Known Antimicrobials Versus Nortriptyline in Candida albicans: Repositioning an Old Drug for New Targets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Testing Drug Working Concentrations
2.3. Identification of Mutants Sensitive or Tolerant to Drugs during Biofilm Formation
2.4. Identification of Mutants Sensitive or Tolerant to Drugs after Biofilm Growth
2.5. Glutathione and Nitric Oxide Assays
2.6. Measurements of Reactive Oxygen Species (ROS) and Cells Viability by Flow Cytometry
2.7. Isolation of RNA and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.8. Ames Test
2.9. Statistical and Bioinformatics Analysis
3. Results
3.1. Determination of GPs: Identification of Mutants Sensitive to NOR
3.2. Identification of Mutants Tolerant to NOR
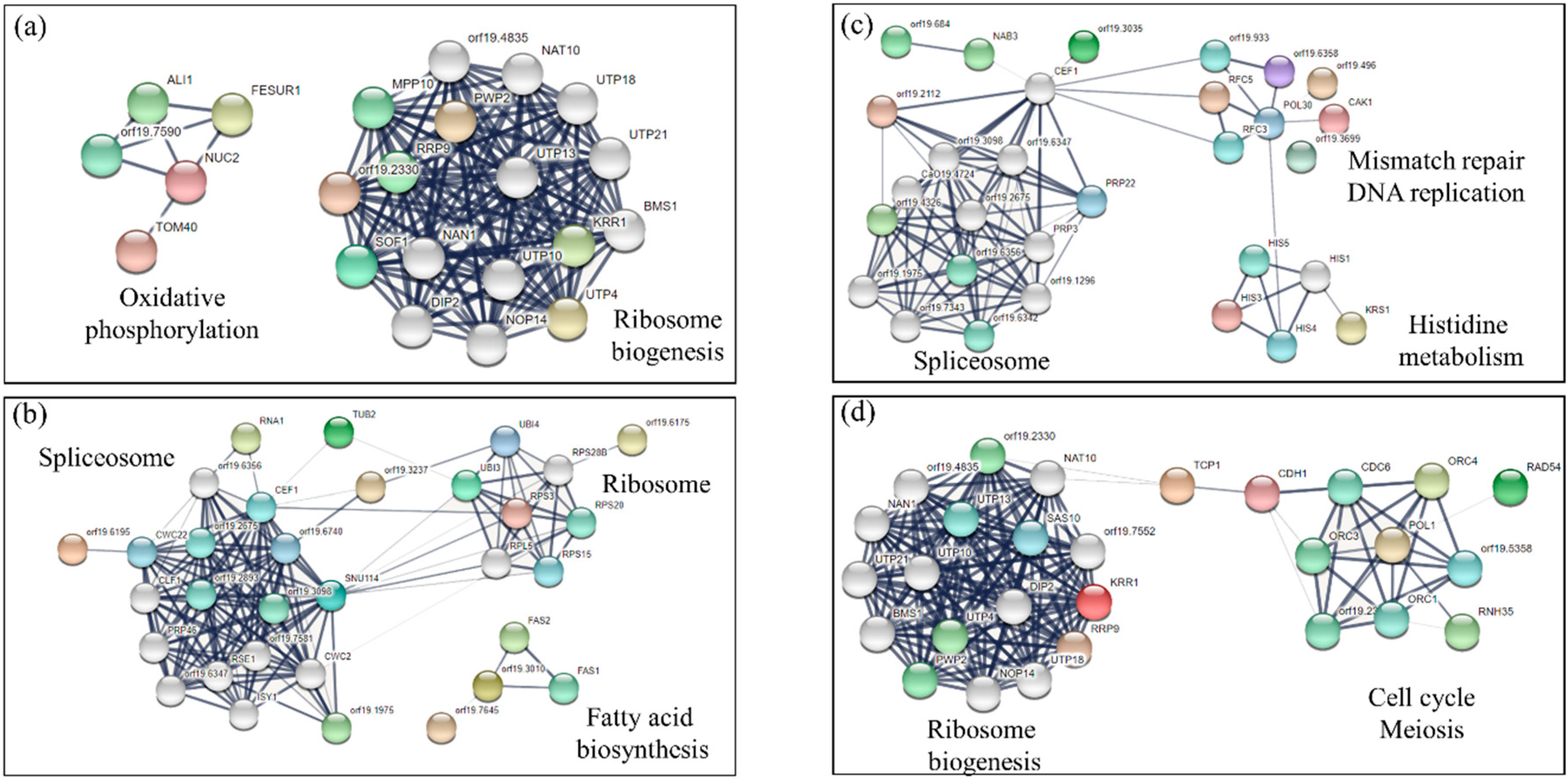
3.3. Analysis of Sensitive and Tolerant Mutants to More Than One Drug
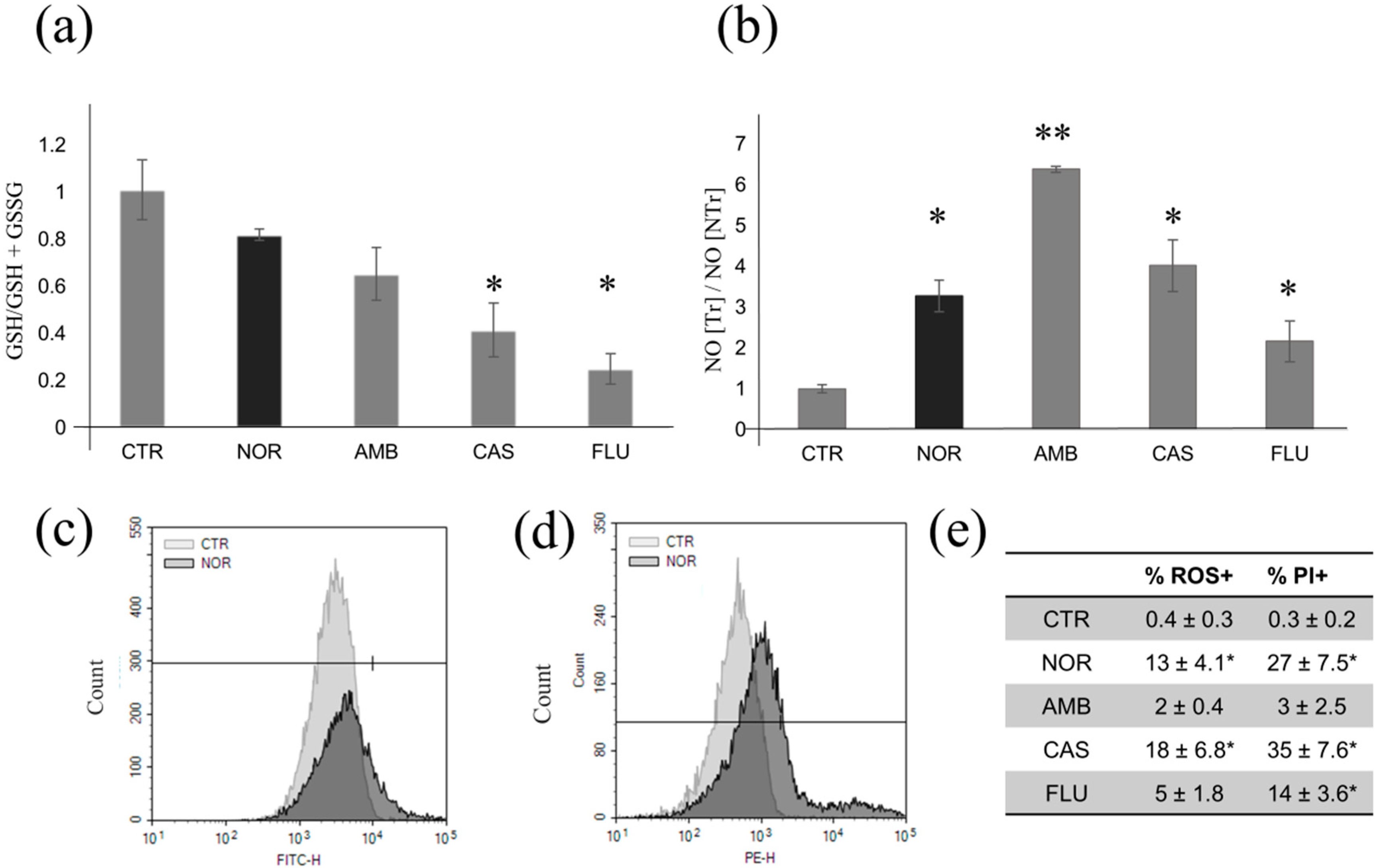
3.4. Determination of Oxidative and Nitrosative Stress Induced by Different Drugs
3.5. DNA Damage
3.6. Expression Analysis of Transporter Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wisplinghoff, H.; Ebbers, J.; Geurtz, L.; Stefanik, D.; Major, Y.; Edmond, M.B.; Wenzel, R.P.; Seifert, H. Nosocomial bloodstream infections due to Candida spp. in the USA: Species distribution, clinical features and antifungal susceptibilities. Int. J. Antimicrob. Agents 2014, 43, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-S.; Sun, W.; Xu, M.; Shen, M.; Khraiwesh, M.; Sciotti, R.J.; Zheng, W. Repurposing Screen Identifies Unconventional Drugs with Activity Against Multidrug Resistant Acinetobacter Baumannii. Front. Cell. Infect. Microbiol. 2019, 8. [Google Scholar] [CrossRef]
- Koromina, M.; Pandi, M.T.; Patrinos, G.P. Rethinking Drug Repositioning and Development with Artificial Intelligence, Machine Learning, and Omics. Omics J. Integr. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Mosolygó, T.; Kincses, A.; Csonka, A.; Tönki, Á.S.; Witek, K.; Sanmartín, C.; Marc, M.A.; Handzlik, J.; Kiec-Kononowicz, K.; Dominguez-Alvarez, E.; et al. Selenocompounds as Novel Antibacterial Agents and Bacterial Efflux Pump Inhibitors. Molecules 2019, 24, 1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, S.; Melander, R.J.; Brackett, C.; Scott, A.J.; Chandler, C.E.; Nguyen, C.; Minrovic, B.M.; Harrill, S.E.; Ernst, R.K.; Manoil, C.; et al. Small Molecule Potentiation of Gram-positive Selective Antibiotics Against Acinetobacter baumannii. ACS Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Oliveira, I.M.; Borges, A.; Borges, F.; Simões, M. Repurposing ibuprofen to control Staphylococcus aureus biofilms. Eur. J. Med. Chem. 2019, 197–205. [Google Scholar] [CrossRef]
