Cloacal Swabs Are Unreliable Sources for Estimating Lower Gastro-Intestinal Tract Microbiota Membership and Structure in Broiler Chickens
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection
2.3. Sample Preparation and Nucleic Acid Isolation
2.4. PCR, Library Preparation, and Sequencing for 16S rRNA Analysis
2.5. Bioinformatic Pipeline for Microbiota Evaluation
2.6. Statistical Tests for α and β Diversity
3. Results
3.1. Sampling Location Yields a Variability in Sequencing Depth
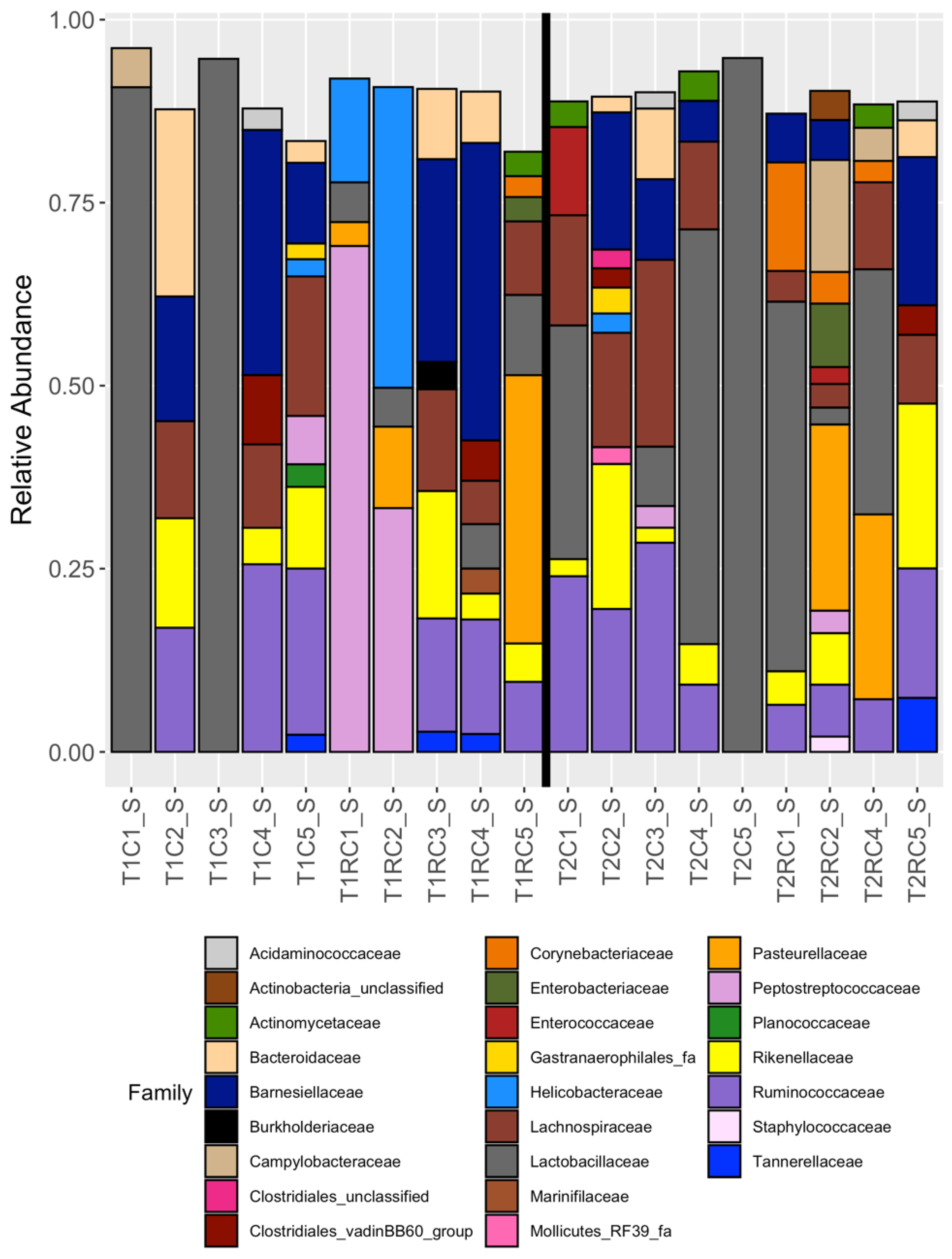
3.2. Broad Differences between Cecal and Cloacal Microbiota Members
3.3. Richness and Diversity Differences between the Cecal and Cloacal Samples
3.4. Cloacal Swabs do not Reflect the Community Structure Inferred from Cecal Samples
4. Discussion
4.1. High Variability of Cloacal Microbiota
4.2. Resolution of Microbial Community Differences between Diets
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- OECD. Food and Agriculture Organization of the United Nations. OECD-FAO Agricultural Outlook 2018-2027; OECD-FAO Agricultural Outlook; OECD Publishing: Paris, France, 2018; ISBN 9789264062030. [Google Scholar]
- Torok, V.A.; Hughes, R.J.; Ophel-Keller, K.; Ali, M.; MacAlpine, R. Influence of different litter materials on cecal microbiota colonization in broiler chickens. Poult. Sci. 2009, 88, 2474–2481. [Google Scholar] [CrossRef] [PubMed]
- Torok, V.A.; Ophel-Keller, K.; Loo, M.; Hughes, R.J. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl. Environ. Microbiol. 2008, 74, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.H. The effect of microbiome modulation on the intestinal health of poultry. Anim. Feed Sci. Technol. 2018, 250, 32–40. [Google Scholar] [CrossRef]
- Teirlynck, E.; Gussem, M.D.E.; Dewulf, J.; Haesebrouck, F.; Ducatelle, R.; Van Immerseel, F. Morphometric evaluation of “dysbacteriosis” in broilers. Avian Pathol. 2011, 40, 139–144. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef]
- Torok, V.A.; Hughes, R.J.; Mikkelsen, L.L.; Perez-Maldonado, R.; Balding, K.; MacAlpine, R.; Percy, N.J.; Ophel-Keller, K. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl. Environ. Microbiol. 2011, 77, 5868–5878. [Google Scholar] [CrossRef]
- Han, Z.; Willer, T.; Li, L.; Pielsticker, C.; Rychlik, I.; Velge, P.; Kaspers, B.; Rautenschlein, S. Influence of the Gut Microbiota Composition on Campylobacter jejuni Colonization in Chickens. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef]
- Biasato, I.; Ferrocino, I.; Biasibetti, E.; Grego, E.; Dabbou, S.; Sereno, A.; Gai, F.; Gasco, L.; Schiavone, A.; Cocolin, L.; et al. Modulation of intestinal microbiota, morphology and mucin composition by dietary insect meal inclusion in free-range chickens. BMC Vet. Res. 2018, 14, 383. [Google Scholar] [CrossRef]
- Liew, W.-P.-P.; Mohd-Redzwan, S. Mycotoxin: Its impact on gut health and microbiota. Front. Cell Infect. Microbiol. 2018, 8, 60. [Google Scholar] [CrossRef]
- Brisbin, J.T.; Gong, J.; Sharif, S. Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim. Health Res. Rev. 2008, 9, 101–110. [Google Scholar] [CrossRef]
- Yegani, M.; Korver, D.R. Factors affecting intestinal health in poultry. Poult. Sci. 2008, 87, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic species in the modulation of gut microbiota: An overview. Biomed. Res. Int. 2018, 2018, 9478630. [Google Scholar] [CrossRef] [PubMed]
