Implementation of Multispectral Imaging (MSI) for Microbiological Quality Assessment of Poultry Products
Abstract
1. Introduction
2. Materials and Methods
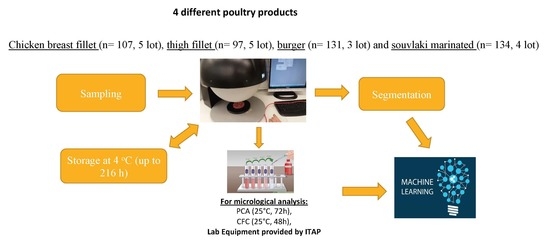
2.1. Experimental Design
2.2. Microbiological Analysis
2.3. Spectra Acquisition
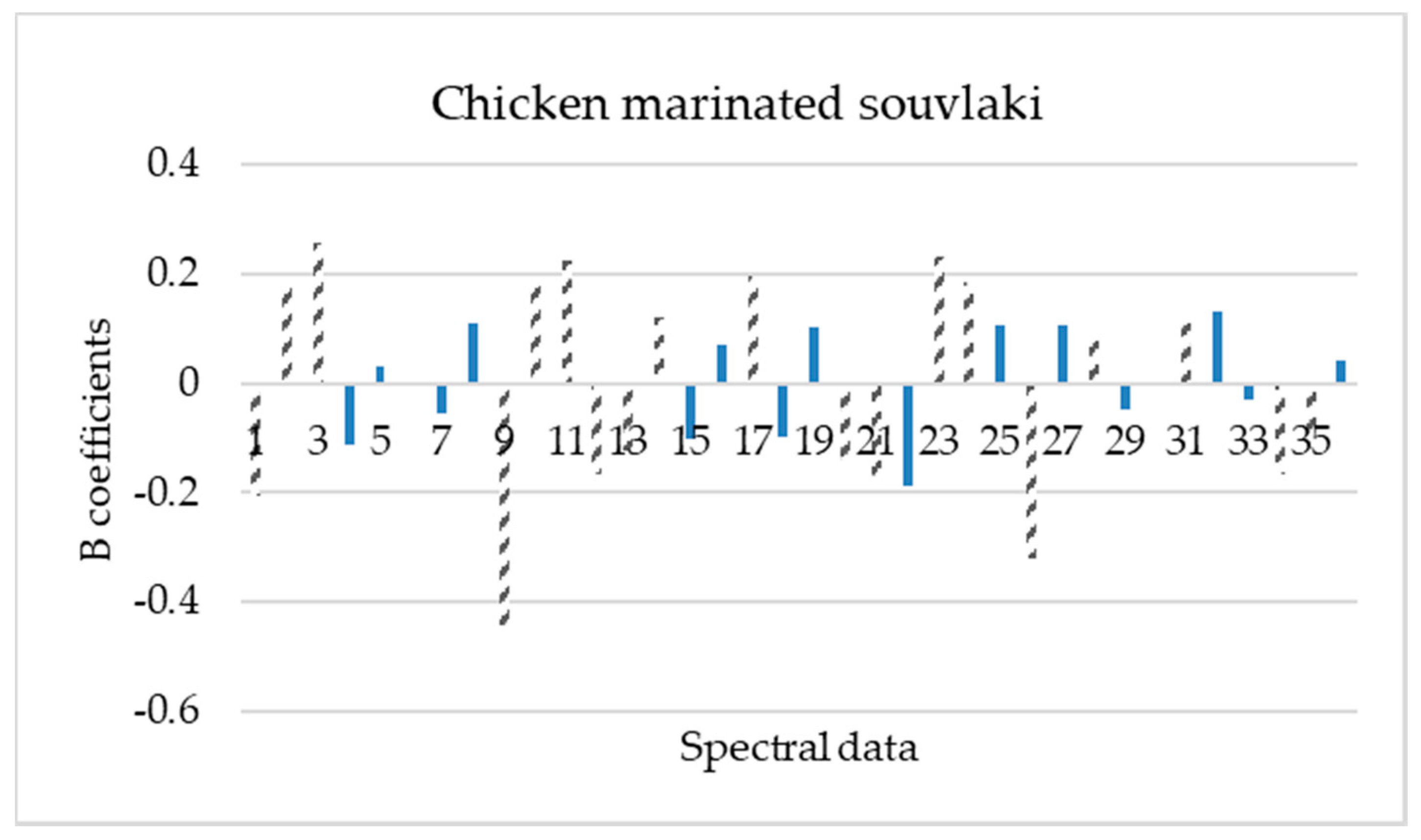
2.4. Data Pre-Processing and Model Development
3. Results
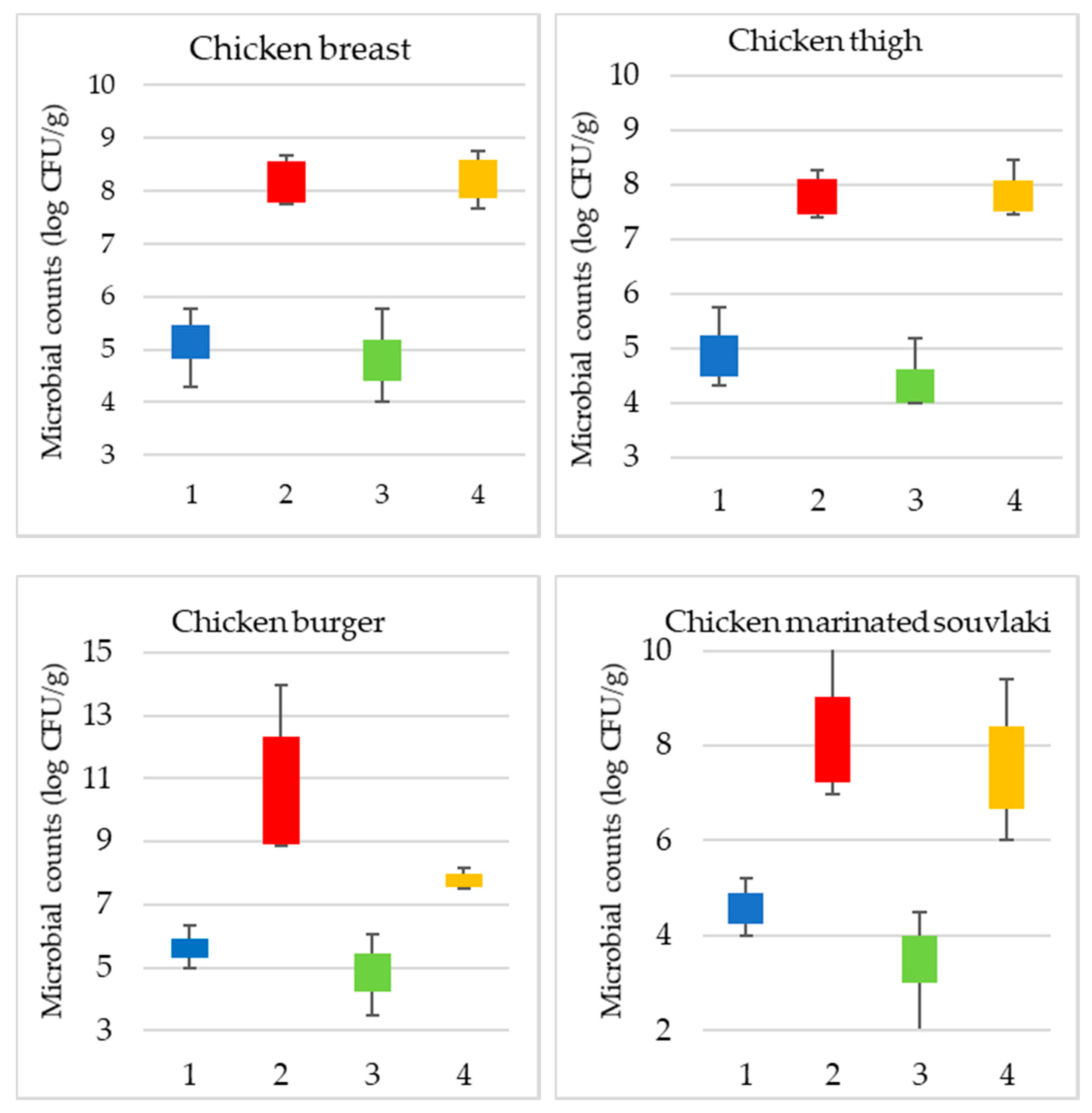
3.1. Microbiological Analysis
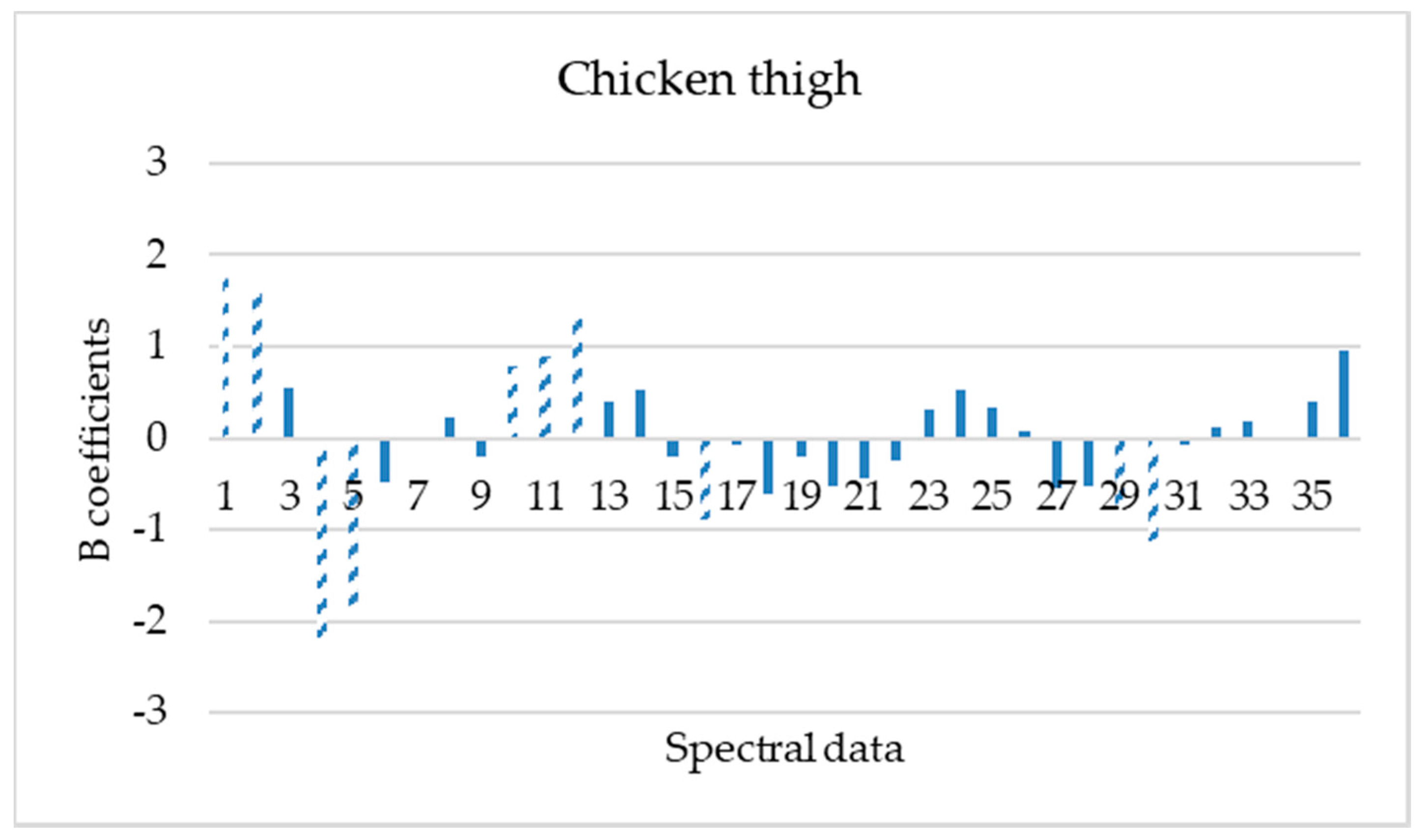
3.2. Spectral Measurements
3.3. PLS-R Model Performance
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- FAO. Gateway to Poultry Production and Products. Available online: http://www.fao.org/poultry-production-products/products-processing/en/ (accessed on 15 March 2020).
