Biofilm Eradication by Symmetrical Selenoesters for Food-Borne Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Reagents and Media
2.3. Bacterial Strains
2.4. Cell Line
2.5. Determination of Minimum Inhibitory Concentrations by Microdilution Method
2.6. Cytotoxicity Assay
2.7. Resistance Modulation Assay
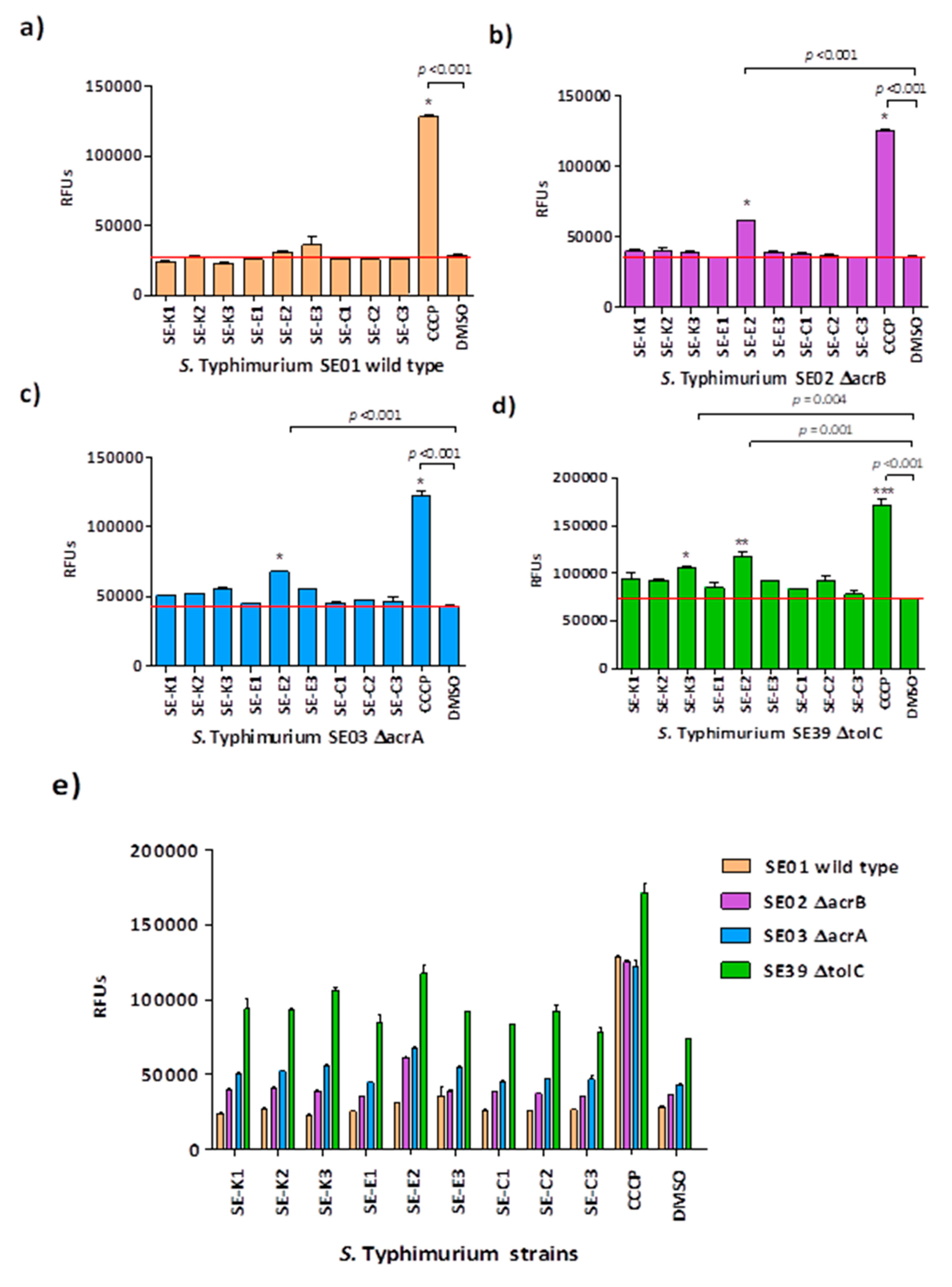
2.8. Real-Time Ethidium Bromide Accumulation Assay
2.9. Measuring Biofilm Formation Using Crystal Violet
2.10. Quorum Sensing (QS) Assay
2.11. Statistical Analysis
3. Results
3.1. Determination of Minimum Inhibitory Concentrations by Microdilution Method
3.2. Resistance Modulation Assay
3.3. Ethidium Bromide Accumulation Assay
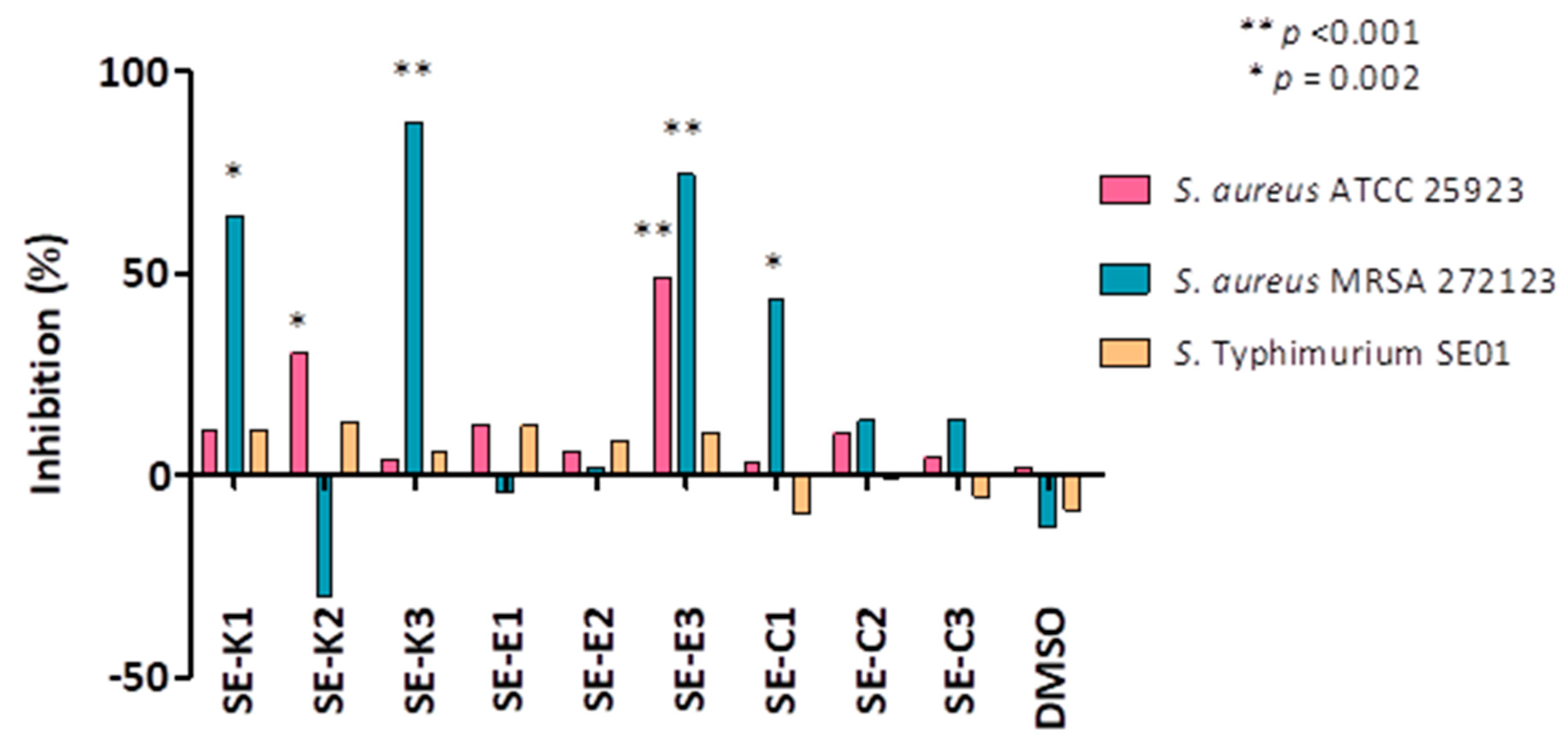
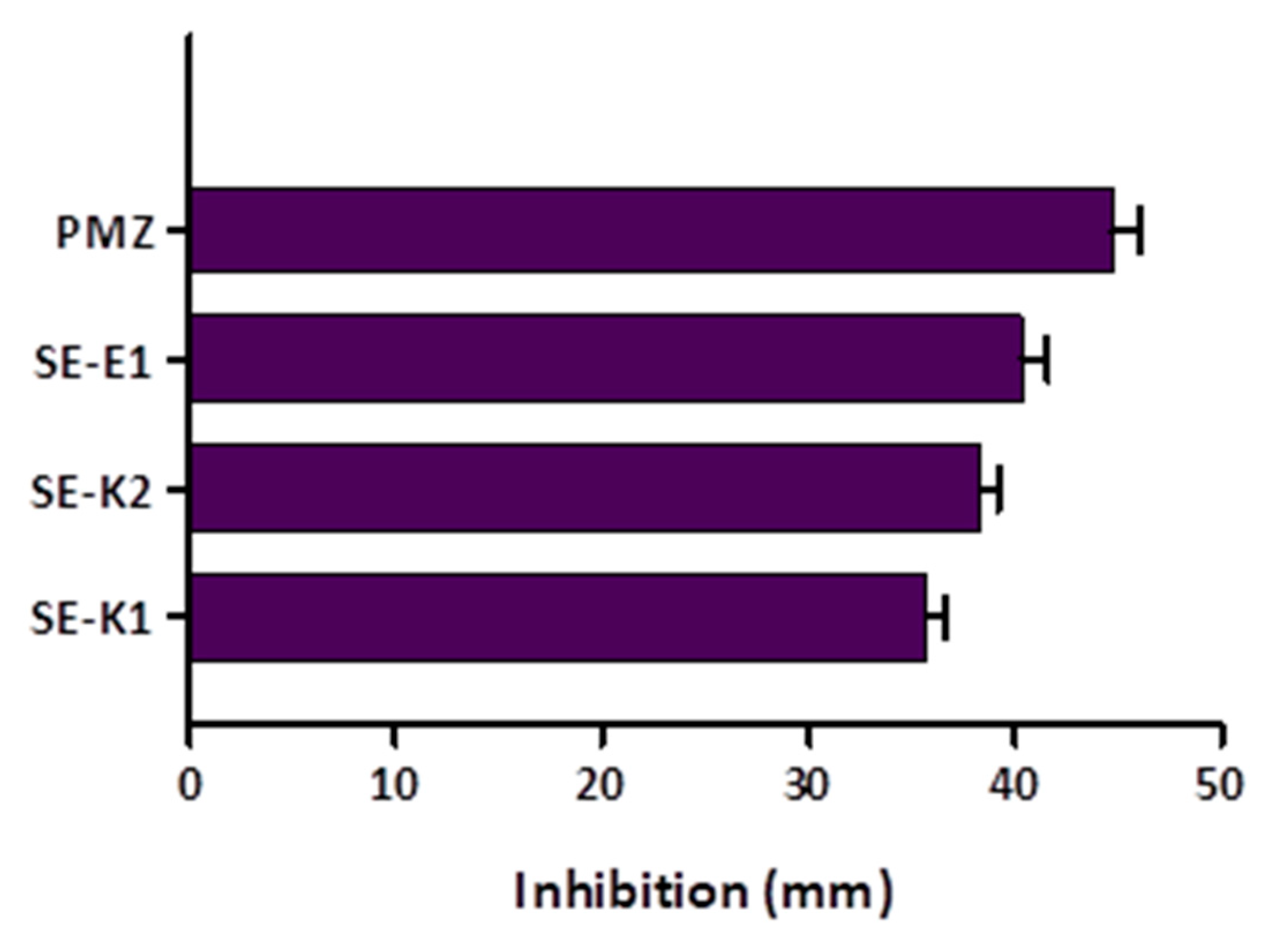
3.4. Measuring Biofilm Formation Using Crystal Violet
3.5. Quorum Sensing (QS) Assay
3.6. Cytotoxicity Assay on Normal Human Fibroblasts
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Conflicts of Interest
References
- Carlet, J.; Pulcini, C.; Piddock, L.J. Antibiotic resistance: A geopolitical issue. Clin. Microbiol. Infect. 2014, 20, 949–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.A.; Liu, Q.; Elbourne, L.D.H.; Ahmad, I.; Sharples, D.; Naidu, V.; Chan, C.L.; Li, L.; Harborne, S.P.D.; Pokhrel, A.; et al. Pacing across the membrane: The novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 2018, 169, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Colclough, A.L.; Alav, I.; Whittle, E.E.; Pugh, H.L.; Darby, E.M.; Legood, S.W.; McNeil, H.E.; Blair, J.M. RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol. 2020, 15, 143–157. [Google Scholar] [CrossRef]
