The Role of Gene Elongation in the Evolution of Histidine Biosynthetic Genes
Abstract
:1. Introduction
1.1. Gene Duplication and Gene Fusion Events
1.2. The Gene Elongation Mechanism
1.3. Histidine Biosynthesis and Its Evolution
2. Materials and Methods
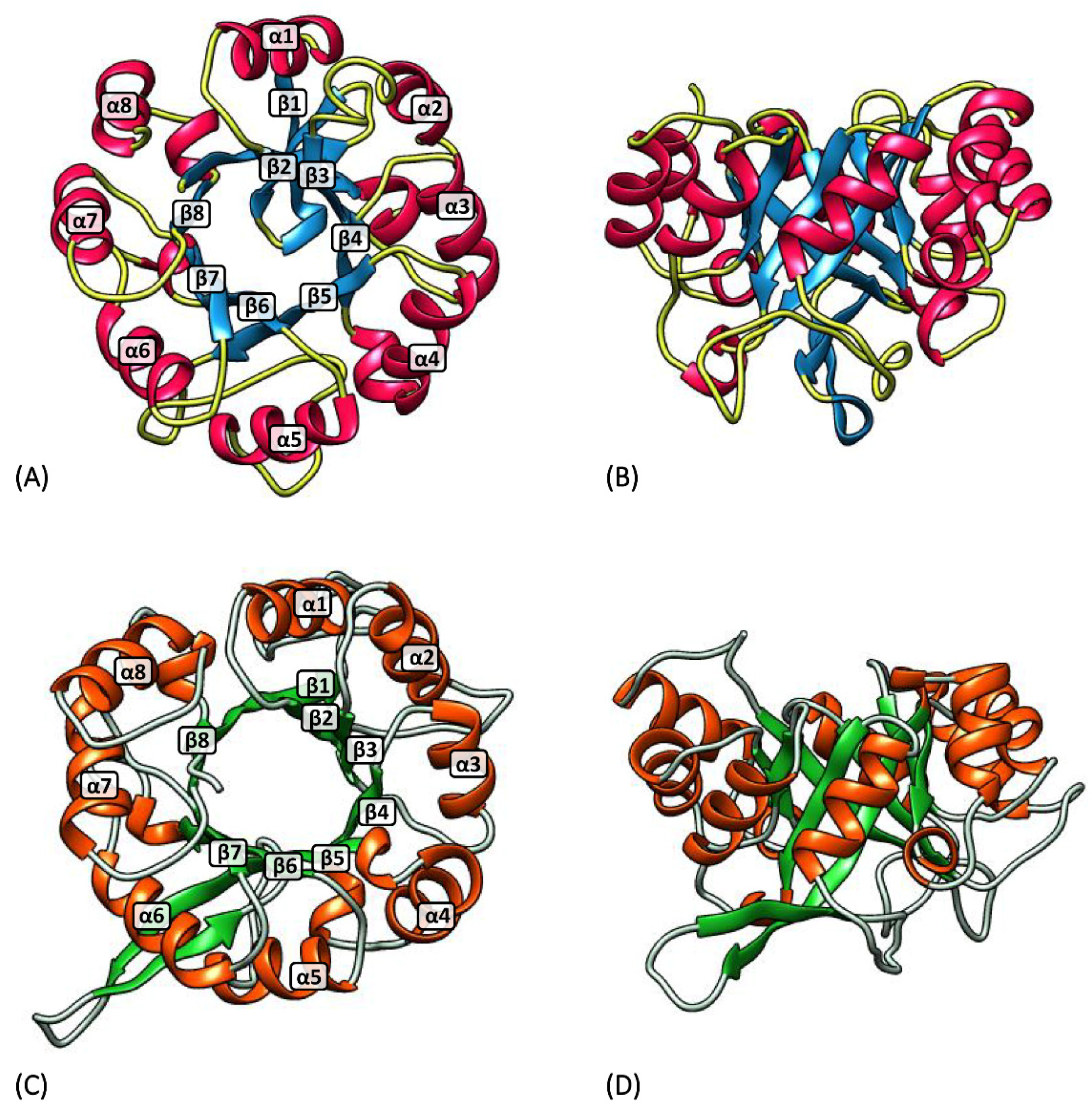
2.1. Three-Dimensional Structure Prediction
2.2. PDB Search for Protein Structures
- -
- 1QO2, 2CFF, 2W79—Thermotoga maritima;
- -
- 1VZW, 2VEP, 2X30, 5DN1—Streptomyces coelicolor;
- -
- 2AGK—Saccharomyces cerevisiae;
- -
- 2Y85, 2Y88, 2Y89, 3ZS4—Mycobacterium tuberculosis;
- -
- 4AXK—Corynebacterium efficiens;
- -
- 4GJ1—Campylobacter jejuni;
- -
- 4TX9, 4U28—Streptomyces sviceus;
- -
- 4W9T, 4X9S—Streptomyces sp.;
- -
- 4WD0—Paenarthrobacter aurescens;
- -
- 4X2R—Actinomyces urogenitalis;
- -
- 5AB3, 5ABT, 5G4W, 5G5I, 5G1T, 5G1Y, 5G4E, 5G2H, 5G2I, 5G2W, 5AC6, 5AC7, 5AC8, 5AHF, 5A5W, 5AHE, 5AHI, 5L9F—Salmonella enterica;