- Hieke, A.-S.C.; Hubert, S.M.; Athrey, G. Circadian disruption and divergent microbiota acquisition under extended photoperiod regimens in chicken. PeerJ 2019, 7, e6592. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, V.; Flórez, M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: A review. Poult. Sci. 2018, 97, 1006–1021. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, M.J.; Constantinidou, C.; Cogan, T.A.; Bedford, M.R.; Penn, C.W.; Pallen, M.J. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS ONE 2014, 9, e91941. [Google Scholar] [CrossRef]
- Waite, D.W.; Taylor, M.W. Characterizing the avian gut microbiota: Membership, driving influences, and potential function. Front. Microbiol. 2014, 5, 223. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Zhong, T.; Pandya, Y.; Joerger, R.D. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 2002, 68, 124–137. [Google Scholar] [CrossRef]
- Clench, M.H.; Mathias, J.R. The Avian Cecum: A Review on. Wilson Bull. 1994, 1087, 93–121. [Google Scholar]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef]
- Wei, S.; Morrison, M.; Yu, Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013, 92, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Brangenberg, N.; McInnes, C.; Connolly, J.H.; Rogers, L.E. Absence of Salmonella and Campylobacter Species in Fecal and Cloacal Swab Samples from Kakapo (Strigops habroptilus) on Codfish Island, New Zealand. J. Avian Med. Surg. 2003, 17, 203–205. [Google Scholar] [CrossRef]
- Arnold, M.E.; Slomka, M.J.; Coward, V.J.; Mahmood, S.; Raleigh, P.J.; Brown, I.H. Evaluation of the pooling of swabs for real-time PCR detection of low titre shedding of low pathogenicity avian influenza in turkeys. Epidemiol. Infect. 2013, 141, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.-J.; Yang, X.-X.; Xia, X.; Anderson, B.D.; Heil, G.L.; Qian, Y.-H.; Lu, B.; Cao, W.-C.; Gray, G.C. Comparison of commercial influenza A virus assays in detecting avian influenza H7N9 among poultry cloacal swabs, China. J. Clin. Virol. 2014, 59, 242–245. [Google Scholar] [CrossRef]
- Ferreri, L.M.; Ortiz, L.; Geiger, G.; Barriga, G.P.; Poulson, R.; Gonzalez-Reiche, A.S.; Crum, J.A.; Stallknecht, D.; Moran, D.; Cordon-Rosales, C.; et al. Improved detection of influenza A virus from blue-winged teals by sequencing directly from swab material. Ecol. Evol. 2019, 9, 6534–6546. [Google Scholar] [CrossRef]
- Velkers, F.C.; Blake, D.P.; Graat, E.A.M.; Vernooij, J.C.M.; Bouma, A.; de Jong, M.C.M.; Stegeman, J.A. Quantification of Eimeria acervulina in faeces of broilers: Comparison of McMaster oocyst counts from 24h faecal collections and single droppings to real-time PCR from cloacal swabs. Vet. Parasitol. 2010, 169, 1–7. [Google Scholar] [CrossRef]
- McLendon, B.L.; Cox, N.A.; Cosby, D.E.; Montiel, E.R.; Russell, S.M.; Hofacre, C.L.; Landrum, M.A.; Jackson, J.S.; Wilson, J.L. Detecting Campylobacter coli in young chicks using two different cloacal swab techniques. J. Appl. Poult. Res. 2018, 27, 223–227. [Google Scholar] [CrossRef]
- Glendinning, L.; Watson, K.A.; Watson, M. Development of the duodenal, ileal, jejunal and caecal microbiota in chickens. Anim. Microbiomed. 2019, 1, 17. [Google Scholar] [CrossRef]