- Baston, O.; Barna, O. Raw chicken leg and breast sensory evaluation. J. Food Sci. Technol. 2010, 11, 25–30. [Google Scholar]
- Dawson, P.L.; Chaves, B.D.; Northcutt, J.K.; Han, I.Y. Quality and shelf life of fresh chicken breasts subjected to crust freezing with and without skin. J. Food Qual. 2013, 36, 361–368. [Google Scholar] [CrossRef]
- Nychas, G.J.E.; Panagou, E.Z.; Mohareb, F. Novel approaches for food safety management and communication. Curr. Opin. Food Sci. 2016, 12, 13–20. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Non-invasive analytical technology for the detection of contamination, adulteration, and authenticity of meat, poultry, and fish: A review. Anal. Chim. Acta 2015, 853, 19–29. [Google Scholar] [CrossRef]
- Chen, Q.; Cai, J.; Wan, X.; Zhao, J. Application of linear/non-linear classification algorithms in discrimination of pork storage time using Fourier transform near infrared (FT-NIR) spectroscopy. LWT-Food Sci. Technol. 2011, 44, 2053–2058. [Google Scholar] [CrossRef]
- van den Berg, F.; Lyndgaard, C.B.; Sørensen, K.M.; Engelsen, S.B. Process analytical technology in the food industry. Trends Food Sci. Technol. 2013, 31, 27–35. [Google Scholar] [CrossRef]
- Cullen, P.J.; O’Donnell, C.P.; Fagan, C.C. Process Analytical Technology for the Food Industry, 1st ed.; Springer: New York, NY, USA, 2014; pp. 1–5. [Google Scholar] [CrossRef]
- Grassi, S.; Alamprese, C. Advances in NIR spectroscopy applied to process analytical technology in food industries. Curr. Opin. Food Sci. 2018, 22, 17–21. [Google Scholar] [CrossRef]
- Teena, M.; Manickavasagan, A.; Mothershaw, A.; El Hadi, S.; Jayas, D.S. Potential of machine vision techniques for detecting fecal and microbial contamination of food products: A review. Food Bioprocess Tech. 2013, 6, 1621–1634. [Google Scholar] [CrossRef]
- Alamprese, C.; Amigo, J.M.; Casiraghi, E.; Engelsen, S.B. Identification and quantification of turkey meat adulteration in fresh, frozen-thawed and cooked minced beef by FT-NIR spectroscopy and chemometrics. Meat Sci. 2016, 121, 175–181. [Google Scholar] [CrossRef]
- Cai, J.; Chen, Q.; Wan, X.; Zhao, J. Determination of total volatile basic nitrogen (TVB-N) content and Warner–Bratzler shear force (WBSF) in pork using Fourier transform near infrared (FT-NIR) spectroscopy. Food Chem. 2011, 126, 1354–1360. [Google Scholar] [CrossRef]
- Qin, J.; Chao, K.; Kim, M.S.; Lu, R.; Burks, T.F. Hyperspectral and multispectral imaging for evaluating food safety and quality. J. Food Eng. 2013, 118, 157–171. [Google Scholar] [CrossRef]
- Liu, D.; Sun, D.W.; Zeng, X.A. Recent advances in wavelength selection techniques for hyperspectral image processing in the food industry. Food Bioproc. Tech. 2014, 7, 307–323. [Google Scholar] [CrossRef]
- Xiong, Z.; Xie, A.; Sun, D.W.; Zeng, X.A.; Liu, D. Applications of hyperspectral imaging in chicken meat safety and quality detection and evaluation: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1287–1301. [Google Scholar] [CrossRef]
- Ghasemi-Varnamkhasti, M.; Mohtasebi, S.S.; Siadat, M. Biomimetic-based odor and taste sensing systems to food quality and safety characterization: An overview on basic principles and recent achievements. J. Food Eng. 2010, 100, 377–387. [Google Scholar] [CrossRef]
- Huffman, B.; Mazrouei, R.; Bevelheimer, J.; Shavezipur, M. Three-Dimensional Biomimetic Biosensors for Food Safety Applications. In Proceedings of the ASME 2017 International Design Engineering Technical Conferences and Computers and Information in Engineering Conference, American Society of Mechanical Engineers Digital Collection, Cleveland, OH, USA, 6–9 August 2017. [Google Scholar] [CrossRef]