- -
- 5L6U—Salmonella heidelberg.
- -
- 1GPW, 1THF, 1VH7, 2A0N, 2WJZ, 3ZR4, 4EWN, 4FX7, 5TQL, 6VDG—Thermotoga maritima;
- -
- 1H5Y—Pyrobaculum aerophilum;
- -
- 1JVN, 1OX4, 1OX5, 1OX6—Saccharomyces cerevisiae;
- -
- 1KA9—Thermus thermophilus.
- -
- 1RHY—Filobasidiella (Cryptococcus) neoformans;
- -
- 2AE8—Staphylococcus aureus;
- -
- 2F1D, 4MU0, 4MU1, 4MU3, 4MU4, 4QNJ, 4QNK, 5ELW, 5EKW, 5EL9, 6EZJ—Arabidopsis thaliana;
- -
- 4GQU, 4LOM, 4LPF, 5XDS, 5ZQN—Mycobacterium tuberculosis;
- -
- 5DNL, 5DNX—Pyrococcus furiosus;
- -
- 6EZM—Saccharomyces cerevisiae;
- -
- 6FWH—Acanthamoeba castellanii.
- -
- 1K75, 1KAE, 1KAH, 1KAR—Escherichia coli;
- -
- 4G07, 4G09—Brucella suis;
- -
- 4GIC—Methylococcus capsulatus;
- -
- 5VLB, 5VLC, 5VLD—Medicago truncatula;
- -
- 6AN0—Elizabethkingia anophelis.
