Candida albicans Adaptation on Simulated Human Body Fluids under Different pH
Abstract
:1. Introduction
2. Materials and Methods
2.1. Simulated Body Fluids
2.2. Organism and Growth Conditions
2.3. Planktonic Growth Analysis
2.3.1. Growth Curves Determination
2.3.2. Colony Forming Unit (CFU) Quantification
2.3.3. Metabolic Activity Determination
2.3.4. Filaments Enumeration
2.4. Biofilm Growth Analysis
2.4.1. Biofilm Formation
2.4.2. Biofilm Characterization
CFUs Quantification
Metabolic Activity Determination
Total Biomass Quantification
Filaments Enumeration
2.5. Statistical Analysis
3. Results
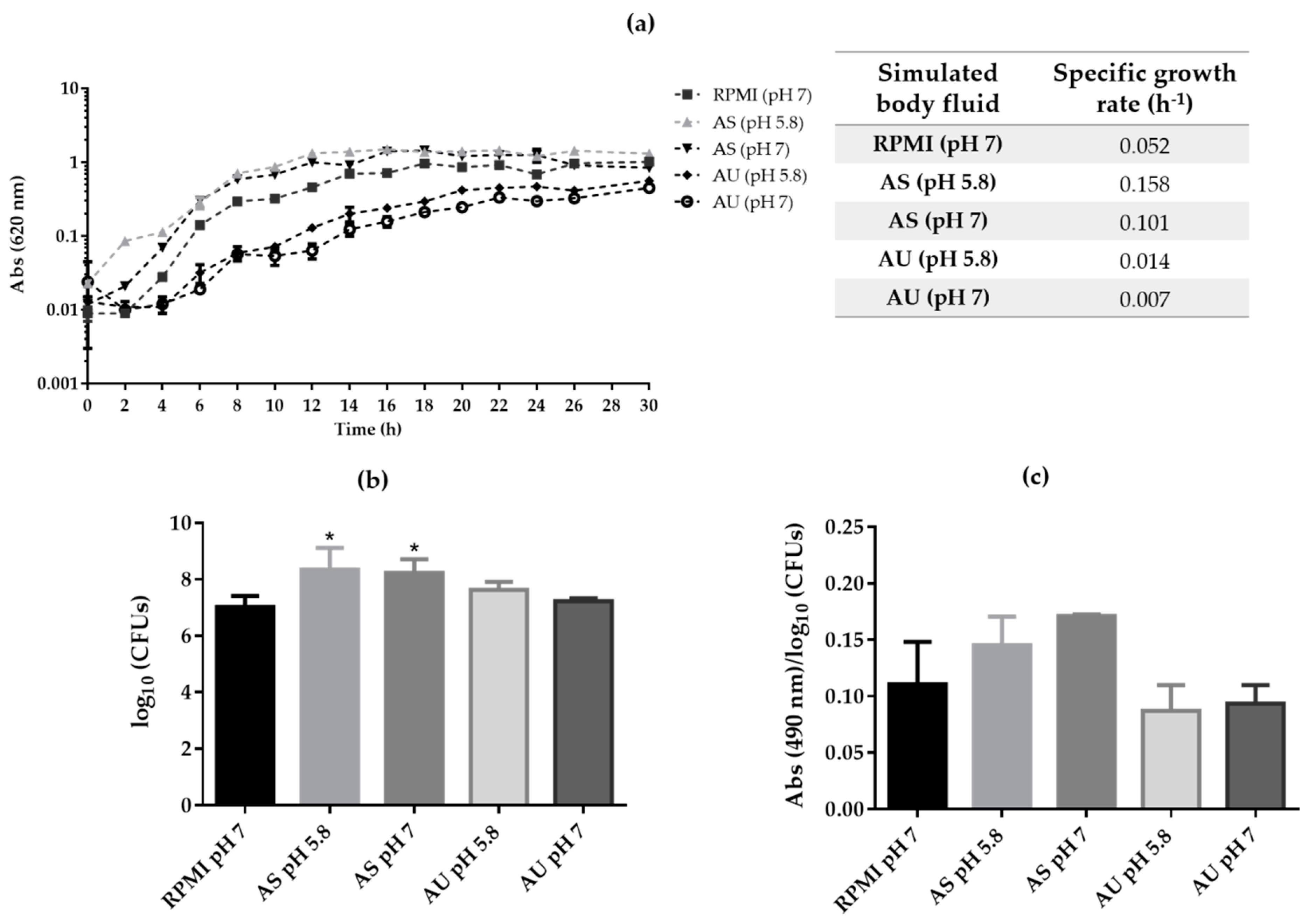
3.1. Candida albicans Planktonic Growth on Artificial Urine and Saliva
3.2. Candida albicans Biofilm Formation on Artificial Urine and Saliva
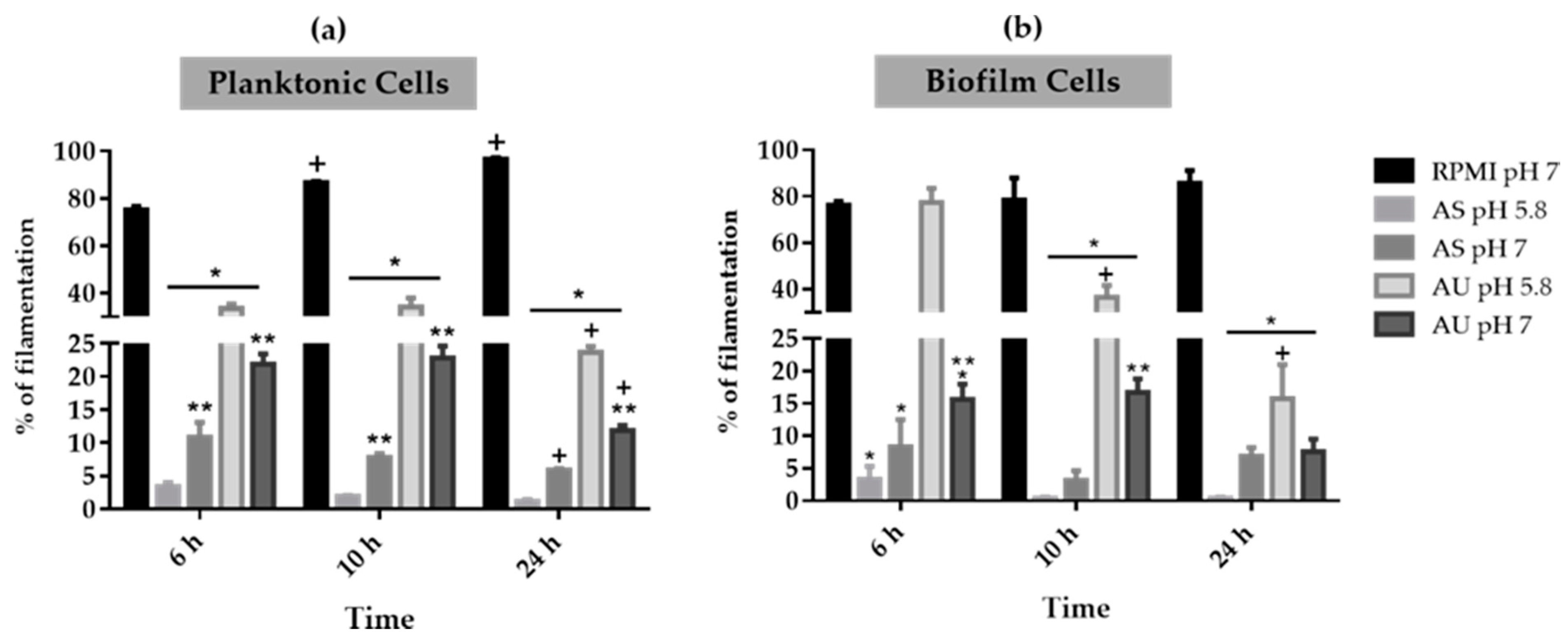
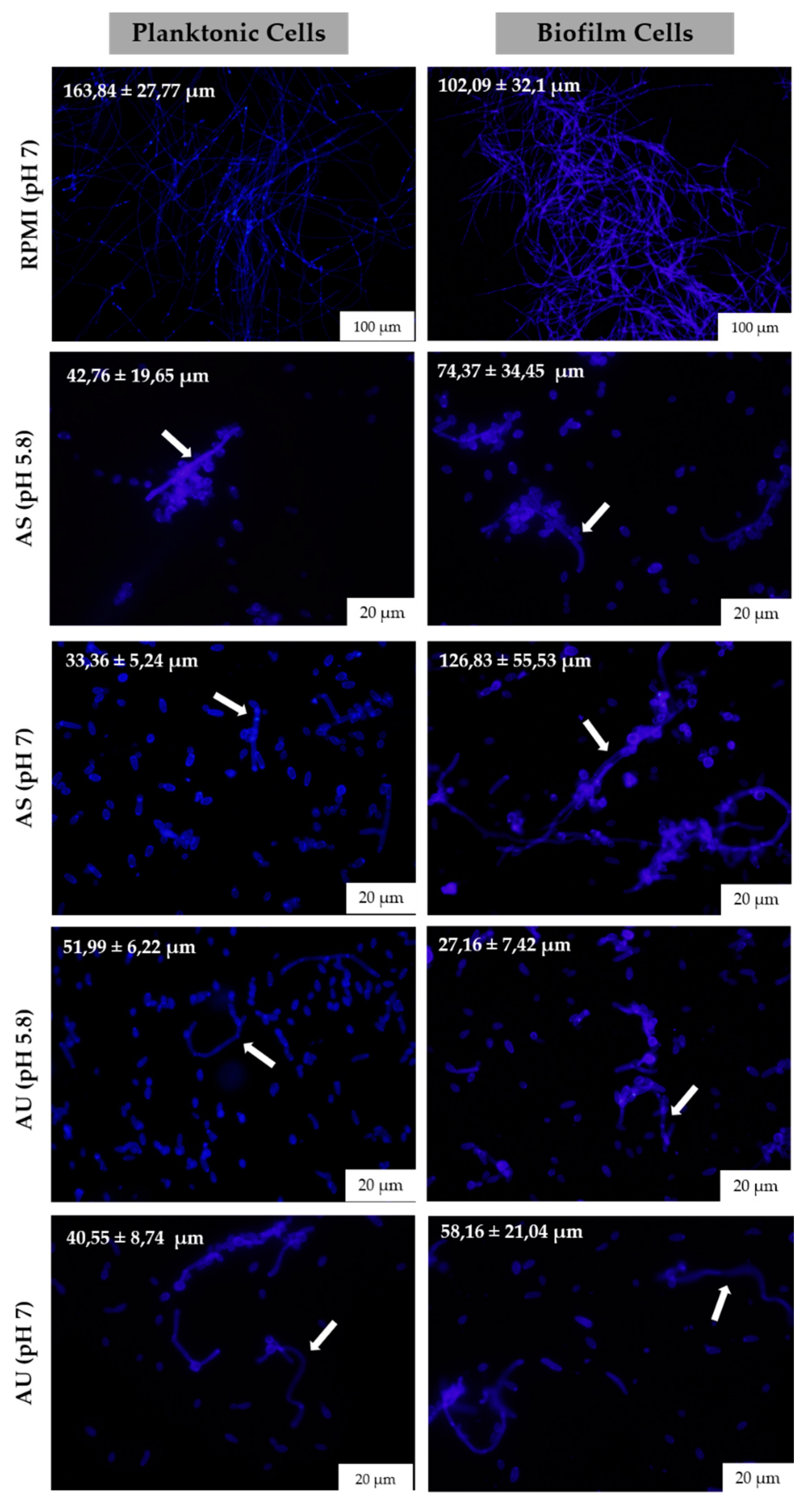
3.3. Candida albicans Filamentation on Artificial Urine and Saliva
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Horn, D.L.; Neofytos, D.; Anaissie, E.J.; Fishman, J.A.; Steinbach, W.J.; Olyaei, A.J.; Marr, K.A.; Pfaller, M.A.; Chang, C.H.; Webster, K.M. Epidemiology and outcomes of candidemia in 2019 patients: Data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 2009, 48, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Yapar, N. Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 2014, 10, 95–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, B.; Ferreia, C.; Alves, C.T.; Henriques, M.; Azeredo, J.; Silva, S. Vulvovaginal candidiasis: Epidemiology, microbiology and risk factors. Crit. Rev. Microbiol. 2016, 42, 905–927. [Google Scholar] [CrossRef] [Green Version]
- Koehler, P.; Stecher, M.; Cornely, O.A.; Koehler, D.; Vehreschild, M.J.G.T.; Bohlius, J.; Wisplinghoff, H.; Vehreschild, J.J. Morbidity and mortality of candidaemia in Europe: An epidemiologic meta-analysis. Clin. Microbiol. Infect. 2019, 25, 1200–1212. [Google Scholar] [CrossRef] [PubMed]
