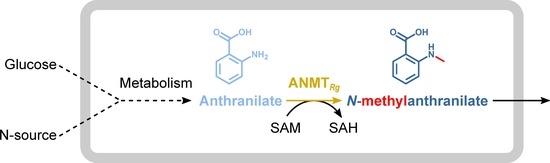
Fermentative N-Methylanthranilate Production by Engineered Corynebacterium glutamicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Fed-Batch Cultivation
2.3. Molecular Genetic Techniques and Strain Construction
2.4. Quantification of Amino Acids and Organic Acids
3. Results
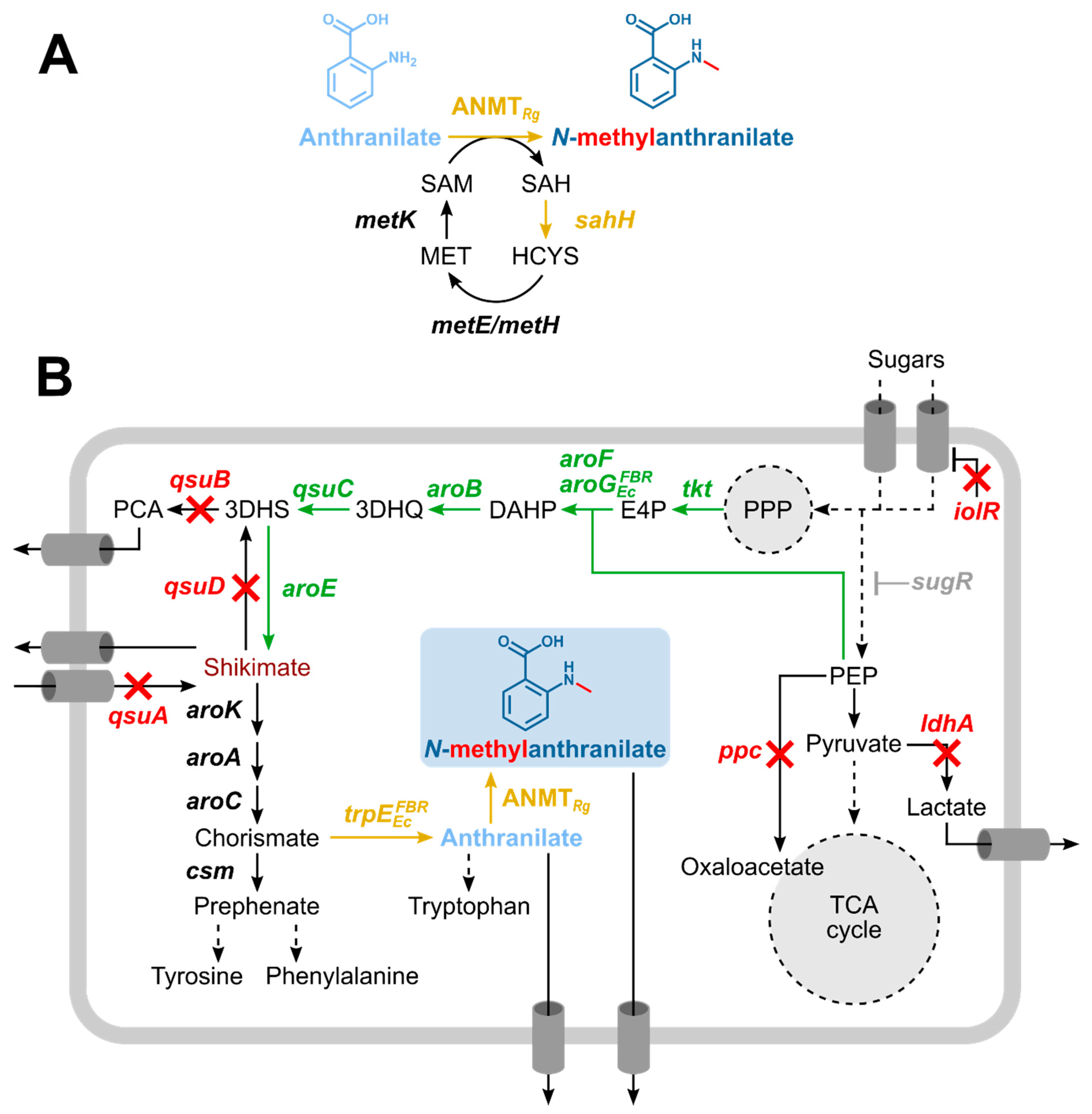
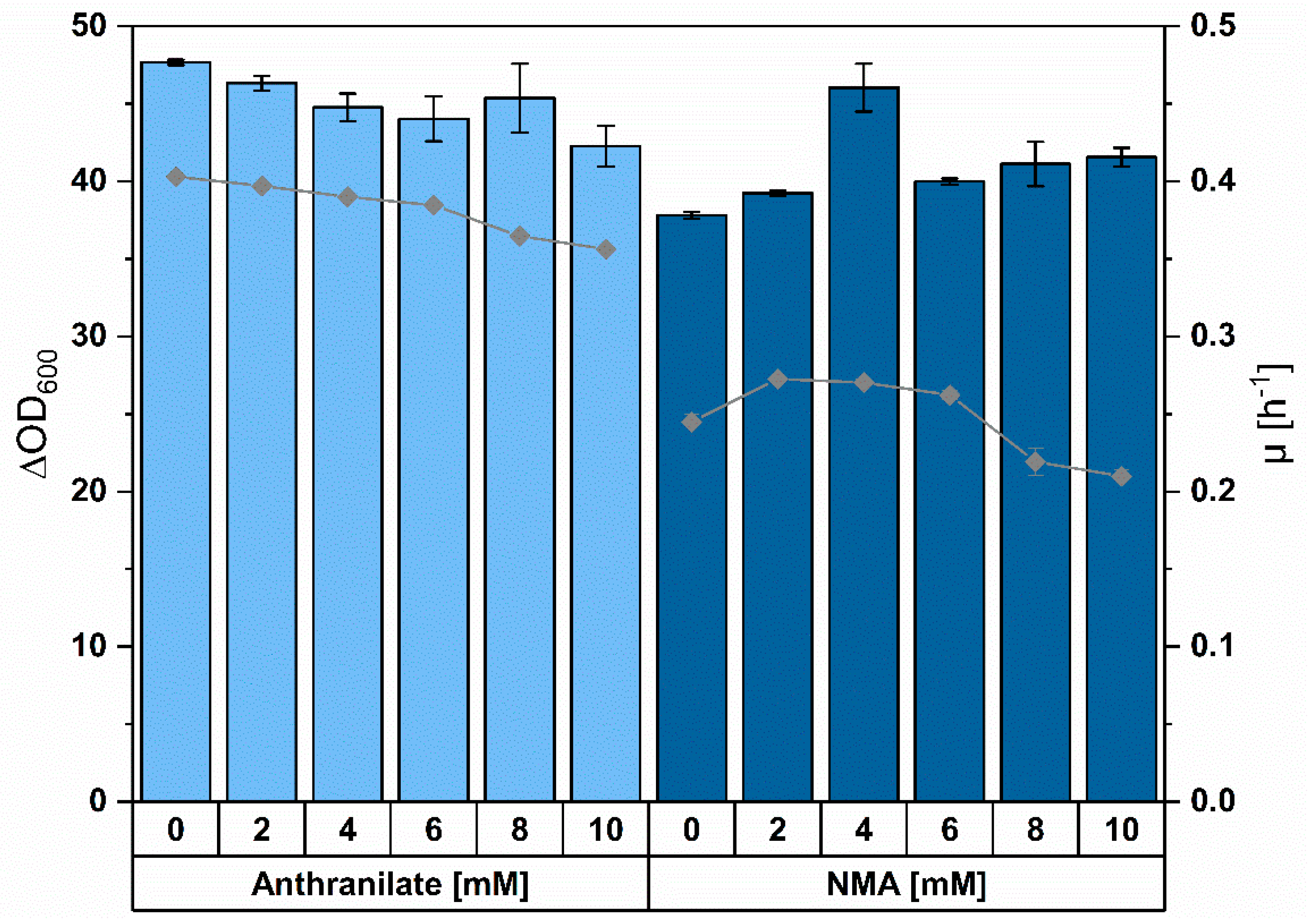
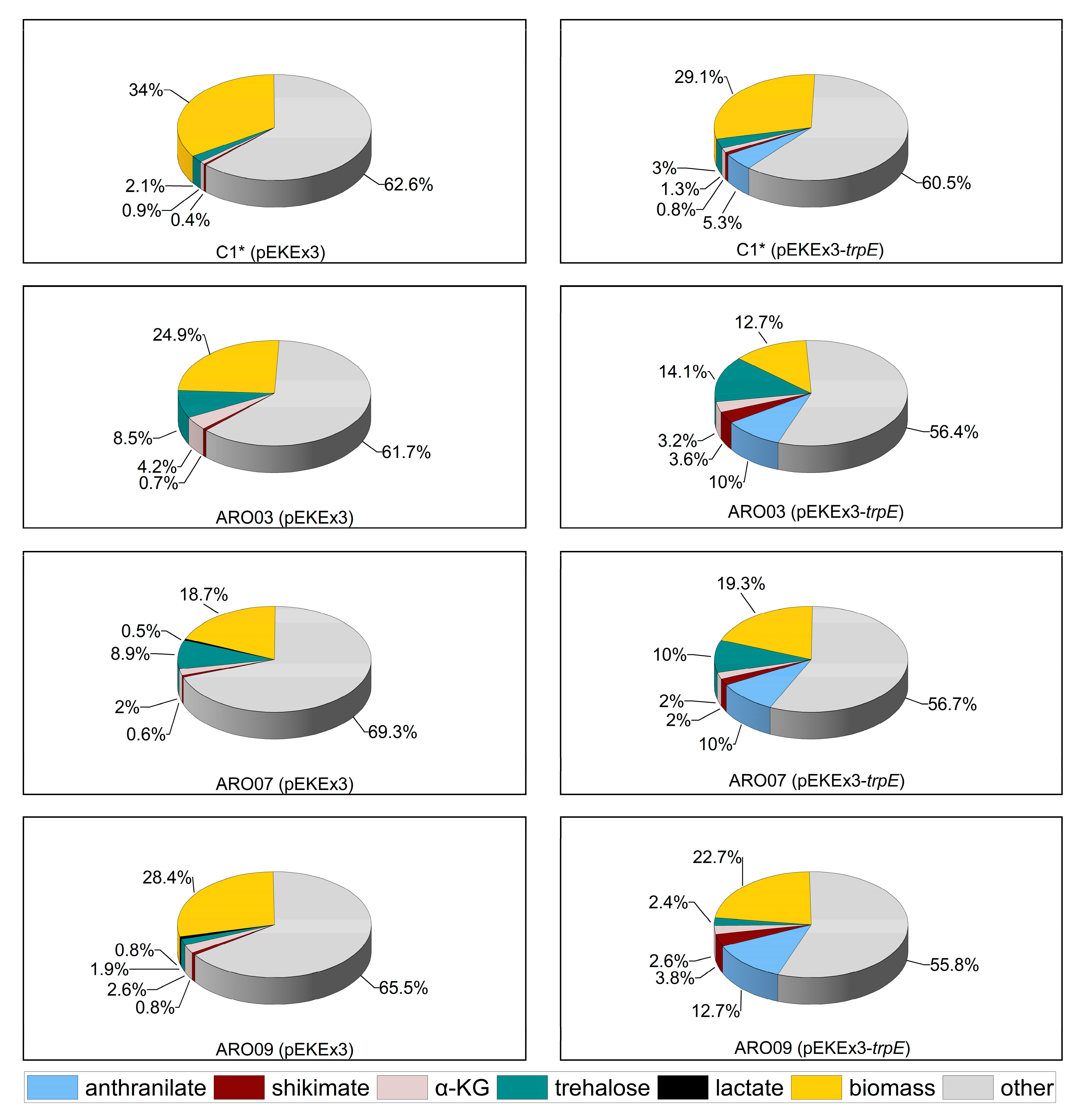
3.1. Corynebacterium glutamicum as Suitable Host for NMA Production
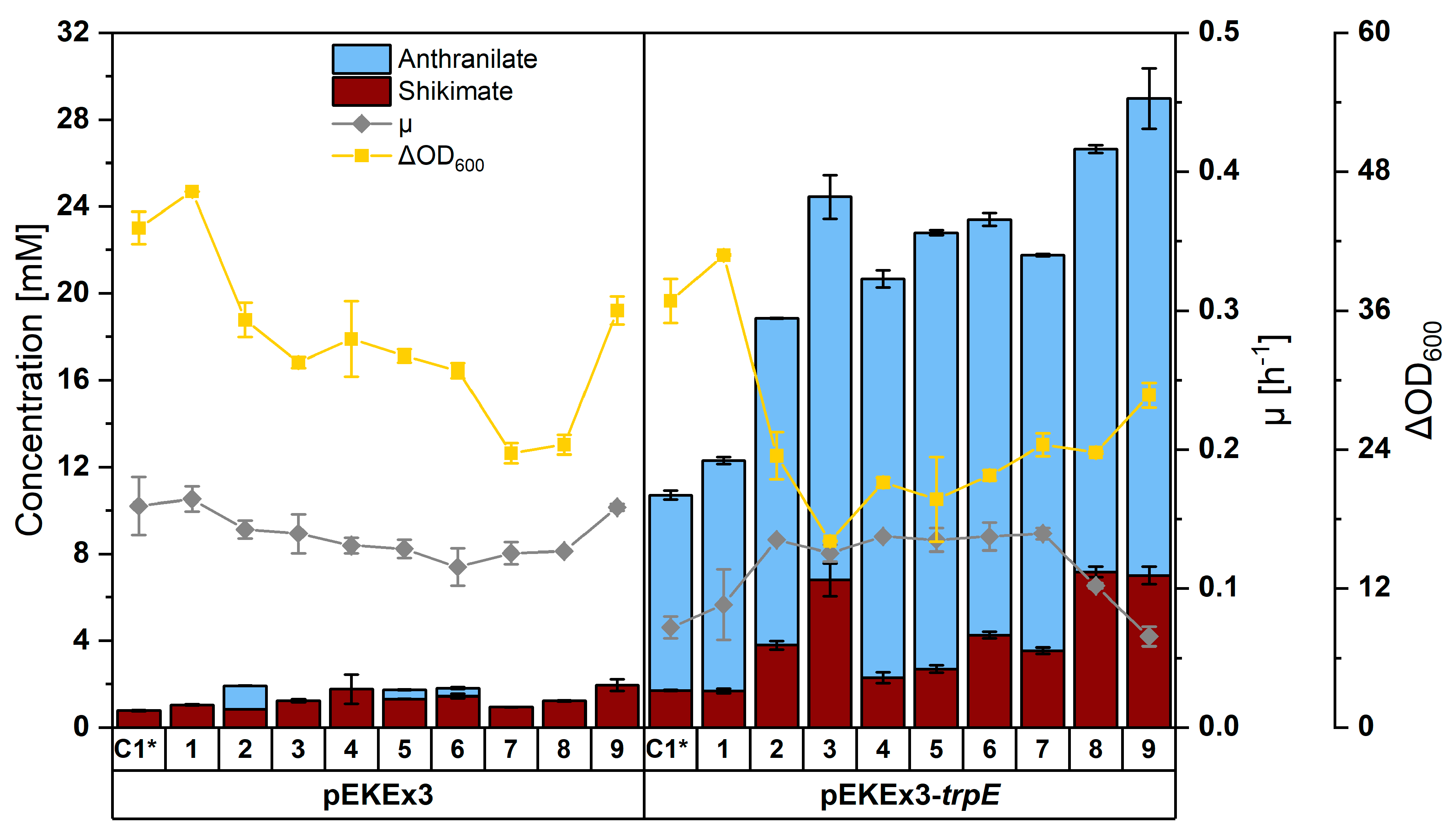
3.2. Construction of a C. glutamicum Platform Strain for Production of Anthranilate
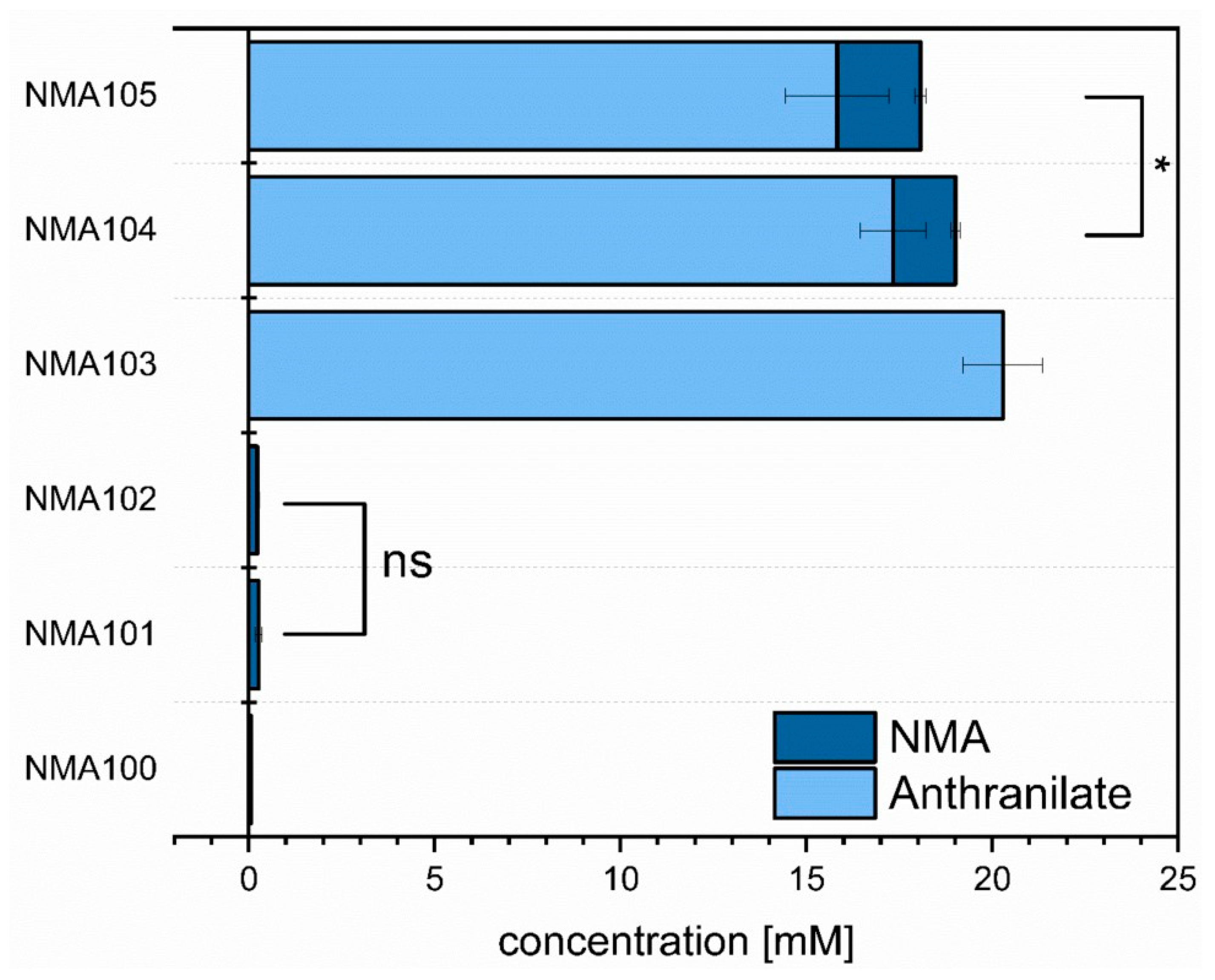
3.3. Establishing Fermentative Production of NMA by C. glutamicum
3.4. Fed-Batch Production of NMA in Bioreactors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, L.-K.; Abdelwahab, H.; del Campo, J.S.M.; Mehra-Chaudhary, R.; Sobrado, P.; Tanner, J.J. The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase. J. Boil. Chem. 2016, 291, 21553–21562. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Yu, Y.; Shen, Y.; Liu, Q.; Zhao, Z.; Sharma, R.; Reiter, R.J. Melatonin Synthesis and Function: Evolutionary History in Animals and Plants. Front. Endocrinol. 2019, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Scanlan, J.; Song, L.; Crombie, A.T.; Rahman, T.; Schäfer, H.; Murrell, J.C. γ-Glutamylmethylamide Is an Essential Intermediate in the Metabolism of Methylamine by Methylocella silvestris. Appl. Environ. Microbiol. 