Epstein-Barr Virus Exploits the Secretory Pathway to Release Virions
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and EBV Lytic Cycle Induction
2.2. siRNA Treatments
2.3. Western Blotting
2.4. Immunofluorescence Staining
2.5. Evaluation of EBV Virion Secretion by Flow Cytometry
2.6. Statistical Analysis
3. Results
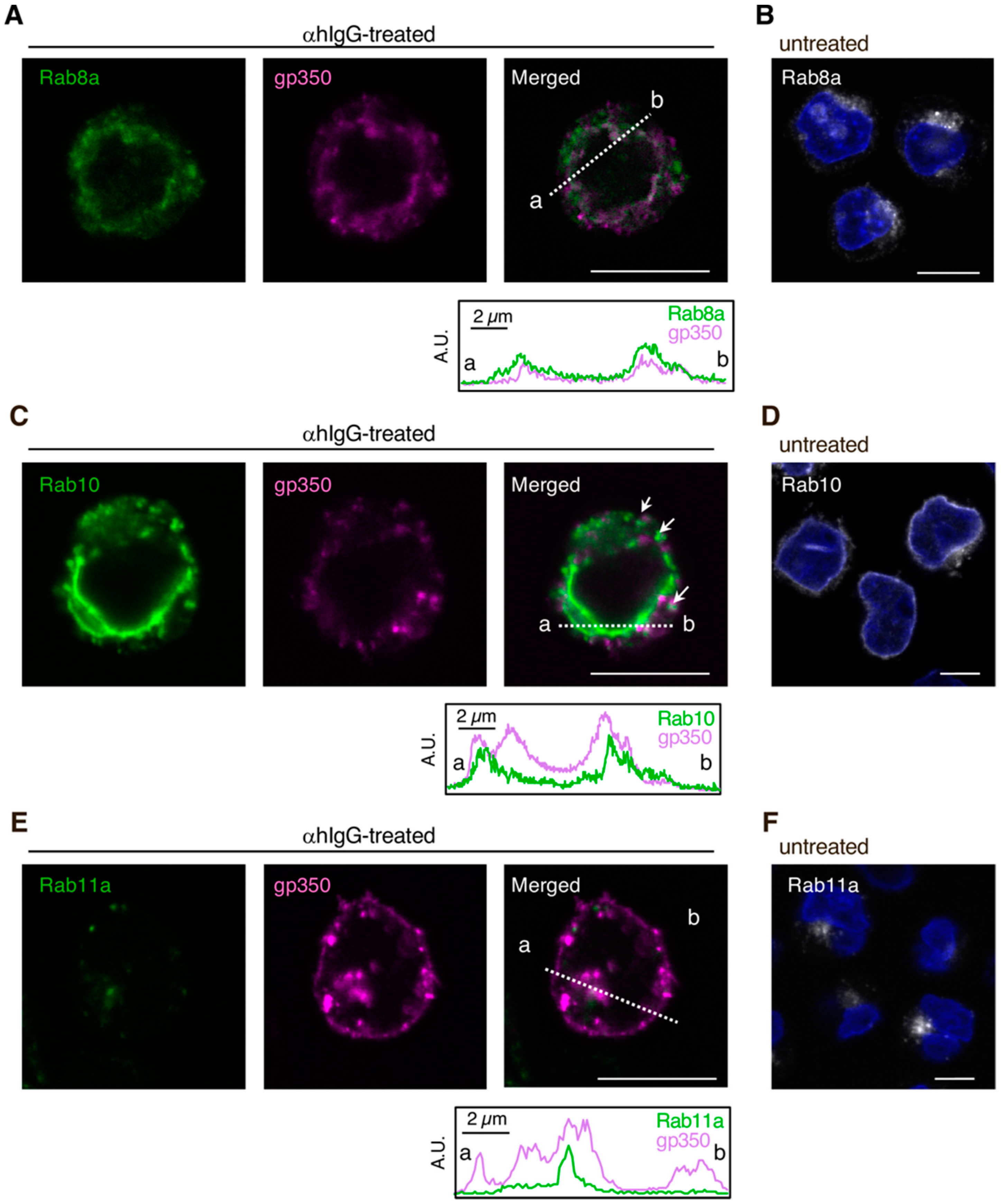
3.1. EBV Structural Proteins Partially Co-Localize with Cellular Secretory Vesicles
3.2. Downregulation of Rab Proteins Promotes the Intracellular Accumulation of EBV Structural Proteins
3.3. Downregulation of Rab Proteins Inhibits the Release of Infectious EBV Virions
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Longnecker, R.K.E.; Cohen, J. Epstein-Barr virus. In Fields Virology, 6th ed.; Knipe, M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1898–1959. [Google Scholar]
- Henaff, D.; Radtke, K.; Lippe, R. Herpesviruses exploit several host compartments for envelopment. Traffic 2012, 13, 1443–1449. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Zhou, S.; Gao, S.; Deng, H. Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell 2019, 10, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Baines, J.D. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 2011, 9, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.C.; Liao, Y.T.; Juan, Y.T.; Cheng, Y.Y.; Su, M.T.; Su, Y.Z.; Liu, H.C.; Tsai, C.H.; Lee, C.P.; Chen, M.R. The Novel Nuclear Targeting and BFRF1-Interacting Domains of BFLF2 Are Essential for Efficient Epstein-Barr Virus Virion Release. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Muller, Y.A.; Hage, S.; Alkhashrom, S.; Hollriegl, T.; Weigert, S.; Dolles, S.; Hof, K.; Walzer, S.A.; Egerer-Sieber, C.; Conrad, M.; et al. High-resolution crystal structures of two prototypical beta- and gamma-herpesviral nuclear egress complexes unravel the determinants of subfamily specificity. J. Biol. Chem. 2020, 295, 3189–3201. [Google Scholar] [CrossRef]
- Zhu, Z.; Gershon, M.D.; Hao, Y.; Ambron, R.T.; Gabel, C.A.; Gershon, A.A. Envelopment of varicella-zoster virus: Targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 1995, 69, 7951–7959. [Google Scholar] [CrossRef]
- Wisner, T.W.; Johnson, D.C. Redistribution of cellular and herpes simplex virus proteins from the trans-golgi network to cell junctions without enveloped capsids. J. Virol. 2004, 78, 11519–11535. [Google Scholar] [CrossRef]
- Sugimoto, K.; Uema, M.; Sagara, H.; Tanaka, M.; Sata, T.; Hashimoto, Y.; Kawaguchi, Y. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 2008, 82, 5198–5211. [Google Scholar] [CrossRef]
- Hogue, I.B.; Bosse, J.B.; Hu, J.R.; Thiberge, S.Y.; Enquist, L.W. Cellular mechanisms of alpha herpesvirus egress: Live cell fluorescence microscopy of pseudorabies virus exocytosis. PLoS Pathog. 2014, 10, e1004535. [Google Scholar] [CrossRef]
- Hambleton, S.; Gershon, M.D.; Gershon, A.A. The role of the trans-Golgi network in varicella zoster virus biology. Cell. Mol. Life Sci. 2004, 61, 3047–3056. [Google Scholar] [CrossRef]