- Vasconcelos, H.; Saraiva, C.; de Almeida, J.M. Evaluation of the spoilage of raw chicken breast fillets using Fourier transform infrared spectroscopy in tandem with chemometrics. Food Bioproc. Tech. 2014, 7, 2330–2341. [Google Scholar] [CrossRef]
- Estelles-Lopez, L.; Ropodi, A.; Pavlidis, D.; Fotopoulou, J.; Gkousari, C.; Peyrodie, A.; Panagou, E.Z.; Nychas, G.-J.E.; Mohareb, F. An automated ranking platform for machine learning regression models for meat spoilage prediction using multi-spectral imaging and metabolic profiling. Food Res. Int. 2017, 99, 206–215. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Fengou, L.C.; Manthou, E.; Lianou, A.; Panagou, E.Z.; Nychas, G.J.E. A unified spectra analysis workflow for the assessment of microbial contamination of ready-to-eat green salads: Comparative study and application of non-invasive sensors. Comput. Electron. Agric. 2018, 155, 212–219. [Google Scholar] [CrossRef]
- Feng, C.H.; Makino, Y.; Oshita, S.; Martín, J.F.G. Hyperspectral imaging and multispectral imaging as the novel techniques for detecting defects in raw and processed meat products: Current state-of-the-art research advances. Food Control 2018, 84, 165–176. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Pavlidis, D.; Panagou, E.; Nychas, G.J. Exploiting multispectral imaging for non-invasive contamination assessment and mapping of meat samples. Talanta 2016, 161, 606–614. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Papadopoulou, O.; Carstensen, J.M.; Nychas, G.J.E. Potential of multispectral imaging technology for rapid and non-destructive determination of the microbiological quality of beef filets during aerobic storage. Int. J. Food Microbiol. 2014, 174, 1–11. [Google Scholar] [CrossRef]
- Pu, H.; Kamruzzaman, M.; Sun, D.W. Selection of feature wavelengths for developing multispectral imaging systems for quality, safety and authenticity of muscle foods-a review. Trends Food Sci. Technol. 2015, 45, 86–104. [Google Scholar] [CrossRef]
- Yang, Y.; Zhuang, H.; Yoon, S.C.; Wang, W.; Jiang, H.; Jia, B. Rapid classification of intact chicken breast fillets by predicting principal component score of quality traits with visible/near-infrared spectroscopy. Food Chem. 2018, 244, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.Z.; Sun, D.W. Determination of total viable count (TVC) in chicken breast fillets by near-infrared hyperspectral imaging and spectroscopic transforms. Talanta 2013, 105, 244–249. [Google Scholar] [CrossRef]
- Feng, Y.Z.; El Masry, G.; Sun, D.W.; Scannell, A.G.; Walsh, D.; Morcy, N. Near-infrared hyperspectral imaging and partial least squares regression for rapid and reagentless determination of Enterobacteriaceae on chicken fillets. Food Chem. 2013, 138, 1829–1836. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.Z.; Sun, D.W. Near-infrared hyperspectral imaging in tandem with partial least squares regression and genetic algorithm for non-destructive determination and visualization of Pseudomonas loads in chicken fillets. Talanta 2013, 109, 74–83. [Google Scholar] [CrossRef]