2.3. Amino Acid Sequence Alignment
3. Results and Discussion
3.1. hisA and hisF Gene Elongation
3.2. hisB
3.3. hisD
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- McLachlan, A.D. Gene duplication and the origin of repetitive protein structures. Cold Spring Harb. Symp. Quant. Biol. 1987, 52, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Brilli, M.; Fani, R. The origin and evolution of eucaryal HIS7 genes: From metabolon to bifunctional proteins? Gene 2004, 339, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Fani, R.; Fondi, M. Origin and evolution of metabolic pathways. Phys. Life Rev. 2009, 6, 23–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. B 1994, 256, 119–124. [Google Scholar]
- Ohno, S. Evolution by Gene Duplication; Springer Science & Business Media: New York, NY, USA, 1970. [Google Scholar]
- Yanai, I.; Wolf, Y.I.; Koonin, E.V. Evolution of gene fusions: Horizontal transfer versus independent events. Genome Biol. 2002, 3, research0024-1. [Google Scholar]
- Adman, E.T.; Sieker, L.C.; Jensen, L.H. Structure of a bacterial ferredoxin. J. Biol. Chem. 1973, 248, 3987–3996. [Google Scholar]
- Tang, J.; James, M.N.G.; Hsu, I.N.; Jenkins, J.A.; Blundell, T.L. Structural evidence for gene duplication in the evolution of the acid proteases. Nature 1978, 271, 618–621. [Google Scholar] [CrossRef]
- Nyunoya, H.; Lusty, C.J. The carB gene of Escherichia coli: A duplicated gene coding for the large subunit of carbamoyl-phosphate synthetase. PNAS 1983, 80, 4629–4633. [Google Scholar] [CrossRef] [Green Version]
- Rubin, R.A.; Levy, S.B.; Heinrikson, R.L.; Kézdy, F.J. Gene duplication in the evolution of the two complementing domains of Gram-negative bacterial tetracycline efflux proteins. Gene 1990, 87, 7–13. [Google Scholar] [CrossRef]
- Gupta, R.S.; Singh, B. Cloning of the HSP70 gene from Halobacterium marismortui: Relatedness of archaebacterial HSP70 to its eubacterial homologs and a model for the evolution of the HSP70 gene. J. Bacteriol. 1992, 174, 4594–4605. [Google Scholar] [CrossRef] [Green Version]
- Reizer, J.; Saier, M.H. Modular multidomain phosphoryl transfer proteins of bacteria. Curr. Opin. Struc. Biol. 1997, 7, 407–415. [Google Scholar] [CrossRef]
- Fani, R.; Liò, P.; Chiarelli, I.; Bazzicalupo, M. The evolution of the histidine biosynthetic genes in prokaryotes: A common ancestor for the hisA and hisF genes. J. Mol. Evol. 1994, 38, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Fani, R.; Brilli, M.; Fondi, M.; Lió, P. The role of gene fusions in the evolution of metabolic pathways: The histidine biosynthesis case. BMC Evol. Biol. 2007, 7, S4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulis-Horn, R.K.; Persicke, M.; Kalinowski, J. Histidine biosynthesis, its regulation and biotechnological application in Corynebacterium glutamicum. Microb. Biotechnol. 2014, 7, 5–25. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Fani, R.; Liò, P.; Lazcano, A.; Bazzicalupo, M.; Carlomagno, M.S.; Bruni, C.B. Histidine biosynthetic pathway and genes: Structure, regulation, and evolution. Microbiol. Rev. 1996, 60, 44–69. [Google Scholar] [CrossRef]
- Barbosa, J.A.; Sivaraman, J.; Li, Y.; Larocque, R.; Matte, A.; Schrag, J.D.; Cygler, M. Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase. PNAS 2002, 99, 1859–1864. [Google Scholar] [CrossRef] [Green Version]
- Winkler, M.E.; Ramos-Montañez, S.M.I.R.L.A. Biosynthesis of histidine. EcoSal Plus 2009, 3. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Lazcano, A.; Oró, J. The enhancement activities of histidyl-histidine in some prebiotic reactions. J. Mol. Evol. 1990, 31, 445–452. [Google Scholar] [CrossRef]
- Shen, C.; Mills, T.; Oró, J. Prebiotic synthesis of histidyl-histidine. J. Mol. Evol. 1990, 31, 175–179. [Google Scholar] [CrossRef]