- Granzow, H.; Weiland, F.; Jons, A.; Klupp, B.G.; Karger, A.; Mettenleiter, T.C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: A reassessment. J. Virol. 1997, 71, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Gershon, A.A.; Sherman, D.L.; Zhu, Z.; Gabel, C.A.; Ambron, R.T.; Gershon, M.D. Intracellular transport of newly synthesized varicella-zoster virus: Final envelopment in the trans-Golgi network. J. Virol. 1994, 68, 6372–6390. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, E.M.; Jarosinski, K.W.; Jackson, W.; Carpenter, J.E.; Grose, C. Exocytosis of Varicella-Zoster Virus Virions Involves a Convergence of Endosomal and Autophagy Pathways. J. Virol. 2016, 90, 8673–8685. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.A.; Dasgupta, A.; Wilson, D.W. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: Role for organelle acidification in assembly of infectious particles. J. Virol. 2001, 75, 1236–1251. [Google Scholar] [CrossRef] [PubMed]
- Tooze, J.; Hollinshead, M.; Reis, B.; Radsak, K.; Kern, H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 1993, 60, 163–178. [Google Scholar]
- Sanchez, V.; Greis, K.D.; Sztul, E.; Britt, W.J. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: Characterization of a potential site of virus assembly. J. Virol. 2000, 74, 975–986. [Google Scholar] [CrossRef]
- Mori, Y.; Koike, M.; Moriishi, E.; Kawabata, A.; Tang, H.; Oyaizu, H.; Uchiyama, Y.; Yamanishi, K. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 2008, 9, 1728–1742. [Google Scholar] [CrossRef]
- Homman-Loudiyi, M.; Hultenby, K.; Britt, W.; Soderberg-Naucler, C. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-golgi network 46, and mannosidase II. J. Virol. 2003, 77, 3191–3203. [Google Scholar] [CrossRef]
- Fraile-Ramos, A.; Pelchen-Matthews, A.; Kledal, T.N.; Browne, H.; Schwartz, T.W.; Marsh, M. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 2002, 3, 218–232. [Google Scholar] [CrossRef]
- Das, S.; Vasanji, A.; Pellett, P.E. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 2007, 81, 11861–11869. [Google Scholar] [CrossRef]
- Seigneurin, J.M.; Vuillaume, M.; Lenoir, G.; De-The, G. Replication of Epstein-Barr virus: Ultrastructural and immunofluorescent studies of P3HR1-superinfected Raji cells. J. Virol. 1977, 24, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Longnecker, R. The Epstein-Barr virus glycoprotein 110 carboxy-terminal tail domain is essential for lytic virus replication. J. Virol. 1997, 71, 4092–4097. [Google Scholar] [CrossRef] [PubMed]
- Lake, C.M.; Hutt-Fletcher, L.M. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 2000, 74, 11162–11172. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, J.S.; Rabanus, J.P.; Petersen, V.; Greenspan, D. Fine structure of EBV-infected keratinocytes in oral hairy leukoplakia. J. Oral. Pathol. Med. 1989, 18, 565–572. [Google Scholar] [CrossRef]
- Peng, L.; Ryazantsev, S.; Sun, R.; Zhou, Z.H. Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure 2010, 18, 47–58. [Google Scholar] [CrossRef]
- Yanagi, Y.; Masud, H.; Watanabe, T.; Sato, Y.; Goshima, F.; Kimura, H.; Murata, T. Initial Characterization of the Epstein(-)Barr Virus BSRF1 Gene Product. Viruses 2019, 11, 285. [Google Scholar] [CrossRef]
- Orenstein, J.M.; Alkan, S.; Blauvelt, A.; Jeang, K.T.; Weinstein, M.D.; Ganem, D.; Herndier, B. Visualization of human herpesvirus type 8 in Kaposi’s sarcoma by light and transmission electron microscopy. AIDS 1997, 11, F35–F45. [Google Scholar] [CrossRef]