- Ye, X.; Iino, K.; Zhang, S. Monitoring of bacterial contamination on chicken meat surface using a novel narrowband spectral index derived from hyperspectral imagery data. Meat Sci. 2016, 122, 25–31. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Rapid and non-destructive detection of chicken adulteration in minced beef using visible near-infrared hyperspectral imaging and machine learning. J. Food Eng. 2016, 170, 8–15. [Google Scholar] [CrossRef]
- Yang, C.C.; Chao, K.; Chen, Y.R.; Early, H.L. Systemically diseased chicken identification using multispectral images and region of interest analysis. Comput. Electron. Agric. 2005, 49, 255–271. [Google Scholar] [CrossRef]
- Cho, B.K.; Chen, Y.R.; Kim, M.S. Multispectral detection of organic residues on poultry processing plant equipment based on hyperspectral reflectance imaging technique. Comput. Electron. Agric. 2007, 57, 177–189. [Google Scholar] [CrossRef]
- Park, B.; Lawrence, K.C.; Windham, W.R.; Smith, D.P. Performance of hyperspectral imaging system for poultry surface fecal contaminant detection. J. Food Eng. 2006, 75, 340–348. [Google Scholar] [CrossRef]
- Yang, C.C.; Chao, K.; Chen, Y.R.; Kim, M.S.; Early, H.L. Simple multispectral image analysis for systemically diseased chicken identification. Trans ASABE 2006, 49, 245–257. [Google Scholar] [CrossRef][Green Version]
- Chao, K.; Yang, C.C.; Chen, Y.R.; Kim, M.S.; Chan, D.E. Hyperspectral-multispectral line-scan imaging system for automated poultry carcass inspection applications for food safety. Poult. Sci. 2007, 86, 2450–2460. [Google Scholar] [CrossRef] [PubMed]
- Barni, M.; Cappellini, V.; Mecocci, A. Colour-based detection of defects on chicken meat. Image Vision Comput. 1997, 15, 549–556. [Google Scholar] [CrossRef]
- Nakariyakul, S.; Casasent, D.P. Fast feature selection algorithm for poultry skin tumor detection in hyperspectral data. J. Food Eng. 2009, 94, 358–365. [Google Scholar] [CrossRef]
- Dixit, Y.; Casado-Gavalda, M.P.; Cama-Moncunill, R.; Cama-Moncunill, X.; Markiewicz-Keszycka, M.; Cullen, P.J.; Sullivan, C. Developments and challenges in online NIR spectroscopy for meat processing. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1172–1187. [Google Scholar] [CrossRef]
- i Furnols, M.F.; Gispert, M. Comparison of different devices for predicting the lean meat percentage of pig carcasses. Meat Sci. 2009, 83, 443–446. [Google Scholar] [CrossRef]
- De Marchi, M.; Riovanto, R.; Penasa, M.; Cassandro, M. At-line prediction of fatty acid profile in chicken breast using near infrared reflectance spectroscopy. Meat Sci. 2012, 90, 653–657. [Google Scholar] [CrossRef]
- Sørensen, K.M.; Petersen, H.; Engelsen, S.B. An on-line near-infrared (NIR) transmission method for determining depth profiles of fatty acid composition and iodine value in porcine adipose fat tissue. Appl. Spectrosc. 2012, 66, 218–226. [Google Scholar] [CrossRef]
- Prieto, N.; Dugan, M.E.R.; Juárez, M.; López-Campos, Ó.; Zijlstra, R.T.; Aalhus, J.L. Using portable near-infrared spectroscopy to predict pig subcutaneous fat composition and iodine value. Can. J. Anim. Sci. 2017, 98, 221–229. [Google Scholar] [CrossRef]
- Raab, V.; Bruckner, S.; Beierle, E.; Kampmann, Y.; Petersen, B.; Kreyenschmidt, J. Generic model for the prediction of remaining shelf life in support of cold chain management in pork and poultry supply chains. J. Chain Netw. Sci. 2008, 8, 59–73. [Google Scholar] [CrossRef]
- Souza, V.G.; Pires, J.R.; Vieira, É.T.; Coelhoso, I.M.; Duarte, M.P.; Fernando, A.L. Shelf life assessment of fresh poultry meat packaged in novel bionanocomposite of chitosan/montmorillonite incorporated with ginger essential oil. Coatings 2018, 8, 177. [Google Scholar] [CrossRef]