- Sardi, J.C.; Scorzoni, L.; Bernardi, T.; Fusco-Almeida, A.M.; Mendes Giannini, M.J. Candida species: Current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 2013, 62, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, P.; Behzadi, E.; Ranjbar, R. Urinary tract infections and Candida albicans. Cent. Eur. J. Urol. 2015, 68, 96–101. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Verma, R.; Murari, A.; Agrawal, A. Oral candidiasis: An overview. J. Oral Maxillofac. Pathol. 2014, 18, 81–85. [Google Scholar]
- Williams, D.; Lewis, M. Pathogenesis and treatment of oral candidosis. J. Oral Microbiol. 2011, 3, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Sawant, B.; Khan, T. Recent advances in delivery of antifungal agents for therapeutic management of candidiasis. Biomed. Pharmacother. 2017, 96, 1478–1490. [Google Scholar] [CrossRef]
- Silva, S.; Henriques, M.; Oliveira, R.; Williams, D.; Azeredo, J. In vitro biofilm activity of non-Candida albicans Candida species. Curr. Microbiol. 2010, 61, 534–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L.; Silva, S.; Ribeiro, B.; Henriques, M.; Azeredo, J. Influence of glucose concentration on the structure and quantity of biofilms formed by Candida parapsilosis. FEMS Yeast Res. 2015, 15, fov043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vylkova, S.; Carman, A.J.; Danhof, H.A.; Collette, J.R.; Zhou, H.; Lorenz, M.C. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2011, 2, e00055–e00111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bensen, E.S.; Martin, S.J.; Li, M.; Berman, J.; Davis, D.A. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 2004, 54, 1335–1351. [Google Scholar] [CrossRef] [PubMed]
- Dewar, S.; Reed, L.C.; Koerner, R.J. Emerging clinical role of pivmecillinam in the treatment of urinary tract infection in the context of multidrug-resistant bacteria. J. Antimicrob. Chemother. 2014, 69, 303–308. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.J.; Budge, S.; Kaloriti, D.; Tillmann, A.; Jacobsen, M.D.; Yin, Z.; Ene, I.V.; Bohovych, I.; Sandai, D.; Kastora, S.; et al. Stress adaptation in a pathogenic fungus. J. Exp. Biol. 2014, 217, 144–155. [Google Scholar] [CrossRef] [Green Version]