- White, H.B. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 1976, 7, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Fondi, M.; Emiliani, G.; Liò, P.; Gribaldo, S.; Fani, R. The evolution of histidine biosynthesis in archaea: Insights into the his genes structure and organization in LUCA. J. Mol. Evol. 2009, 69, 512. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, N.H. The Evolution of Biochemical Syntheses—Retrospect and Prospect. In Evolving Genes and Proteins; Academic Press: Cambridge, MA, USA, 1965; pp. 15–23. [Google Scholar]
- Fani, R.; Liò, P.; Lazcano, A. Molecular evolution of the histidine biosynthetic pathway. J. Mol. Evol. 1995, 41, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A User-friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTALW: Improving the sensitivity of progressive multiple sequence through weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Lang, D.; Thoma, R.; Henn-Sax, M.; Sterner, R.; Wilmanns, M. Structural evidence for evolution of the β/α barrel scaffold by gene duplication and fusion. Science 2000, 289, 1546–1550. [Google Scholar] [CrossRef]
- Jürgens, C.; Strom, A.; Wegener, D.; Hettwer, S.; Wilmanns, M.; Sterner, R. Directed evolution of a (βα)8-barrel enzyme to catalyze related reactions in two different metabolic pathways. PNAS 2000, 97, 9925–9930. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Salazar, A.; Becerra, A.; Lazcano, A. Evolutionary convergence in the biosyntheses of the imidazole moieties of histidine and purines. PLoS ONE 2018, 13, e0196349. [Google Scholar] [CrossRef] [Green Version]
- Gerlt, J.A.; Babbitt, P.C. Barrels in pieces? Nat. Struct. Biol. 2001, 8, 5–7. [Google Scholar] [CrossRef]
- Horowitz, N.H. On the evolution of biochemical syntheses. PNAS 1945, 31, 153. [Google Scholar] [CrossRef] [Green Version]
- Thoma, R.; Obmolova, G.; Lang, D.A.; Schwander, M.; Jenö, P.; Sterner, R.; Wilmanns, M. Efficient expression, purification and crystallisation of two hyperthermostable enzymes of histidine biosynthesis. FEBS Lett. 1999, 454, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gerlt, J.A. New wine from old barrels. Nat. Struct. Biol. 2000, 7, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Reardon, D.; Farber, G.K. The structure and evolution of α/β barrel proteins. FASEB J. 1995, 9, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Petsko, G.A. Design by necessity. Nature 2000, 403, 606–607. [Google Scholar] [CrossRef] [PubMed]
- Yčas, M. On earlier states of the biochemical system. J. Theor. Biol. 1974, 44, 145–160. [Google Scholar] [CrossRef]
- Jensen, R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976, 30, 409–425. [Google Scholar] [CrossRef] [Green Version]
- Barona-Gómez, F.; Hodgson, D.A. Occurrence of a putative ancient-like isomerase involved in histidine and tryptophan biosynthesis. EMBO Rep. 2003, 4, 296–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazcano, A.; Miller, S.L. How long did it take for life to begin and evolve to cyanobacteria? J. Mol. Evol. 1994, 39, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. The universal ancestor. Proc. Natl. Acad. Sci. USA 1998, 95, 6854–6859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.H. Evolution by Gene Duplication and Domain Shuffling. In Molecular Evolution; Sinauer Associates Inc.: Sunderland, MA, USA, 1997. [Google Scholar]
- Bisson, C.; Britton, K.L.; Sedelnikova, S.E.; Rodgers, H.F.; Eadsforth, T.C.; Viner, R.C.; Hawkes, T.R.; Baker, P.J.; Rice, D.W. Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination. Structure 2015, 23, 1236–1245. [Google Scholar] [CrossRef] [Green Version]
- Parker, A.R.; Moore, T.D.; Edman, J.C.; Schwab, J.M.; Davisson, V.J. Cloning, sequence analysis and expression of the gene encoding imidazole glycerol phosphate dehydratase in Cryptococcus neoformans. Gene 1994, 145, 135–138. [Google Scholar] [CrossRef]