- Baltic, T.; Ciric, J.; Lazic, I.B.; Pelic, D.L.; Mitrovic, R.; Djordjevic, V.; Parunovic, N. Packaging as a tool to improve the shelf life of poultry meat. In Proceedings of the 60th International Meat Industry Conference MEATCON2019, Kopaonik, Serbia, 22–25 September 2019; IOP Publishing: Bristol, UK, 2019; Volume 333, p. 012044. [Google Scholar] [CrossRef]
- Porep, J.U.; Kammerer, D.R.; Carle, R. On-line application of near infrared (NIR) spectroscopy in food production. Trends Food Sci. Technol. 2015, 46, 211–230. [Google Scholar] [CrossRef]
- Dissing, B.S.; Papadopoulou, O.S.; Tassou, C.; Ersbøll, B.K.; Carstensen, J.M.; Panagou, E.Z.; Nychas, G.J. Using multispectral imaging for spoilage detection of pork meat. Food Bioproc. Tech. 2013, 6, 2268–2279. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Pavlidis, D.; Nychas, G.J. High throughput multispectral image processing with applications in food science. PLoS ONE 2015, 10, e0140122. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, S.B.; Adler-Nissen, J.; Carstensen, J.M. New vision technology for multidimensional quality monitoring of continuous frying of meat. Food Control 2010, 21, 626–632. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification, 2nd ed.; Wiley-Interscience: New York, NY, USA, 2000; ISBN 978-0-468 471-05669-0. [Google Scholar]
- Carstensen, J.M.; Hansen, J.F. An Apparatus and a Method of Recording an Image of an Object. Patent Family EP1051660 Patent 17, 198, 15 November 2003. [Google Scholar]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Xiaobo, Z.; Jiewen, Z.; Povey, M.J.; Holmes, M.; Hanpin, M. Variables selection methods in near-infrared spectroscopy. Anal. Chim. Acta 2010, 667, 14–32. [Google Scholar] [CrossRef]
- Brereton, R.G.; Lloyd, G.R. Partial least squares discriminant analysis: Taking the magic away. J. Chemom. 2014, 28, 213–225. [Google Scholar] [CrossRef]
- Bi, Y.; Yuan, K.; Xiao, W.; Wu, J.; Shi, C.; Xia, J.; Chu, G.; Zhang, G.; Zhou, G. A local pre-processing method for near-infrared spectra, combined with spectral segmentation and standard normal variate transformation. Anal. Chim. Acta 2016, 909, 30–40. [Google Scholar] [CrossRef]
- Engel, J.; Gerretzen, J.; Szymańska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M. Breaking with trends in pre- processing? Trends Anal. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Rinnan, A.; Berg, F.; Engelsen, B. Review of the most common pre-processing techniques for near-infrared spectra. Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, D.; Murray, I. Identification of animal meat muscles by visible and near infrared reflectance spectroscopy. LWT-Food Sci. Technol. 2004, 37, 447–452. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Sun, D.W.; El Masry, G.; Allen, P. Fast detection and visualization of minced lamb meat adulteration using NIR hyperspectral imaging and multivariate image analysis. Talanta 2013, 103, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Lin, L.C.; Hsu, J.C. Effect of caponization on muscle composition, shear value, ATP related compounds and taste appraisal in Taiwan country chicken cockerels. Asian-Australas. J. Anim. Sci. 2011, 24, 1026–1030. [Google Scholar] [CrossRef]
- Amorim, A.; Rodrigues, S.; Pereira, E.; Valentim, R.; Teixeira, A. Effect of caponisation on physicochemical and sensory characteristics of chickens. Animal 2016, 10, 978–986. [Google Scholar] [CrossRef]