2010, 76, 4530–4537. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, J.; Rechenmacher, F.; Kessler, H. N-Methylation of Peptides and Proteins: An Important Element for Modulating Biological Functions. Angew. Chem. Int. Ed. 2012, 52, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Gazdik, M.; O’Neill, M.; Lopaticki, S.; Lowes, K.N.; Smith, B.J.; Cowman, A.F.; Boddey, J.A.; Gasser, R. The effect of N-methylation on transition state mimetic inhibitors of the Plasmodium protease, plasmepsin V. MedChemComm 2015, 6, 437–443. [Google Scholar] [CrossRef]
- Aurelio, L.; Brownlee, R.T.C.; Hughes, A.B. Synthetic Preparation of N-Methyl-α-amino Acids. Chem. Rev. 2004, 104, 5823–5846. [Google Scholar] [CrossRef]
- de Marco, R.; Leggio, A.; Liguori, A.; Marino, T.; Perri, F.; Russo, N. Site-Selective Methylation ofNβ-Nosyl Hydrazides of N-Nosyl Protected α-Amino Acids. J. Org. Chem. 2010, 75, 3381–3386. [Google Scholar] [CrossRef]
- Belsito, E.; di Gioia, M.L.; Greco, A.; Leggio, A.; Liguori, A.; Perri, F.; Siciliano, C.; Viscomi, M.C. N-Methyl-N-nosyl-β3-amino Acids. J. Org. Chem. 2007, 72, 4798–4802. [Google Scholar] [CrossRef]
- Freidinger, R.M.; Hinkle, J.S.; Perlow, D.S. Synthesis of 9-fluorenylmethyloxycarbonyl-protected N-alkyl amino acids by reduction of oxazolidinones. J. Org. Chem. 1983, 48, 77–81. [Google Scholar] [CrossRef]