- Marzo, T.; Cirri, D.; Pollini, S.; Prato, M.; Fallani, S.; Cassetta, M.I.; Novelli, A.; Rossolini, G.M.; Messori, L. Auranofin and its Analogues Show Potent Antimicrobial Activity against Multidrug-Resistant Pathogens: Structure–Activity Relationships. ChemMedChem 2018, 13, 2448–2454. [Google Scholar] [CrossRef]
- Chen, J.; Korostyshevsky, D.; Lee, S.; Perlstein, E.O. Accumulation of an antidepressant in vesiculogenic membranes of yeast cells triggers autophagy. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Méndez-Galomo, K.S.; González, G.M.; Montoya, A.M.; Becerril-García, M.A.; Solís-Villegas, E.M.; Villanueva-Lozano, H.; Robledo-Leal, E.R.; Trevino-Rangel, R.d.J. In vivo evaluation of the antifungal activity of sertraline against Aspergillus fumigatus. J. Antimicrob. Chemother. 2018. [Google Scholar] [CrossRef] [Green Version]
- Zhai, B.; Wu, C.; Wang, L.; Sachs, S.; Lin, X. The antidepressant sertraline provides a promising therapeutic option for neurotropic cryptococcal infections. Antimicrob. Agents Chemother. 2012, 56, 3758–3766. [Google Scholar] [CrossRef] [Green Version]
- Caldara, M.; Marmiroli, N. Tricyclic antidepressants inhibit Candida albicans growth and biofilm formation. Int. J. Antimicrob. Agents 2018, 52, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Uesono, Y.; Toh-e, A.; Kikuchi, Y.; Terashima, I. Structural analysis of compounds with actions similar to local anesthetics and antipsychotic phenothiazines in yeast. Yeast 2011, 28, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Uesono, Y.; Araki, T.; Toh-E, A. Local Anesthetics, Antipsychotic Phenothiazines, and Cationic Surfactants Shut Down Intracellular Reactions through Membrane Perturbation in Yeast. Biosci. Biotechnol. Biochem. 2008, 72, 2884–2894. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Nislow, C. The yeast deletion collection: A decade of functional genomics. Genetics 2014, 197, 451–465. [Google Scholar] [CrossRef] [Green Version]
- Giaever, G.; Shoemaker, D.D.; Jones, T.W.; Liang, H.; Winzeler, E.A.; Astromoff, A.; Davis, R.W. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 1999, 21, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Hillenmeyer, M.E.; Ericson, E.; Davis, R.W.; Nislow, C.; Koller, D.; Giaever, G. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoepfner, D.; Helliwell, S.B.; Sadlish, H.; Schuierer, S.; Filipuzzi, I.; Brachat, S.; Bhullar, B.; Plikat, U.; Abraharm, Y.; Altorfer, M.; et al. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol. Res. 2014, 169, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Roemer, T.; Jiang, B.; Davison, J.; Ketela, T.; Veillette, K.; Breton, A.; Tandia, F.; Linteau, A.; Sillaots, S.; Marta, C.; et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003, 50, 167–181. [Google Scholar] [CrossRef]