- Hall, R.A. Dressed to impress: Impact of environmental adaptation on the Candida albicans cell wall. Mol. Microbiol. 2015, 97, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.; Azeredo, J. Silicone colonization by non-Candida albicans Candida species in the presence of urine. J. Med. Microbiol. 2010, 59, 747–754. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.; Pires, P.; Monteiro, D.R.; Negri, M.; Gorup, L.F.; Camargo, E.R.; Barbosa, D.B.; Olievira, R.; Williams, D.W.; Henriques, M.; et al. The effect of silver nanoparticles and nystatin on mixed biofilms of Candida glabrata and Candida albicans on acrylic. Med. Mycol. 2013, 51, 178–184. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.W.; Wilson, M.J.; Lewis, M.A.; Potts, A.J. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J. Clin. Microbiol. 1995, 33, 2476–2479. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.; Henriques, M.; Martins, A.; Oliveira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araújo, D.; Henriques, M.; Silva, S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017, 25, 62–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quindós, G. Epidemiology of invasive mycoses: A landscape in continuous change. Rev. Iberoam. Micol. 2018, 35, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Pirofski, L. Host-pathogen interactions: The attributes of virulence. J. Infect. Dis. 2001, 184, 337–344. [Google Scholar] [CrossRef]
- Saville, S.P.; Lazzell, A.L.; Monteagudo, C.; Lopez-Ribot, J.L. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during Infection. Eukaryot. Cell. 2003, 2, 1053–1060. [Google Scholar] [CrossRef] [Green Version]