- di Gioia, M.L.; Leggio, A.; Malagrinò, F.; Romio, E.; Siciliano, C.; Liguori, A. N-Methylated α-Amino Acids and Peptides: Synthesis and Biological Activity. Mini-Rev. Med. Chem. 2016, 16, 1. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, A.; Monaim, S.A.H.A.; Jad, Y.E.; El-Faham, A.; de la Torre, B.G.; Albericio, F. N-methylation in amino acids and peptides: Scope and limitations. Biopolymers 2018, 109, e23110. [Google Scholar] [CrossRef] [PubMed]
- Hyslop, J.F.; Lovelock, S.L.; Watson, A.J.B.; Sutton, P.W.; Roiban, G.-D. N-Alkyl-α-amino acids in Nature and their biocatalytic preparation. J. Biotechnol. 2019, 293, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Mindt, M.; Walter, T.; Risse, J.M.; Wendisch, V. Fermentative Production of N-Methylglutamate From Glycerol by Recombinant Pseudomonas putida. Front. Bioeng. Biotechnol. 2018, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Mindt, M.; Hannibal, S.; Heuser, M.; Risse, J.M.; Sasikumar, K.; Nampoothiri, K.M.; Wendisch, V. Fermentative Production of N-Alkylated Glycine Derivatives by Recombinant Corynebacterium glutamicum Using a Mutant of Imine Reductase DpkA From Pseudomonas putida. Front. Bioeng. Biotechnol. 2019, 7, 232. [Google Scholar] [CrossRef]
- Lee, H.L.; Kim, S.-Y.; Kim, E.J.; Han, D.Y.; Kim, B.-G.; Ahn, J.-H. Synthesis of Methylated Anthranilate Derivatives Using Engineered Strains of Escherichia coli. J. Microbiol. Biotechnol. 2019, 29, 839–844. [Google Scholar] [CrossRef]