- Santos, A.; Galdino, A.C.M.; Mello, T.P.; Ramos, L.S.; Branquinha, M.H.; Bolognese, A.M.; Columbano Neto, J.; Roudbary, M. What are the advantages of living in a community? A microbial biofilm perspective! Mem. Inst. Oswaldo Cruz 2018, 113, e180212. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, Y.; Liu, Y.; Shi, X.; Jiang, M.; Lin, Y.; Qiu, Y.; Zhang, Q.; Chen, Q.; Zhou, L.; et al. Clonal Expansion of Biofilm-Forming Salmonella Typhimurium ST34 with Multidrug-Resistance Phenotype in the Southern Coastal Region of China. Front. Microbiol. 2017, 8, 2090. [Google Scholar] [CrossRef] [Green Version]
- Hennekinne, J.A.; De Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef] [Green Version]
- Knowles, J.R.; Roller, S.; Murray, D.B.; Naidu, A.S. Antimicrobial action of carvacrol at different stages of dual-species biofilm development by Staphylococcus aureus and Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 2005, 71, 797–803. [Google Scholar] [CrossRef] [Green Version]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Saxena, P.; Joshi, Y.; Rawat, K.; Bisht, R. Biofilms: Architecture, Resistance, Quorum Sensing and Control Mechanisms. Indian J. Microbiol. 2019, 59, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Holden, J.A.; Heath, D.E.; O’Brien-Simpson, N.M.; O’Connor, A.J. Engineering highly effective antimicrobial selenium nanoparticles through control of particle size. Nanoscale 2019, 11, 14937–14951. [Google Scholar] [CrossRef] [PubMed]