- Sant’Ana, A.S.; Franco, B.D.; Schaffner, D.W. Modeling the growth rate and lag time of different strains of Salmonella enterica and Listeria monocytogenes in ready-to-eat lettuce. Food Microbiol. 2012, 30, 267–273. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.J.E. Rapid detection of frozen-then-thawed minced beef using multispectral imaging and Fourier transform infrared spectroscopy. Meat Sci. 2018, 135, 142–147. [Google Scholar] [CrossRef]
- Alomar, D.; Gallo, C.; Castaneda, M.; Fuchslocher, R. Chemical and discriminant analysis of bovine meat by near infrared reflectance spectroscopy (NIRS). Meat Sci. 2003, 63, 441–450. [Google Scholar] [CrossRef]
- Boulesteix, A.L.; Strimmer, K. Partial least squares: A versatile tool for the analysis of high-dimensional genomic data. Brief. Bioinform. 2006, 8, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.J. Data mining derived from food analyses using non-invasive/non-destructive analytical techniques; determination of food authenticity, quality & safety in tandem with computer science disciplines. Trends Food Sci. Tech. 2016, 50, 11–25. [Google Scholar] [CrossRef]
- Park, B.; Lawrence, K.C.; Windham, W.R.; Chen, Y.R.; Chao, K. Discriminant analysis of dual-wavelength spectral images for classifying poultry carcasses. Comput. Electron. Agric. 2002, 33, 219–231. [Google Scholar] [CrossRef]

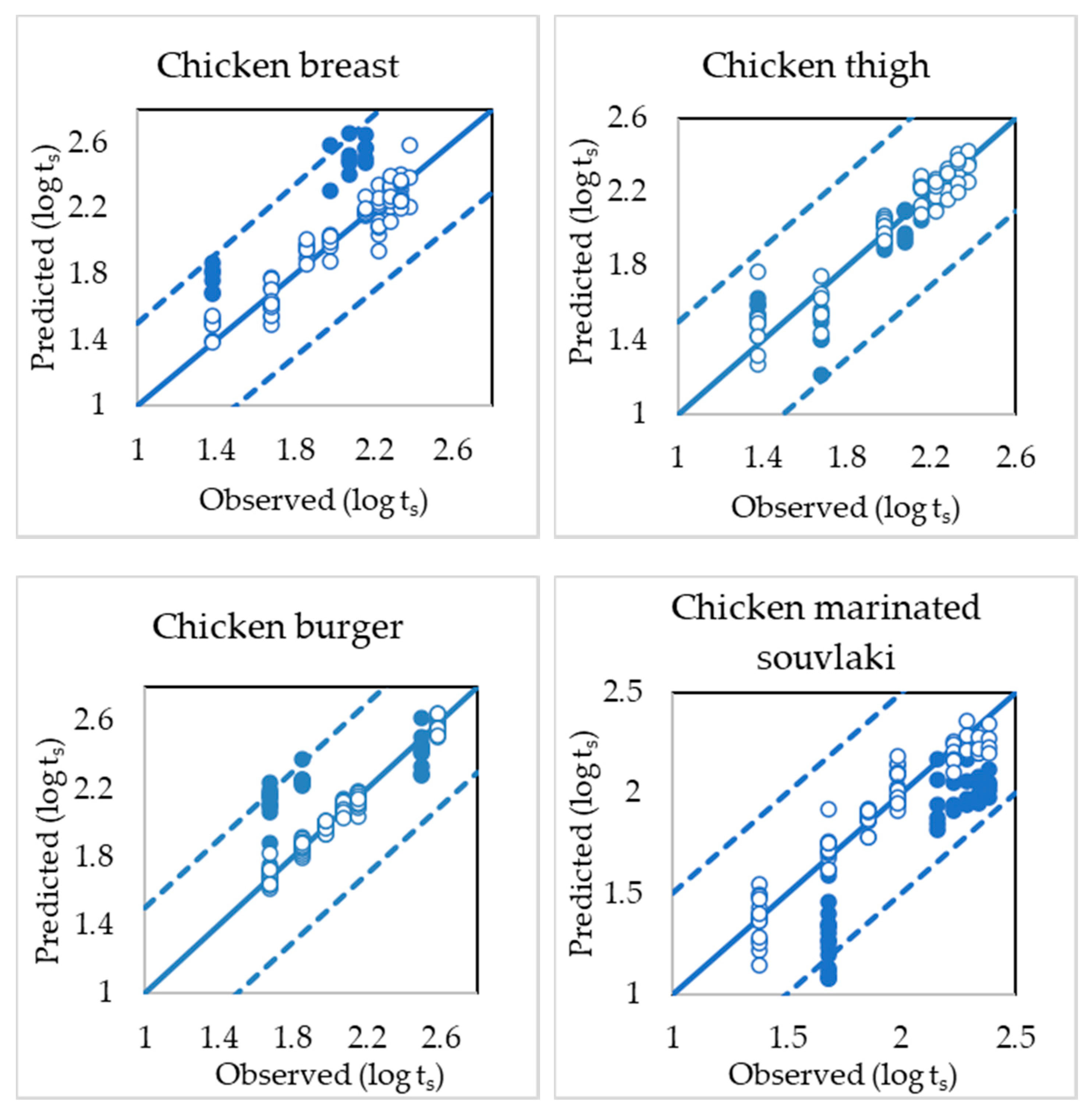


| Poultry Product | Stage of Modelling | No of Samples | Slope | Offset | r (Correlation Coefficient) | RMSE |
|---|---|---|---|---|---|---|
| Chicken Breast | Calibration | 82 | 0.933 | 0.138 | 0.966 | 0.076 |
| FCV 1 | 82 | 0.916 | 0.173 | 0.953 | 0.091 | |
| Prediction | 22 | 1.150 | 0.055 | 0.886 | 0.383 | |
| Chicken Thigh | Calibration | 67 | 0.953 | 0.097 | 0.976 | 0.065 |
| FCV | 67 | 0.933 | 0.136 | 0.957 | 0.088 | |
| Prediction | 30 | 0.854 | 0.243 | 0.859 | 0.160 | |
| Chicken Burger | Calibration | 87 | 0.982 | 0.035 | 0.991 | 0.033 |
| FCV | 87 | 0.968 | 0.063 | 0.987 | 0.040 | |
| Prediction | 44 | 0.513 | 1.172 | 0.778 | 0.285 | |
| Chicken Marinated Souvlaki | Calibration | 91 | 0.962 | 0.073 | 0.981 | 0.067 |
| FCV | 91 | 0.954 | 0.092 | 0.964 | 0.093 | |
| Prediction | 43 | 1.183 | 0.650 | 0.934 | 0.348 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spyrelli, E.D.; Doulgeraki, A.I.; Argyri, A.A.; Tassou, C.C.; Panagou, E.Z.; Nychas, G.-J.E. Implementation of Multispectral Imaging (MSI) for Microbiological Quality Assessment of Poultry Products. Microorganisms 2020, 8, 552. https://doi.org/10.3390/microorganisms8040552
Spyrelli ED, Doulgeraki AI, Argyri AA, Tassou CC, Panagou EZ, Nychas G-JE. Implementation of Multispectral Imaging (MSI) for Microbiological Quality Assessment of Poultry Products. Microorganisms. 2020; 8(4):552. https://doi.org/10.3390/microorganisms8040552
Chicago/Turabian StyleSpyrelli, Evgenia D., Agapi I. Doulgeraki, Anthoula A. Argyri, Chrysoula C. Tassou, Efstathios Z. Panagou, and George-John E. Nychas. 2020. "Implementation of Multispectral Imaging (MSI) for Microbiological Quality Assessment of Poultry Products" Microorganisms 8, no. 4: 552. https://doi.org/10.3390/microorganisms8040552
APA StyleSpyrelli, E. D., Doulgeraki, A. I., Argyri, A. A., Tassou, C. C., Panagou, E. Z., & Nychas, G.-J. E. (2020). Implementation of Multispectral Imaging (MSI) for Microbiological Quality Assessment of Poultry Products. Microorganisms, 8(4), 552. https://doi.org/10.3390/microorganisms8040552