- Rohde, B.; Hans, J.; Martens, S.; Baumert, A.; Hunziker, P.; Matern, U. Anthranilate N-methyltransferase, a branch-point enzyme of acridone biosynthesis. Plant J. 2007, 53, 541–553. [Google Scholar] [CrossRef]
- Mugford, S.T.; Louveau, T.; Melton, R.; Qi, X.; Bakht, S.; Hill, L.; Tsurushima, T.; Honkanen, S.; Rosser, S.J.; Lomonossoff, G.P.; et al. Modularity of plant metabolic gene clusters: A trio of linked genes that are collectively required for acylation of triterpenes in oat. Plant Cell 2013, 25, 1078–1092. [Google Scholar] [CrossRef]
- Baumert, A.; Schmidt, J.; Gröger, D. Synthesis and mass spectral analysis of coenzyme a thioester of anthranilic acid and itsN-methyl derivative involved in acridone alkaloid biosynthesis. Phytochem. Anal. 1993, 4, 165–170. [Google Scholar] [CrossRef]
- Michael, J.P. Acridone Alkaloids. In The Alkaloids: Chemistry and Biology; Elsevier BV: Amsterdam, The Netherlands, 2017; pp. 1–108. [Google Scholar]
- Piboonprai, K.; Khumkhrong, P.; Khongkow, M.; Yata, T.; Ruangrungsi, N.; Chansriniyom, C.; Iempridee, T. Anticancer activity of arborinine from Glycosmis parva leaf extract in human cervical cancer cells. Biochem. Biophys. Res. Commun. 2018, 500, 866–872. [Google Scholar] [CrossRef]
- Flavours and Fragrances; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2007.