- Staicu, L.C.; Oremland, R.S.; Tobe, R.; Mihara, H. Bacteria versus selenium: A view from the inside out. In Selenium in Plants; Springer: Cham, Switzerland, 2017; pp. 79–108. [Google Scholar]
- Verma, P. A review on synthesis and their antibacterial activity of Silver and Selenium nanoparticles against biofilm forming Staphylococcus aureus. World J. Pharm. Pharmaceut. Sci. 2015, 4, 652–677. [Google Scholar]
- Mosolygo, T.; Kincses, A.; Csonka, A.; Tonki, A.S.; Witek, K.; Sanmartin, C.; Marc, M.A.; Handzlik, J.; Kiec-Kononowicz, K.; Dominguez-Alvarez, E.; et al. Selenocompounds as Novel Antibacterial Agents and Bacterial Efflux Pump Inhibitors. Molecules 2019, 24, 1487. [Google Scholar] [CrossRef] [Green Version]
- Witek, K.; Nasim, M.J.; Bischoff, M.; Gaupp, R.; Arsenyan, P.; Vasiljeva, J.; Marc, M.A.; Olejarz, A.; Latacz, G.; Kiec-Kononowicz, K.; et al. Selenazolinium Salts as “Small Molecule Catalysts” with High Potency against ESKAPE Bacterial Pathogens. Molecules 2017, 22, 2174. [Google Scholar] [CrossRef] [Green Version]
- Spengler, G.; Kincses, A.; Mosolygo, T.; Marc, M.A.; Nove, M.; Gajdacs, M.; Sanmartin, C.; McNeil, H.E.; Blair, J.M.A.; Dominguez-Alvarez, E. Antiviral, Antimicrobial and Antibiofilm Activity of Selenoesters and Selenoanhydrides. Molecules 2019, 24, 4264. [Google Scholar] [CrossRef] [Green Version]
- Medina Cruz, D.; Mi, G.; Webster, T.J. Synthesis and characterization of biogenic selenium nanoparticles with antimicrobial properties made by Staphylococcus aureus, methicillin-resistant Staphylococcus aureus (MRSA), Escherichia coli, and Pseudomonas aeruginosa. J. Biomed. Mater. Res. A 2018, 106, 1400–1412. [Google Scholar] [CrossRef]