- Merlob, P.; Schaefer, C. Psychotropic drugs. Drugs Dur. Pregnancy Lact. Treat. Options Risk Assess. Third Ed. 2015. [Google Scholar] [CrossRef]
- Pagano, L.; Caldara, M.; Villani, M.; Zappettini, A.; Marmiroli, N.; Marmiroli, M. In Vivo-In Vitro Comparative Toxicology of Cadmium Sulphide Quantum Dots in the Model Organism Saccharomyces cerevisiae. Nanomaterials 2019, 9, 512. [Google Scholar] [CrossRef] [Green Version]
- de Mello Silva Oliveira, N.; Reis Resende, M.; Alexandre Morales, D.; de ragão Umbuzeiro, G.; Boriollo, M.F.G. In vitro mutagenicity assay (Ames test) and phytochemical characterization of seeds oil of Helianthus annuus Linné (sunflower). Toxicol. Rep. 2016, 3, 733–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, R.A.; Bates, S.; Lenardon, M.D.; MacCallum, D.M.; Wagener, J.; Lowman, D.W.; Kruppa, M.D.; Williams, D.L.; Odds, F.C.; Brown, A.J.P.; et al. The Mnn2 Mannosyltransferase Family Modulates Mannoprotein Fibril Length, Immune Recognition and Virulence of Candida albicans. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Kusch, H.; Engelmann, S.; Bode, R.; Albrecht, D.; Morschhäuser, J.; Hecker, M. A proteomic view of Candida albicans yeast cell metabolism in exponential and stationary growth phases. Int. J. Med. Microbiol. 2008, 298, 291–318. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Biggest Threats and Data | Antibiotic/Antimicrobial Resistance | CDC. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 26 November 2018).
- Sucher, A.J.; Chahine, E.B.; Balcer, H.E. Echinocandins: The newest class of antifungals. Ann. Pharmacother. 2009, 43, 1647–1657. [Google Scholar] [CrossRef]
- Uppuluri, P.; Srinivasan, A.; Ramasubramanian, A.; Lopez-Ribot, J.L. Effects of Fluconazole, Amphotericin B, and Caspofungin on Candida albicans Biofilms under Conditions of Flow and on Biofilm Dispersion. Antimicrob. Agents Chemother. 2011, 55, 3591–3593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila, T.V.M.; Ishida, K.; de Souza, W.; Prousis, K.; Calogeropoulou, T.; Rozental, S. Effect of alkylphospholipids on Candida albicans biofilm formation and maturation. J. Antimicrob. Chemother. 2013, 68, 113–125. [Google Scholar] [CrossRef]
- Khan, M.S.A.; Ahmad, I. Antibiofilm activity of certain phytocompounds and their synergy with fluconazole against Candida albicans biofilms. J. Antimicrob. Chemother. 2012, 67, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of fluconazole resistance in Candida albicans biofilms: Phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 2003, 71, 4333–4340. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Yeh, P.J. Suppressive drug combinations and their potential to combat antibiotic resistance. J. Antibiot. (Tokyo) 2017, 70, 1033–1042. [Google Scholar] [CrossRef]