- Radulović, N.S.; Miltojevic, A.; McDermott, M.; Waldren, S.; Parnell, J.A.; Pinheiro, M.M.G.; Fernandes, P.D.; Menezes, F.D.S. Identification of a new antinociceptive alkaloid isopropyl N-methylanthranilate from the essential oil of Choisya ternata Kunth. J. Ethnopharmacol. 2011, 135, 610–619. [Google Scholar] [CrossRef]
- Pinheiro, M.M.G.; Miltojevic, A.; Radulović, N.S.; Abdul-Wahab, I.R.; Boylan, F.; Fernandes, P.D. Anti-Inflammatory Activity of Choisya ternata Kunth Essential Oil, Ternanthranin, and Its Two Synthetic Analogs (Methyl and Propyl N-Methylanthranilates). PLoS ONE 2015, 10, e0121063. [Google Scholar] [CrossRef] [PubMed]
- Wendisch, V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020, 58, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M. Sugar transport systems in Corynebacterium glutamicum: Features and applications to strain development. Appl. Microbiol. Biotechnol. 2012, 96, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Blombach, B.; Seibold, G. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl. Microbiol. Biotechnol. 2010, 86, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Polen, T.; Schluesener, D.; Poetsch, A.; Bott, M.; Wendisch, V. Characterization of citrate utilization in Corynebacterium glutamicum by transcriptome and proteome analysis. FEMS Microbiol. Lett. 2007, 273, 109–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arndt, A.; Auchter, M.; Ishige, T.; Wendisch, V.F.; Eikmanns, B.J. Ethanol catabolism in Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 2008, 15, 222–233. [Google Scholar] [CrossRef]
- Jorge, J.M.P.; Leggewie, C.; Wendisch, V. A new metabolic route to produce gamma-aminobutyric acid by Corynebacterium glutamicum from glucose. Amino Acids 2016, 48, 2519–2531. [Google Scholar] [CrossRef]
- Jorge, J.M.; Pérez-García, F.; Wendisch, V. A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 2017, 245, 1701–1709. [Google Scholar] [CrossRef]
- Shin, J.; Park, S.H.; Oh, Y.H.; Choi, J.W.; Lee, M.H.; Cho, J.S.; Jeong, K.J.; Joo, J.C.; Yu, J.; Park, S.J.; et al. Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid. Microb. Cell Fact. 2016, 15, 174. [Google Scholar] [CrossRef]
- Pérez-García, F.; Risse, J.M.; Friehs, K.; Wendisch, V. Fermentative production of L-pipecolic acid from glucose and alternative carbon sources. Biotechnol. J. 2017, 12, 1600646. [Google Scholar] [CrossRef]
- Pérez-García, F.; Peters-Wendisch, P.; Wendisch, V. Engineering Corynebacterium glutamicum for fast production of l-lysine and l-pipecolic acid. Appl. Microbiol. Biotechnol. 2016, 100, 8075–8090. [Google Scholar] [CrossRef] [PubMed]
- Mindt, M.; Risse, J.M.; Gruß, H.; Sewald, N.; Eikmanns, B.J.; Wendisch, V. One-step process for production of N-methylated amino acids from sugars and methylamine using recombinant Corynebacterium glutamicum as biocatalyst. Sci. Rep. 2018, 8, 12895. [Google Scholar] [CrossRef] [PubMed]
- Mindt, M.; Heuser, M.; Wendisch, V. Xylose as preferred substrate for sarcosine production by recombinant Corynebacterium glutamicum. Bioresour. Technol. 2019, 281, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Purwanto, H.S.; Kang, M.-S.; Ferrer, L.; Han, S.-S.; Lee, J.-Y.; Kim, H.-S.; Lee, J.-H. Rational engineering of the shikimate and related pathways in Corynebacterium glutamicum for 4-hydroxybenzoate production. J. Biotechnol. 2018, 282, 92–100. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Marienhagen, J. Corynebacterium glutamicum as platform to produce hydroxybenzoic acids. Microb. Cell Factories 2018, 17, 70. [Google Scholar] [CrossRef]
- Okai, N.; Miyoshi, T.; Takeshima, Y.; Kuwahara, H.; Ogino, C.; Kondo, A. Production of protocatechuic acid by Corynebacterium glutamicum expressing chorismate-pyruvate lyase from Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 100, 135–145. [Google Scholar] [CrossRef]
- Veldmann, K.H.; Dachwitz, S.; Risse, J.M.; Lee, J.-H.; Sewald, N.; Wendisch, V. Bromination of L-tryptophan in a Fermentative Process with Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2019, 7, 219. [Google Scholar] [CrossRef]
- Veldmann, K.H.; Minges, H.; Sewald, N.; Lee, J.-H.; Wendisch, V. Metabolic engineering of Corynebacterium glutamicum for the fermentative production of halogenated tryptophan. J. Biotechnol. 2019, 291, 7–16. [Google Scholar] [CrossRef]
- Luo, Z.W.; Cho, J.S.; Lee, S.Y. Microbial production of methyl anthranilate, a grape flavor compound. Proc. Natl. Acad. Sci. USA 2019, 116, 10749–10756. [Google Scholar] [CrossRef]
- Baumgart, M.; Unthan, S.; Kloß, R.; Radek, A.; Polen, T.; Tenhaef, N.; Müller, M.F.; Küberl, A.; Siebert, D.; Brühl, N.; et al. Corynebacterium glutamicum Chassis C1*: Building and Testing a Novel Platform Host for Synthetic Biology and Industrial Biotechnology. ACS Synth. Boil. 2017, 7, 132–144. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Boil. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Eggeling, L.; Bott, M. Handbook of Corynebacterium glutamicum; CRC Press: Boca Raton, FL, USA, 2005; ISBN 978-1-4200-3969-6. [Google Scholar]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Biotechnology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Codon Usage Database. Available online: https://www.kazusa.or.jp/codon/ (accessed on 20 May 2020).