- Khiralla, G.M.; Bahig, A.E.D. Antimicrobial and antibiofilm effects of selenium nanoparticles on some foodborne pathogens. LWT Food Sci. Technol. 2015, 63, 1001–1007. [Google Scholar] [CrossRef]
- La Cruz-Claure, D.; María, L.; Cèspedes-Llave, A.A.; Ulloa, M.T.; Benito-Lama, M.; Domínguez-Álvarez, E.; Bastida, A. Inhibition–Disruption of Candida glabrata Biofilms: Symmetrical Selenoesters as Potential Anti-Biofilm Agents. Microorganisms 2019, 7, 664. [Google Scholar] [CrossRef] [Green Version]
- Domínguez Álvarez, E.; Spengler, G.; Jacob, C.; Sanmartín Grijalba, M.C. Selenoester-Containing Compounds for Use in the Treatment of Microbial Infections or Colorectal Cancer. European Patent EP18382693, 28 September 2018. [Google Scholar]
- Wray, C.; Sojka, W.J. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 1978, 25, 139–143. [Google Scholar] [CrossRef]
- Blair, J.M.; La Ragione, R.M.; Woodward, M.J.; Piddock, L.J. Periplasmic adaptor protein AcrA has a distinct role in the antibiotic resistance and virulence of Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2009, 64, 965–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eaves, D.J.; Ricci, V.; Piddock, L.J. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: Role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 2004, 48, 1145–1150. [Google Scholar] [CrossRef] [Green Version]