- Nanbo, A.; Noda, T.; Ohba, Y. Epstein-Barr Virus Acquires Its Final Envelope on Intracellular Compartments With Golgi Markers. Front. Microbiol. 2018, 9, 454. [Google Scholar] [CrossRef]
- Roberts, K.L.; Baines, J.D. Myosin Va enhances secretion of herpes simplex virus 1 virions and cell surface expression of viral glycoproteins. J. Virol. 2010, 84, 9889–9896. [Google Scholar] [CrossRef]
- Takada, K.; Horinouchi, K.; Ono, Y.; Aya, T.; Osato, T.; Takahashi, M.; Hayasaka, S. An Epstein-Barr virus-producer line Akata: Establishment of the cell line and analysis of viral DNA. Virus Genes 1991, 5, 147–156. [Google Scholar] [CrossRef]
- Shimizu, N.; Tanabe-Tochikura, A.; Kuroiwa, Y.; Takada, K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: Malignant phenotypes of BL cells are dependent on EBV. J. Virol. 1994, 68, 6069–6073. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Terada, H.; Kachi, K.; Takada, K.; Matsuda, T. Roles of cell signaling pathways in cell-to-cell contact-mediated Epstein-Barr virus transmission. J. Virol. 2012, 86, 9285–9296. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Kawanishi, E.; Yoshida, R.; Yoshiyama, H. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J. Virol. 2013, 87, 10334–10347. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Kachi, K.; Yoshiyama, H.; Ohba, Y. Epstein-Barr virus exploits host endocytic machinery for cell-to-cell viral transmission rather than a virological synapse. J. Gen. Virol. 2016, 97, 2989–3006. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Inoue, K.; Adachi-Takasawa, K.; Takada, K. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 2002, 21, 954–965. [Google Scholar] [CrossRef]
- Delecluse, H.J.; Hilsendegen, T.; Pich, D.; Zeidler, R.; Hammerschmidt, W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 1998, 95, 8245–8250. [Google Scholar] [CrossRef]
- Chiu, Y.F.; Sugden, B.; Chang, P.J.; Chen, L.W.; Lin, Y.J.; Lan, Y.C.; Lai, C.H.; Liou, J.Y.; Liu, S.T.; Hung, C.H. Characterization and intracellular trafficking of Epstein-Barr virus BBLF1, a protein involved in virion maturation. J. Virol. 2012, 86, 9647–9655. [Google Scholar] [CrossRef]
- Klein, E.; Klein, G.; Nadkarni, J.S.; Nadkarni, J.J.; Wigzell, H.; Clifford, P. Surface IgM-kappa specificity on a Burkitt lymphoma cell in vivo and in derived culture lines. Cancer Res. 1968, 28, 1300–1310. [Google Scholar]
- Takada, K.; Ono, Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 1989, 63, 445–449. [Google Scholar] [CrossRef]
- Takada, K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt lymphoma lines. Int. J. Cancer 1984, 33, 27–32. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Geilinger, K. Monoclonal antibodies against the major glycoprotein (gp350/220) of Epstein-Barr virus neutralize infectivity. Proc. Natl. Acad. Sci. USA 1980, 77, 5307–5311. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Sirkis, D.W.; Schekman, R. Protein sorting at the trans-Golgi network. Annu. Rev. Cell Dev. Biol. 2014, 30, 169–206. [Google Scholar] [CrossRef]
