecule production | |||
| TNRHNPHHLHHV (peptide) | Inhibit LuxS enzyme activity | Inhibit AI-2 production | [106] |
| MT-DADMe-ImmA | Picomolar inhibitor | Inhibit AI-2 production | [107] |
| (2-nitrophenyl) methanol derivatives | Inhibit enzyme of signal molecule biosynthesis | Inhibit the production of signaling molecules | [108] |
| FabI derivatives | Inhibit enzyme activity | Inhibit the production of signaling molecules | [109] |
| Degradation of signal molecule | |||
| The aiiA gene in Bacillus sp. 240B1 | The aiiA gene encodes AHL degrading enzyme | Enzymes degrade AHL signaling molecules | [103] |
| Thermostable AHL lactonase (AidB) | Hydrolyzing the ester bond of the HSL ring | Degrade AHL | [110] |
| A recombinant strain, named BbMomL | MomL causes loss of AHL | Degrade AHL | [111] |
| Crude extracts from Lb. crustorum ZHG 2-1 | Crude extract can degrade AHL | Degrade AHL by 37.1–87.6% at 3 sub-MICs | [112] |
| Imidazole | Degrade AI-2 | Inhibit AI-2 function | [113] |
| Externally added addition of ATP and LsrK | Phosphorylation and degradation of AI-2 | Reduced QS reaction | [114] |
| Inhibition of signal molecule conduction or binding to receptors | |||
| Medicinal herb extracts (MHE) | MHE as a competitive agent | Inhibit QS system | [115] |
| Flavonoids compounds | Reduce QS signal concentration | Inhibit QS signal | [116] |
| D-galactose | As an inhibitor of AI-2 activity | Inhibit AI-2 activity | [117] |
| A small peptide 5906 | Prevents homodimer formation | Inhibit LuxS activity | [118] |
| Haloquinone analogs | Block endogenous Wnt-driven transcription | Inhibit Wnt/β-Catenin signaling | [119] |
| 2H-pyran-3(6H)-one derivatives | As an inhibitor | Inhibit signaling pathway | [120] |
| Alkyl-Quinoxalin-2(1H)-One derivatives | The QS inhibition of these compounds | Reduce or inhibit biofilm formation | [121] |
| N-(3-oxododecanoyl) homoserine lactone derivatives | Block the binding site of the QS molecule | Inhibit the formation of biofilms and increase the antibiotic sensitivity | [122] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Yu, Z.; Ding, T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms 2020, 8, 425. https://doi.org/10.3390/microorganisms8030425
Zhao X, Yu Z, Ding T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms. 2020; 8(3):425. https://doi.org/10.3390/microorganisms8030425
Chicago/Turabian StyleZhao, Xihong, Zixuan Yu, and Tian Ding. 2020. "Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria" Microorganisms 8, no. 3: 425. https://doi.org/10.3390/microorganisms8030425
APA StyleZhao, X., Yu, Z., & Ding, T. (2020). Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms, 8(3), 425. https://doi.org/10.3390/microorganisms8030425