- Hawkes, T.R.; Thomas, P.G.; Edwards, L.S.; Rayner, S.J.; Wilkinson, K.W.; Rice, D.W. Purification and characterization of the imidazoleglycerol-phosphate dehydratase of Saccharomyces cerevisiae from recombinant Escherichia coli. Biochem. J. 1995, 306, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Mano, J.; Hatano, M.; Koizumi, S.; Tada, S.; Hashimoto, M.; Scheidegger, A. Purification and properties of a monofunctional imidazoleglycerol-phosphate dehydratase from wheat. Plant Physiol. 1993, 103, 733–739. [Google Scholar] [CrossRef] [Green Version]
- Tada, S.; Volrath, S.; Guyer, D.; Scheidegger, A.; Ryals, J.; Ohta, D.; Ward, E. Isolation and characterization of cDNAs encoding imidazoleglycerolphosphate dehydratase from Arabidopsis thaliana. Plant Physiol. 1994, 105, 579–583. [Google Scholar] [CrossRef]
- Sinha, S.C.; Chaudhuri, B.N.; Burgner, J.W.; Yakovleva, G.; Davisson, V.J.; Smith, J.L. Crystal structure of imidazole glycerol-phosphate dehydratase: Duplication of an unusual fold. J. Biol. Chem. 2004, 279, 15491–15498. [Google Scholar] [CrossRef] [Green Version]
- Agapova, Y.K.; Timofeev, V.I.; Komolov, A.S. Molecular Dynamics Study of Triazole Derivative Binding to the Active Site of Imidazole Glycerol Phosphate Dehydratase from Mycobacterium tuberculosis. Crystallogr. Rep. 2019, 64, 608–610. [Google Scholar] [CrossRef]
- Glynn, S.E.; Baker, P.J.; Sedelnikova, S.E.; Davies, C.L.; Eadsforth, T.C.; Levy, C.W.; Rodgers, H.F.; Blackburn, G.M.; Hawkes, T.R.; Viner, R.; et al. Structure and mechanism of imidazoleglycerol-phosphate dehydratase. Structure 2005, 13, 1809–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, A.; Ward, E.; Beck, J.; Tada, S.; Chang, J.Y.; Scheidegger, A.; Ryals, J. Structural and functional conservation of histidinol dehydrogenase between plants and microbes. PNAS 1991, 88, 4133–4137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grubmeyer, C.; Skiadopoulos, M.; Senior, A.E. L-histidinol dehydrogenase, a Zn2+-metalloenzyme. Arch. Biochem. Biophys. 1989, 272, 311–317. [Google Scholar] [CrossRef]
- D’ambrosio, K.; Lopez, M.; Dathan, N.A.; Ouahrani-Bettache, S.; Köhler, S.; Ascione, G.; Monti, S.M.; Winum, J.Y.; De Simone, G. Structural basis for the rational design of new anti-Brucella agents: The crystal structure of the C366S mutant of L-histidinol dehydrogenase from Brucella suis. Biochimie 2014, 97, 114–120. [Google Scholar] [CrossRef]
- Ruszkowski, M.; Dauter, Z. Structures of Medicago truncatula L-histidinol dehydrogenase show rearrangements required for NAD+ binding and the cofactor positioned to accept a hydride. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]










© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Duca, S.; Chioccioli, S.; Vassallo, A.; Castronovo, L.M.; Fani, R. The Role of Gene Elongation in the Evolution of Histidine Biosynthetic Genes. Microorganisms 2020, 8, 732. https://doi.org/10.3390/microorganisms8050732
Del Duca S, Chioccioli S, Vassallo A, Castronovo LM, Fani R. The Role of Gene Elongation in the Evolution of Histidine Biosynthetic Genes. Microorganisms. 2020; 8(5):732. https://doi.org/10.3390/microorganisms8050732
Chicago/Turabian StyleDel Duca, Sara, Sofia Chioccioli, Alberto Vassallo, Lara Mitia Castronovo, and Renato Fani. 2020. "The Role of Gene Elongation in the Evolution of Histidine Biosynthetic Genes" Microorganisms 8, no. 5: 732. https://doi.org/10.3390/microorganisms8050732
APA StyleDel Duca, S., Chioccioli, S., Vassallo, A., Castronovo, L. M., & Fani, R. (2020). The Role of Gene Elongation in the Evolution of Histidine Biosynthetic Genes. Microorganisms, 8(5), 732. https://doi.org/10.3390/microorganisms8050732