- Maruo, S.; Yang, L.; Takada, K. Roles of Epstein-Barr virus glycoproteins gp350 and gp25 in the infection of human epithelial cells. J. Gen. Virol. 2001, 82, 2373–2383. [Google Scholar] [CrossRef]
- Nemerow, G.R.; Mold, C.; Schwend, V.K.; Tollefson, V.; Cooper, N.R. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: Sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 1987, 61, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Stenmark, H. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 2015, 128, 3171–3176. [Google Scholar] [CrossRef] [PubMed]
- Wandinger-Ness, A.; Zerial, M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef]
- Takahashi, S.; Kubo, K.; Waguri, S.; Yabashi, A.; Shin, H.W.; Katoh, Y.; Nakayama, K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 2012, 125, 4049–4057. [Google Scholar] [CrossRef]
- Wilcke, M.; Johannes, L.; Galli, T.; Mayau, V.; Goud, B.; Salamero, J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network. J. Cell Biol. 2000, 151, 1207–1220. [Google Scholar] [CrossRef]
- Welz, T.; Wellbourne-Wood, J.; Kerkhoff, E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 2014, 24, 407–415. [Google Scholar] [CrossRef]
- Henson, B.W.; Perkins, E.M.; Cothran, J.E.; Desai, P. Self-assembly of Epstein-Barr virus capsids. J. Virol. 2009, 83, 3877–3890. [Google Scholar] [CrossRef][Green Version]
- van Grunsven, W.M.; Spaan, W.J.; Middeldorp, J.M. Localization and diagnostic application of immunodominant domains of the BFRF3-encoded Epstein-Barr virus capsid protein. J. Infect. Dis. 1994, 170, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Behnia, R.; Munro, S. Organelle identity and the signposts for membrane traffic. Nature 2005, 438, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Nishikawa, J.; Takada, K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 1998, 72, 4371–4378. [Google Scholar] [CrossRef] [PubMed]
- Agosto, L.M.; Uchil, P.D.; Mothes, W. HIV cell-to-cell transmission: Effects on pathogenesis and antiretroviral therapy. Trends Microbiol. 2015, 23, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Jolly, C.; Kashefi, K.; Hollinshead, M.; Sattentau, Q.J. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 2004, 199, 283–293. [Google Scholar] [CrossRef]
- Jolly, C.; Welsch, S.; Michor, S.; Sattentau, Q.J. The regulated secretory pathway in CD4(+) T cells contributes to human immunodeficiency virus type-1 cell-to-cell spread at the virological synapse. PLoS Pathog. 2011, 7, e1002226. [Google Scholar] [CrossRef]




© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanbo, A. Epstein-Barr Virus Exploits the Secretory Pathway to Release Virions. Microorganisms 2020, 8, 729. https://doi.org/10.3390/microorganisms8050729
Nanbo A. Epstein-Barr Virus Exploits the Secretory Pathway to Release Virions. Microorganisms. 2020; 8(5):729. https://doi.org/10.3390/microorganisms8050729
Chicago/Turabian StyleNanbo, Asuka. 2020. "Epstein-Barr Virus Exploits the Secretory Pathway to Release Virions" Microorganisms 8, no. 5: 729. https://doi.org/10.3390/microorganisms8050729
APA StyleNanbo, A. (2020). Epstein-Barr Virus Exploits the Secretory Pathway to Release Virions. Microorganisms, 8(5), 729. https://doi.org/10.3390/microorganisms8050729