- Buckley, A.M.; Webber, M.A.; Cooles, S.; Randall, L.P.; La Ragione, R.M.; Woodward, M.J.; Piddock, L.J. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 2006, 8, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Ballantine, J.; Beer, R.; Crutchley, D.; Dodd, G.; Palmer, D. The synthesis of violacein and related compounds. Proc. Chem. Soc. Lond. 1958, 8, 232–233. [Google Scholar]
- Kothari, V.; Sharma, S.; Padia, D. Recent research advances on Chromobacterium violaceum. Asian Pac. J. Trop Med. 2017, 10, 744–752. [Google Scholar] [CrossRef]
- CLSI. Susceptibility Testing Process. In Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 10th ed.; Christopher, P.J., Polgar, E.P., Eds.; Clinical and Laboratory Standards Institute: Wayne, MI, USA, 2015; Volume 32, pp. 15–49. [Google Scholar]
- Gajdacs, M.; Spengler, G. The Role of Drug Repurposing in the Development of Novel Antimicrobial Drugs: Non-Antibiotic Pharmacological Agents as Quorum Sensing-Inhibitors. Antibiotics (Basel) 2019, 8, 270. [Google Scholar] [CrossRef] [Green Version]
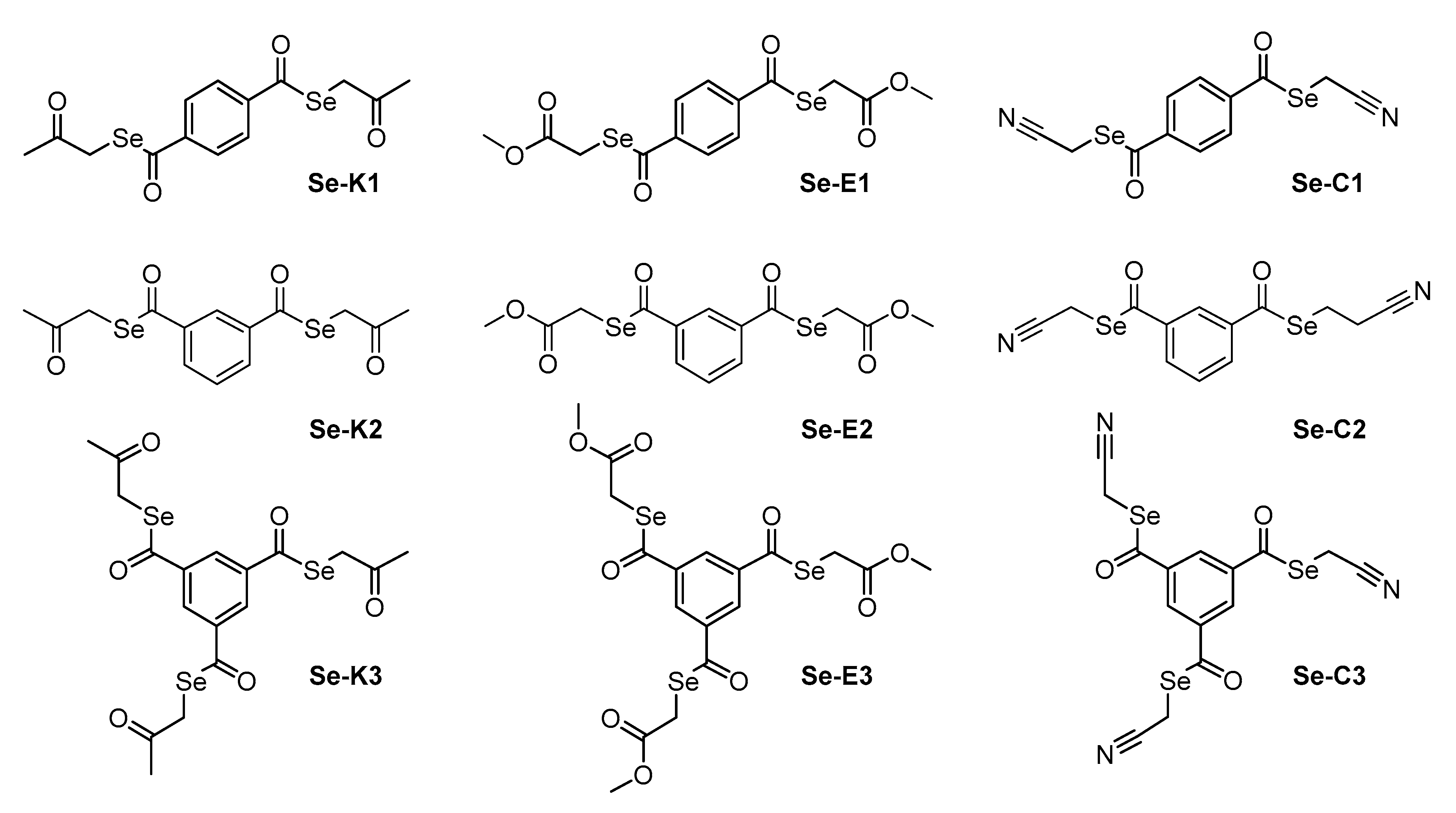

| MIC Determination (µM) | ||||||
|---|---|---|---|---|---|---|
| Compounds | S. aureus ATCC 25923 | S. aureus MRSA 272123 | S. Typhimurium SE01 Wild-Type | S. Typhimurium SE02 ΔacrB | S. Typhimurium SE03 ΔacrA | S. Typhimurium SE39 ΔtolC |
| Se-K1 | 0.39 | 1.56 | 50 | 50 | 50 | 50 |
| Se-K2 | 0.39 | 1.56 | 50 | 50 | 50 | 100 |
| Se-K3 | 0.39 | 0.78 | 50 | 25 | 25 | 50 |
| Se-E1 | 12.5 | 100 | >100 | >100 | >100 | >100 |
| Se-E2 | 12.5 | 100 | >100 | >100 | >100 | >100 |
| Se-E3 | 12.5 | 100 | >100 | >100 | >100 | >100 |
| Se-C1 | 6.25 | 50 | 25 | 25 | 25 | 25 |