- Delattin, N.; De Brucker, K.; Vandamme, K.; Meert, E.; Marchand, A.; Chaltin, P.; Cammue, B.P.A.; Thevissen, K. Repurposing as a means to increase the activity of amphotericin B and caspofungin against Candida albicans biofilms. J. Antimicrob. Chemother. 2014, 69, 1035–1044. [Google Scholar] [CrossRef] [Green Version]
- LaFleur, M.D.; Sun, L.; Lister, I.; Keating, J.; Nantel, A.; Long, L.; Ghannoum, M.; North, J.; Lee, R.E.; Coleman, K.; et al. Potentiation of azole antifungals by 2-adamantanamine. Antimicrob. Agents Chemother. 2013, 57, 3585–3592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marmiroli, N.; Tedeschi, F.; Sabatini, M.A.; Truzzi, G.; Ferrari, C.; Puglisi, P.P. Relationship between growth inhibition and mitochondrial function in petite-negative yeasts. II. Effects of central nervous system drugs upon pathogenic and non-pathogenic Candida species. Biol. Cell 1985, 53, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Peralta, M.A.; da Silva, M.A.; Ortega, M.G.; Cabrera, J.L.; Paraje, M.G. Usnic Acid Activity on Oxidative and Nitrosative Stress of Azole-Resistant Candida albicans Biofilm. Planta Med. 2017, 83, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Shingu-Vazquez, M.; Traven, A. Mitochondria and Fungal Pathogenesis: Drug Tolerance, Virulence, and Potential for Antifungal Therapy. Eukaryot. Cell 2011, 10, 1376–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnani, T.; Soriani, F.M.; Martins Vde, P.; Policarpo, A.C.; Sorgi, C.A.; Faccioli, L.H.; Curti, C.; Uyemura, S.A. Silencing of mitochondrial alternative oxidase gene of Aspergillus fumigatus enhances reactive oxygen species production and killing of the fungus by macrophages. J. Bioenerg. Biomembr. 2008, 40, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Donaghey, F.; Helming, K.; McCarthy, M.; Rogers, S.; Austriaco, N. Deletion of AIF1 but not of YCA1/MCA1 protects Saccharomyces cerevisiae and Candida albicans cells from caspofungin-induced programmed cell death. Microb. Cell 2014, 15, 58–63. [Google Scholar] [CrossRef] [Green Version]
- Datta, S.; Kundu, S.; Ghosh, P.; De, S.; Ghosh, A.; Chatterjee, M. Correlation of oxidant status with oxidative tissue damage in patients with rheumatoid arthritis. Clin. Rheumatol. 2014, 33, 1557–1564. [Google Scholar] [CrossRef]
- de Oliveira, G.A.; Cheng, R.Y.S.; Ridnour, L.A.; Basudhar, D.; Somasundaram, V.; McVicar, D.W.; Monteiro, H.P.; Wink, D.A. Inducible Nitric Oxide Synthase in the Carcinogenesis of Gastrointestinal Cancers. Antioxid. Redox Signal. 2017, 26, 1059–1077. [Google Scholar] [CrossRef]