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef] [PubMed]
- De Novo DNA: The Future of Genetic Systems Design and Engineering. Available online: https://www.denovodna.com/software/design_rbs_calculator (accessed on 22 May 2020).
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Heider, S.A.E.; Peters-Wendisch, P.; Netzer, R.; Stafnes, M.; Brautaset, T.; Wendisch, V. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2013, 98, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Blombach, B.; Riester, T.; Wieschalka, S.; Ziert, C.; Youn, J.-W.; Wendisch, V.; Eikmanns, B.J. Corynebacterium glutamicum Tailored for Efficient Isobutanol Production. Appl. Environ. Microbiol. 2011, 77, 3300–3310. [Google Scholar] [CrossRef]
- Engels, V.; Wendisch, V. The DeoR-Type Regulator SugR Represses Expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 2007, 189, 2955–2966. [Google Scholar] [CrossRef]
- Stansen, C.; Uy, D.; Delaunay, S.; Eggeling, L.; Goergen, J.-L.; Wendisch, V. Characterization of a Corynebacterium glutamicum Lactate Utilization Operon Induced during Temperature-Triggered Glutamate Production. Appl. Environ. Microbiol. 2005, 71, 5920–5928. [Google Scholar] [CrossRef]
- Kirchner, O.; Tauch, A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 287–299. [Google Scholar] [CrossRef]
- Wendisch, V.; Lee, J.-H. Metabolic Engineering in Corynebacterium Glutamicum; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2020; pp. 287–322. [Google Scholar]
- Kikuchi, Y.; Tsujimoto, K.; Kurahashi, O. Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl. Environ. Microbiol. 1997, 63, 761–762. [Google Scholar] [CrossRef]
- Lee, J.-H.; Wendisch, V. Production of amino acids—Genetic and metabolic engineering approaches. Bioresour. Technol. 2017, 245, 1575–1587. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Tanaka, Y.; Suda, M.; Jojima, T.; Inui, M. Enhanced Glucose Consumption and Organic Acid Production by Engineered Corynebacterium glutamicum Based on Analysis of a pfkB1 Deletion Mutant. Appl. Environ. Microbiol. 2016, 83. [Google Scholar] [CrossRef] [PubMed]
- Sander, T.; Farke, N.; Diehl, C.; Kuntz, M.; Glatter, T.; Link, H. Allosteric Feedback Inhibition Enables Robust Amino Acid Biosynthesis in E. coli by Enforcing Enzyme Overabundance. Cell Syst. 2019, 8, 66–75. [Google Scholar] [CrossRef]
- Neshat, A.; Mentz, A.; Rückert, C.; Kalinowski, J. Transcriptome sequencing revealed the transcriptional organization at ribosome-mediated attenuation sites in Corynebacterium glutamicum and identified a novel attenuator involved in aromatic amino acid biosynthesis. J. Biotechnol. 2014, 190, 55–63. [Google Scholar] [CrossRef]
- Draths, K.M.; Pompliano, D.L.; Conley, D.L.; Frost, J.W.; Berry, A.; Disbrow, G.L.; Staversky, R.J.; Lievense, J.C. Biocatalytic synthesis of aromatics from D-glucose: The role of transketolase. J. Am. Chem. Soc. 1992, 114, 3956–3962. [Google Scholar] [CrossRef]
- Klaffl, S.; Brocker, M.; Kalinowski, J.; Eikmanns, B.J.; Bott, M. Complex Regulation of the Phosphoenolpyruvate Carboxykinase Gene pck and Characterization of Its GntR-Type Regulator IolR as a Repressor of myo-Inositol Utilization Genes in Corynebacterium glutamicum. J. Bacteriol. 2013, 195, 4283–4296. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.E.; Kang, D.H.; Kim, Y.I.; You, S.K.; Han, S.O. GntR-Type Transcriptional Regulator PckR Negatively Regulates the Expression of Phosphoenolpyruvate Carboxykinase in Corynebacterium glutamicum. J. Bacteriol. 2012, 194, 2181–2188. [Google Scholar] [CrossRef][Green Version]
- Choi, G.-S.; Choo, H.J.; Kim, B.-G.; Ahn, J.-H. Synthesis of acridone derivatives via heterologous expression of a plant type III polyketide synthase in Escherichia coli. Microb. Cell Fact. 2020, 19, 1–11. [Google Scholar] [CrossRef]
- Choo, H.J.; Ahn, J.-H. Synthesis of Three Bioactive Aromatic Compounds by Introducing Polyketide Synthase Genes into Engineered Escherichia coli. J. Agric. Food Chem. 2019, 67, 8581–8589. [Google Scholar] [CrossRef]
- Sgobba, E.; Stumpf, A.K.; Vortmann, M.; Jagmann, N.; Krehenbrink, M.; Dirks-Hofmeister, M.E.; Moerschbacher, B.M.; Philipp, B.; Wendisch, V. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Bioresour. Technol. 2018, 260, 302–310. [Google Scholar] [CrossRef]