| Se-C2 | 6.25 | 50 | 25 | 25 | 25 | 25 |
| Se-C3 | 1.56 | 12.5 | 12.5 | 12.5 | 12.5 | 25 |
| MIC Reduction (µM) In Brackets, the X-Fold Reduction of MIC Is Presented | |||||
|---|---|---|---|---|---|
| Compounds | S. aureus ATCC 25923 with | S. aureus MRSA 272123 with | |||
| TET | CIP | TET | CIP | ||
| − | 0.88 | 1.06 | 14.06 | 33.99 | |
| Se-K1 | 0.88 | 1.06 | 3.51 (4) | 16.99 (2) | |
| Se-K2 | 0.88 | 1.06 | 7.03 (2) | 33.99 | |
| Se-K3 | 0.88 | 1.06 | 7.03 (2) | 33.99 | |
| Se-E1 | 0.88 | 1.06 | 0.88 (16) | 33.99 | |
| Se-E2 | 0.88 | 1.06 | 1.76 (8) | 33.99 | |
| Se-E3 | 0.44 (2) | 1.06 | 0.44 (32) | 16.99 (2) | |
| Se-C1 | 0.88 | 1.06 | 0.88 (16) | 33.99 | |
| Se-C2 | 0.88 | 1.06 | 0.44 (32) | 33.99 | |
| Se-C3 | 0.88 | 1.06 | 3.51 (4) | 33.99 | |
| Relative Fluorescence Index (RFI) | ||||||
|---|---|---|---|---|---|---|
| Compounds | S. Typhimurium SE01 Wild-Type | S. Typhimurium SE02 ΔacrB | S. Typhimurium SE03 ΔacrA | S. Typhimurium SE39 ΔtolC | S. aureus ATCC 25923 | S. aureus MRSA 272123 |
| Se-K1 | −0.16 | 0.10 | 0.17 | 0.27 | 0.1 | −0.15 |
| Se-K2 | −0.04 | 0.13 | 0.20 | 0.26 | 0.11 | −0.07 |
| Se-K3 | −0.20 | 0.08 | 0.28 | 0.44 | 0.16 | −0.18 |
| Se-E1 | −0.10 | −0.03 | 0.03 | 0.15 | 0.98 | 0.19 |
| Se-E2 | 0.09 | 0.70 | 0.56 | 0.59 | 0.67 | 0.33 |
| Se-E3 | 0.26 | 0.08 | 0.27 | 0.25 | 4.15 | 0.47 |
| Se-C1 | −0.08 | 0.06 | 0.04 | 0.13 | 0.14 | −0.15 |
| Se-C2 | −0.10 | 0.03 | 0.09 | 0.25 | 0.08 | −0.13 |
| Se-C3 | −0.07 | −0.02 | 0.08 | 0.06 | 0.18 | −0.05 |
| CCCP | 3.50 | 2.46 | 1.81 | 1.32 | 0.52 | − |