- Kers, J.G.; Fischer, E.A.J.; Stegeman, J.A.; Smidt, H.; Velkers, F.C. Comparison of Different Invasive and Non-Invasive Methods to Characterize Intestinal Microbiota throughout a Production Cycle of Broiler Chickens. Microorganisms 2019, 7. [Google Scholar] [CrossRef]
- Vo, A.T.E.; Jedlicka, J.A. Protocols for metagenomic DNA extraction and Illumina amplicon library preparation for faecal and swab samples. Mol. Ecol. Resour. 2014, 14, 1183–1197. [Google Scholar] [CrossRef]
- Gerasimidis, K.; Bertz, M.; Quince, C.; Brunner, K.; Bruce, A.; Combet, E.; Calus, S.; Loman, N.; Ijaz, U.Z. The effect of DNA extraction methodology on gut microbiota research applications. BMC Res. Notes 2016, 9, 365. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, P.-Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019.
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. Available online: https://CRAN.R-project.org/package=vegan (accessed on 12 July 2019).
- Wickham, H. ggplot2 - Elegant Graphics for Data Analysis, 2nd ed.; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Pepler, T. Tpepler/Nonpar: Collection of Methods for Non-Parametric Analysis; R. 2017. Available online: https://rdrr.io/github/tpepler/nonpar/man/nonpar-package.html (accessed on 22 March 2020).
- Cucconi, O. Sulla dimensione ottimale del campione nel controllo statistico di qualità, per variabili, dei prodotti industriali in corso di lavorazione. Calcolo 1966, 2, 177–202. [Google Scholar] [CrossRef]
- Marozzi, M. Some notes on the location–scale Cucconi test. J. Nonparametr. Stat. 2009, 21, 629–647. [Google Scholar] [CrossRef]
- Marozzi, M. The multisample Cucconi test. Stat. Methods Appl. 2014, 23, 209–227. [Google Scholar] [CrossRef]
- Lepage, Y. A combination of Wilcoxon’s and Ansari-Bradley’s statistics. Biometrika 1971, 58, 213–217. [Google Scholar] [CrossRef]
- Marozzi, M. The Lepage location-scale test revisited. FJTS 2008, 24, 137–155. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Schloss, P.D. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2008, 2, 265–275. [Google Scholar] [CrossRef]
- Yue, J.C.; Clayton, M.K. A similarity measure based on species proportions. Commun. Stat. Theory Methods 2005, 34, 2123–2131. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef]
- Montgomery-Smith, S.J.; Schmidt, F.J. Statistical methods for estimating complexity from competition experiments between two populations. J. Theor. Biol. 2010, 264, 1043–1046. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 265–270. [Google Scholar]
- Videvall, E.; Strandh, M.; Engelbrecht, A.; Cloete, S.; Cornwallis, C.K. Measuring the gut microbiome in birds: Comparison of faecal and cloacal sampling. Mol. Ecol. Resour. 2018, 18, 424–434. [Google Scholar] [CrossRef]
- Stanley, D.; Geier, M.S.; Chen, H.; Hughes, R.J.; Moore, R.J. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. 2015, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Kreisinger, J.; Čížková, D.; Kropáčková, L.; Albrecht, T. Cloacal Microbiome Structure in a Long-Distance Migratory Bird Assessed Using Deep 16sRNA Pyrosequencing. PLoS ONE 2015, 10, e0137401. [Google Scholar] [CrossRef] [PubMed]
- Van Veelen, H.P.J.; Falcao Salles, J.; Tieleman, B.I. Multi-level comparisons of cloacal, skin, feather and nest-associated microbiota suggest considerable influence of horizontal acquisition on the microbiota assembly of sympatric woodlarks and skylarks. Microbiome 2017, 5, 156. [Google Scholar] [CrossRef]