- Valentijn-Benz, M.; Nazmi, K.; Brand, H.S.; van’t Hof, W.; Veerman, E.C. Growth of Candida albicans in human saliva is supported by low-molecular-mass compounds. FEMS Yeast Res. 2015, 15, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Davis, D. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 2003, 44, 1–7. [Google Scholar] [CrossRef]
- Selvig, K.; Alspaugh, J.A. pH response pathways in fungi: Adapting to host-derived and environmental signals. Mycobiology 2011, 39, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Van Dijck, P.; Datta, A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007, 71, 348–376. [Google Scholar] [CrossRef] [Green Version]
- Huang, G. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence 2012, 3, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Selmecki, A.; Forche, A.; Berman, J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell. 2010, 9, 991–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkel, J.S.; Mitchell, A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 2011, 9, 109–118. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, A.; Araújo, D.; Ribeiro, E.; Henriques, M.; Silva, S. Candida albicans Adaptation on Simulated Human Body Fluids under Different pH. Microorganisms 2020, 8, 511. https://doi.org/10.3390/microorganisms8040511
Barbosa A, Araújo D, Ribeiro E, Henriques M, Silva S. Candida albicans Adaptation on Simulated Human Body Fluids under Different pH. Microorganisms. 2020; 8(4):511. https://doi.org/10.3390/microorganisms8040511
Chicago/Turabian StyleBarbosa, Ana, Daniela Araújo, Eduarda Ribeiro, Mariana Henriques, and Sónia Silva. 2020. "Candida albicans Adaptation on Simulated Human Body Fluids under Different pH" Microorganisms 8, no. 4: 511. https://doi.org/10.3390/microorganisms8040511
APA StyleBarbosa, A., Araújo, D., Ribeiro, E., Henriques, M., & Silva, S. (2020). Candida albicans Adaptation on Simulated Human Body Fluids under Different pH. Microorganisms, 8(4), 511. https://doi.org/10.3390/microorganisms8040511