| Verapamil | − | − | − | − | − | 0.32 |
| Compound | MRC-5 | |
|---|---|---|
| IC50 (µM) | SD ± | |
| Se-K1 | 0.54 | 0.00 |
| Se-K2 | 1.34 | 0.16 |
| Se-K3 | 0.74 | 0.04 |
| Se-E1 | 77.91 | 15.86 |
| Se-E2 | >100 | − |
| Se-E3 | 76.61 | 9.18 |
| Se-C1 | >100 | − |
| Se-C2 | >100 | − |
| Se-C3 | >100 | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nové, M.; Kincses, A.; Szalontai, B.; Rácz, B.; Blair, J.M.A.; González-Prádena, A.; Benito-Lama, M.; Domínguez-Álvarez, E.; Spengler, G. Biofilm Eradication by Symmetrical Selenoesters for Food-Borne Pathogens. Microorganisms 2020, 8, 566. https://doi.org/10.3390/microorganisms8040566
Nové M, Kincses A, Szalontai B, Rácz B, Blair JMA, González-Prádena A, Benito-Lama M, Domínguez-Álvarez E, Spengler G. Biofilm Eradication by Symmetrical Selenoesters for Food-Borne Pathogens. Microorganisms. 2020; 8(4):566. https://doi.org/10.3390/microorganisms8040566
Chicago/Turabian StyleNové, Márta, Annamária Kincses, Beatrix Szalontai, Bálint Rácz, Jessica M. A. Blair, Ana González-Prádena, Miguel Benito-Lama, Enrique Domínguez-Álvarez, and Gabriella Spengler. 2020. "Biofilm Eradication by Symmetrical Selenoesters for Food-Borne Pathogens" Microorganisms 8, no. 4: 566. https://doi.org/10.3390/microorganisms8040566
APA StyleNové, M., Kincses, A., Szalontai, B., Rácz, B., Blair, J. M. A., González-Prádena, A., Benito-Lama, M., Domínguez-Álvarez, E., & Spengler, G. (2020). Biofilm Eradication by Symmetrical Selenoesters for Food-Borne Pathogens. Microorganisms, 8(4), 566. https://doi.org/10.3390/microorganisms8040566