- Nishimura, A.; Kawahara, N.; Takagi, H. The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem. Biophys. Res. Commun. 2013, 430, 137–143. [Google Scholar] [CrossRef]
- Li, D.D.; Yang, C.C.; Liu, P.; Wang, Y.; Sun, Y. Effect of Nitric Oxide on the Antifungal Activity of Oxidative Stress and Azoles Against Candida albicans. Indian J. Microbiol. 2016. [Google Scholar] [CrossRef] [Green Version]
- Vediyappan, G.; Rossignol, T.; D’Enfert, C. Interaction of Candida albicans biofilms with antifungals: Transcriptional response and binding of antifungals to beta-glucans. Antimicrob. Agents Chemother. 2010, 54, 2096–2111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldara, M.; Graziano, S.; Gullì, M.; Cadonici, S.; Marmiroli, N. Off-target effects of neuroleptics and antidepressants on Saccharomyces cerevisiae. Toxicol. Sci. 2017, 156, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.-C.; Chao, C.-C.; Yang, C.-Y.; Ye, Y.-L.; Liu, F.-C.; Chuang, Y.-J.; Lan, C.-Y. Diverse Hap43-Independent Functions of the Candida albicans CCAAT-Binding Complex. Eukaryot. Cell 2013, 12, 804–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, N.; Li, D.; Zhang, Y.; Killeen, K.; Groutas, W.; Calderone, R. Repurposing an inhibitor of ribosomal biogenesis with broad anti-fungal activity. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Patterson, T.F. Multidrug-resistant candida: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 2017, 216, S445–S451. [Google Scholar] [CrossRef] [Green Version]
- Lacka, I.; Wakieć, R. Multidrug resistance in fungi. Postepy Biochem. 2008, 54, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Barker, K.S.; Rogers, P.D. Recent insights into the mechanisms of antifungal resistance. Curr. Infect. Dis. Rep. 2006, 8, 449–456. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caldara, M.; Marmiroli, N. Known Antimicrobials Versus Nortriptyline in Candida albicans: Repositioning an Old Drug for New Targets. Microorganisms 2020, 8, 742. https://doi.org/10.3390/microorganisms8050742
Caldara M, Marmiroli N. Known Antimicrobials Versus Nortriptyline in Candida albicans: Repositioning an Old Drug for New Targets. Microorganisms. 2020; 8(5):742. https://doi.org/10.3390/microorganisms8050742
Chicago/Turabian StyleCaldara, Marina, and Nelson Marmiroli. 2020. "Known Antimicrobials Versus Nortriptyline in Candida albicans: Repositioning an Old Drug for New Targets" Microorganisms 8, no. 5: 742. https://doi.org/10.3390/microorganisms8050742
APA StyleCaldara, M., & Marmiroli, N. (2020). Known Antimicrobials Versus Nortriptyline in Candida albicans: Repositioning an Old Drug for New Targets. Microorganisms, 8(5), 742. https://doi.org/10.3390/microorganisms8050742