- Sgobba, E.; Wendisch, V. Synthetic microbial consortia for small molecule production. Curr. Opin. Biotechnol. 2020, 62, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Morihara, Y.; Wakayama, M.; Tachiki, T. Theanine Production by Coupled Fermentation with Energy Transfer Using γ-Glutamylmethylamide Synthetase of Methylovorus mays No. 9. Biosci. Biotechnol. Biochem. 2008, 72, 1206–1211. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Tanimoto, Y.; Yamauchi, S.; Tozawa, Y.; Sawayama, S.; Watanabe, Y. Identification and characterization of trans-3-hydroxy-l-proline dehydratase and Δ(1)-pyrroline-2-carboxylate reductase involved in trans-3-hydroxy-l-proline metabolism of bacteria. FEBS Open Biol. 2014, 4, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Wieschalka, S.; Blombach, B.; Eikmanns, B.J. Engineering Corynebacterium glutamicum to produce pyruvate. Appl. Microbiol. Biotechnol. 2012, 94, 449–459. [Google Scholar] [CrossRef]
- Zahoor, A.; Otten, A.; Wendisch, V. Metabolic engineering of Corynebacterium glutamicum for glycolate production. J. Biotechnol. 2014, 192, 366–375. [Google Scholar] [CrossRef]
- Li, P.-P.; Liu, Y.-J.; Liu, S.-J. Genetic and biochemical identification of the chorismate mutase from Corynebacterium glutamicum. Microbiology 2009, 155, 3382–3391. [Google Scholar] [CrossRef][Green Version]
- Maier, W.; Baumert, A.; Gröger, D. Partial Purification and Characterization of S-Adenosyl-L- Methionine: Anthranilic Acid N-Methyltransferase from Ruta Cell Suspension Cultures. J. Plant Physiol. 1995, 145, 1–6. [Google Scholar] [CrossRef]
- Han, G.; Hu, X.; Qin, T.; Li, Y.; Wang, X. Metabolic engineering of Corynebacterium glutamicum ATCC13032 to produce S -adenosyl- l -methionine. Enzym. Microb. Technol. 2016, 83, 14–21. [Google Scholar] [CrossRef]
- Francis, M.M.; Vining, L.C.; Westlake, D.W. Characterization, and regulation of anthranilate synthetase from a chloramphenicol-producing streptomycete. J. Bacteriol. 1978, 134, 10–16. [Google Scholar] [CrossRef]
- Henderson, E.J.; Nagano, H.; Zalkin, H.; Hwang, L.H. The anthranilate synthetase-anthranilate 5-phosphoribosylpyrophosphate phosphoribosyltransferase aggregate. Purification of the aggregate and regulatory properties of anthranilate synthetase. J. Boil. Chem. 1970, 245, 1416–1423. [Google Scholar]
- Caligiuri, M.G.; Bauerle, R. Identification of amino acid residues involved in feedback regulation of the anthranilate synthase complex from Salmonella typhimurium. Evidence for an amino-terminal regulatory site. J. Boil. Chem. 1991, 266, 8328–8335. [Google Scholar]
- Leßmeier, L.; Wendisch, V. Identification of two mutations increasing the methanol tolerance of Corynebacterium glutamicum. BMC Microbiol. 2015, 15, 216. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, L.; Tuyishime, P.; Liu, J.; Zhang, K.; Gao, N.; Zhang, Z.; Ni, X.; Feng, J.; Yuan, Q.; et al. Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum. Commun. Boil. 2020, 3, 217. [Google Scholar] [CrossRef] [PubMed]
- Hennig, G.; Haupka, C.; Brito, L.F.; Rückert, C.; Cahoreau, E.; Heux, S.; Wendisch, V.F. Methanol-Essential Growth of Corynebacterium glutamicum: Adaptive Laboratory Evolution Overcomes Limitation due to Methanethiol Assimilation Pathway. Int. J. Mol. Sci. 2020, 21, 3617. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Khushk, I.; Xiao, Y.; Gao, Q.; Bao, J. Tolerance improvement of Corynebacterium glutamicum on lignocellulose derived inhibitors by adaptive evolution. Appl. Microbiol. Biotechnol. 2017, 102, 377–388. [Google Scholar] [CrossRef] [PubMed]

| Strains | Description | Source |
|---|---|---|
| Corynebacterium glutamicum | ||
| WT | C. glutamicum wild-type strain ATCC13032 | ATCC |
| C1* | Genome-reduced chassis strain derived from | [42] |
| ARO01 | Δvdh::PilvC-aroGD146N mutant of C1* | This work |
| ARO02 | ΔldhA mutant of ARO01 | This work |
| ARO03 | ΔsugR mutant of ARO02 | This work |
| ARO04 | ΔaroR::PilvC-aroF mutant of ARO03 | This work |
| ARO05 | ΔqsuABCD::Ptuf-qsuC mutant of ARO04 | This work |
| ARO06 | Δppc::Psod-aroB mutant of ARO05 | This work |