- Hird, S.M.; Ganz, H.; Eisen, J.A.; Boyce, W.M. The cloacal microbiome of five wild duck species varies by species and influenza A virus infection status. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Todd, D.; Duchatel, J.P.; Bustin, J.C.; Scullion, F.T.; Scullion, M.G.; Scott, A.N.J.; Curry, A.; Ball, N.W.; Smyth, J.A. Detection of pigeon circovirus in cloacal swabs: Implications for diagnosis, epidemiology and control. Vet. Rec. 2006, 159, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Pantin-Jackwood, M.J.; Swayne, D.E.; Suarez, D.L. An evaluation of avian influenza diagnostic methods with domestic duck specimens. Avian Dis. 2009, 53, 276–280. [Google Scholar] [CrossRef]
- Das, A.; Spackman, E.; Pantin-Jackwood, M.J.; Suarez, D.L. Removal of real-time reverse transcription polymerase chain reaction (RT-PCR) inhibitors associated with cloacal swab samples and tissues for improved diagnosis of Avian influenza virus by RT-PCR. J Vet. Diagn Investig. 2009, 21, 771–778. [Google Scholar] [CrossRef]
- Hubert, S.M.; Al-Ajeeli, M.; Bailey, C.A.; Athrey, G. The role of housing environment and dietary protein source on the gut microbiota of chicken. Animals (Basel) 2019, 9, 1085. [Google Scholar] [CrossRef]




| Descriptive Statistic | Cecal Content Reads | Cecal Content Good’s Coverage | Cloacal Swab Reads | Cloacal Swab Good’s Coverage |
|---|---|---|---|---|
| Sample Size | 19 | 19 | 19 | 19 |
| Minimum | 18,106 | 99.61% | 3005 | 98.70% |
| 1st Quartile | 20,312 | 99.70% | 6902 | 99.15% |
| Median | 21,662 | 99.72% | 11,043 | 99.49% |
| Mean | 22,656 | 99.72% | 13,900 | 99.41% |
| 3rd Quartile | 24,515 | 99.74% | 20,366 | 99.72% |
| Maximum | 29,088 | 99.86% | 31,862 | 99.86% |
| IQR | 4204 | 0.043% | 13,464 | 0.576% |
| Range | 10,982 | 0.25% | 28,857 | 1.16% |
| Comparison | KS Test | Lepage Test | Cucconi Test |
|---|---|---|---|
| Cecal Content T1 vs. Cecal Content T2 | D = 0.2569; p-value = 0.5494 | L = 1.1637; p-value = 0.5625 | C = 0.346; p-value = 0.705 |
| Cloacal Swab T1 vs. Cloacal Swab T2 | D = 0.1773; p-value = 0.8199 | L = 1.853; p-value = 0.4033 | C = 0.733; p-value = 0.523 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williams, T.; Athrey, G. Cloacal Swabs Are Unreliable Sources for Estimating Lower Gastro-Intestinal Tract Microbiota Membership and Structure in Broiler Chickens. Microorganisms 2020, 8, 718. https://doi.org/10.3390/microorganisms8050718
Williams T, Athrey G. Cloacal Swabs Are Unreliable Sources for Estimating Lower Gastro-Intestinal Tract Microbiota Membership and Structure in Broiler Chickens. Microorganisms. 2020; 8(5):718. https://doi.org/10.3390/microorganisms8050718
Chicago/Turabian StyleWilliams, Travis, and Giridhar Athrey. 2020. "Cloacal Swabs Are Unreliable Sources for Estimating Lower Gastro-Intestinal Tract Microbiota Membership and Structure in Broiler Chickens" Microorganisms 8, no. 5: 718. https://doi.org/10.3390/microorganisms8050718
APA StyleWilliams, T., & Athrey, G. (2020). Cloacal Swabs Are Unreliable Sources for Estimating Lower Gastro-Intestinal Tract Microbiota Membership and Structure in Broiler Chickens. Microorganisms, 8(5), 718. https://doi.org/10.3390/microorganisms8050718