| ARO07 | ΔPtkt::Ptuf-tkt mutant of ARO06 | This work |
| ARO08 | ΔiolR::Ptuf-aroE mutant of ARO07 | This work |
| ARO09 | ΔsugR::sugR mutant of ARO08 | This work |
| NMA100 | ARO09 carrying pEKEx3 and pGold | This work |
| NMA101 | ARO09 carrying pEKEx3 and pGold-anmt | This work |
| NMA102 | ARO09 carrying pEKEx3 and pGold-anmt-sahH | This work |
| NMA103 | ARO09 carrying pEKEx3-trpEFBR and pGold | This work |
| NMA104 | ARO09 carrying pEKEx3-trpEFBR and pGold-anmt | This work |
| NMA105 | ARO09 carrying pEKEx3-trpEFBR and pGold-anmt-sahH | This work |
| Escherichia coli | ||
| S17-1 | recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 | [45] |
| DH5α | F-thi-1 endA1 hsdr17(r-, m-) supE44 1lacU169 (Φ80lacZ1M15) recA1 gyrA96 | [43] |
| Name | Oligonucleotide Sequence (5′ to 3′) |
|---|---|
| vdh-conf-fw | GACCTCTAGGGCAGCAGTG |
| vdh-conf-rv | CTGTTCAGCGGATTAGCG |
| ldhA-conf-fw | TGATGGCACCAGTTGCGATGT |
| ldhA-conf-rv | CCATGATGCAGGATGGAGTA |
| sugR-conf-fw | CGAGATGCTGTGGTTTTGAG |
| sugR-conf-rv | GCTTATCGGGTGTGGGAATG |
| US-aroR-fw | CCTGCAGGTCGACTCTAGAGCGATGCAGAATAATGCAGTTAG |
| US-aroR-rv | CGGAGCTTGCCTGGGAGTTTGGAACCTTAACACACTTTC |
| PilvC-aroR-fw | GAAAGTGTGTTAAGGTTCCAAACTCCCAGGCAAGCTCCGCGC |
| PilvC-aroR-rv | GAAAAAACCTCCTTTAGTGTGTAGTTAAGTTATGGTGATGGGAGAAAATCTCGCCTTTCG |
| DS-aroR-fw | ATCACCATAACTTAACTACACACTAAAGGAGGTTTTTTCATGAGTTCTCCAGTCTCACTCGAAAAC |
| DS-aroR-rv | GAATTCGAGCTCGGTACCCGGGCAATGCGCAAGCCCTCTGGG |
| aroR-conf-fw | GGAACTCCCGTTGAGGTG |
| aroR-conf-rv | GTGGTACGAGCGCCGATTG |
| US-qsuA-fw | CCTGCAGGTCGACTCTAGAGGTTGGCAGCGCAACCAGTC |
| US-qsuA-rv | CTACTGACACGCTAAAACGCTGTCGATCCTGTTCATCG |
| Ptuf-qsuC-fw | CGATGAACAGGATCGACAGCGTTTTAGCGTGTCAGTAG |
| Ptuf-qsuC-rv | CTGAAGGGCCTCCTTTCTCCTCCTGGACTTCGTGG |
| qsuC-fw | GGAGAAAGGAGGCCCTTCAGATGCCTGGAAAAATTCTCCTCC |
| qsuC-rv | GTCGAGGTTTTACTGACTCTTCTACTTTTTGAGATTTGCCAGG |
| DS-qsuD-fw | CTCAAAAAGTAGAAGAGTCAGTAAAACCTCGACGC |
| DS-qsuD-rv | GAATTCGAGCTCGGTACCCGGGATTTCGCGGATGGGTCTAAGTATG |
| qsu-conf-fw | GTTCGTGGACAAGTGTGGTGG |
| qsu-conf-rv | GTTCGTGGACAAGTGTGGTGG |
| US-ppc-fw | GCCTGCAGGTCGACTCTAGAGCGCTCAGGAAGTGTGCAAGGC |
| US-ppc-rv | GTACTACCCAGCCGGCTGGGGATCCCTACTTTAAACACTCTTTCACATTGAGGGTG |
| Psod-aroB-fw | AATGTGAAAGAGTGTTTAAAGTAGGAAGCGCCTCATCAGCGGTAAC |
| Psod-aroB-rv | CTCCTTTAAAAATAAGTCGCCTACCAAAATCCTTTCGTAGGTTTCCGC |
| aroB-fw | GCGGAAACCTACGAAAGGATTTTGGTAGGCGACTTATTTTTAAAGGAGGTTTTTTATGAGCGCAGTGCAGATTTTC |
| aroB-rv | CTTCTCTCATCCGCCAAAATTAGTGGCTGATTGCCTCATAAG |
| Term-aroB-fw | CTTATGAGGCAATCAGCCACTAATTTTGGCGGATGAGAGAAG |
| Term-aroB-rv | AGTACTACCCAGCCGGCTGGGGATCCAAAAGAGTTTGTAGAAACGC |
| DS-ppc-fw | TGAAAGAGTGTTTAAAGTAGGGATCCCCAGCCGGCTGGGTAGTAC |
| DS-ppc-rv | GAATTCGAGCTCGGTACCCGGGCAGTGGGGAGACAACAGGTCG |
| ppc-conf-fw | CCGTCGGGAAACAGTTCCCC |
| ppc-conf-rv | GCAGACCCGTAAGTCCCTTGC |
| US-tkt-fw | GCATGCCTGCAGGTCGACTCTAGAGTGACCCAGGTGGACGCCAAC |
| US-tkt-rv | GTGGACATTCGCAGGGTAACGGCCAAGGTGTGATCAATCTTAAGTC |
| Ptuf-tkt-fw | GACTTAAGATTGATCACACCTTGGCCGTTACCCTGCGAATGTCCAC |
| Ptuf-tkt-rv | CGTCAAGGTGGTCATCTGAAGGGCCTCCTTTCTGTATGTCCTCCTGGACTTC |
| DS-tkt-fw | CAGGAGGACATACAGAAAGGAGGCCCTTCAGATGACCACCTTGACGCTGTC |
| DS-tkt-rv | GAATTCGAGCTCGGTACCCGGGTGGCGGTACTCAGGGTGTCC |
| tkt-conf-fw | GTTCCCGAATCAATCTTTTTAATG |
| tkt-conf-rv | GACCCTGGCCAAGAGGGCCAGTG |
| US-iolR-fw | GCCTGCAGGTCGACTCTAGAGCGACCCTCACGATCGCATG |
| US-iolR-rv | CTACTGACACGCTAAAACGCGATGTCTCCTTTCGTTGCCC |
| Ptuf-aroE-fw | GGGCAACGAAAGGAGACATCGCGTTTTAGCGTGTCAGTAG |
| Ptuf-aroE-rv | CCCATCTGAAGGGCCTCCTTTCTCCTCCTGGACTTCGTGGTG |
| aroE-fw | GGAGAAAGGAGGCCCTTCAGATGGGTTCTCACATCACTCACCG |
| aroE-rv | CAGAAGGGCTCTTTGGTTTATTTCTTAGTGTTCTTCTGAGATGCCTAAAGACTC |
| DS-iolR-fw | GAGTCTTTAGGCATCTCAGAAGAACACTAAGAAATAAACCAAAGAGCCCTTCTG |
| DS-iolR-rv | GAATTCGAGCTCGGTACCCGGGCGCTCTCCATCCGCTGGAC |
| iolR-conf-fw | CAGATAGAGGAACCCAAGGCG |
| iolR-conf-rv | GGACTTCGTGAGTGCTCGTC |
| sugR_reintegr-fw | CTGCAGGTCGACTCTAGAGCCTGCGCAGGGACCCTAATAAG |
| sugR_reintegr-rv | GAATTCGAGCTCGGTACCCGGGCCTGCAGTAAAAGATTCCCGC |
| x3-trpE-fw | CCTGCAGGTCGACTCTAGAGGAAAGGAGGCCCTTCAGATGCAAACACAAAAACCGACTCTCGAACTG |
| x3-trpE-rv | AAAACGACGGCCAGTGAATTTCAGAAAGTCTCCTGTGCATGATGCGC |
| pGANMT-sahH-fw | ATGAGCTCGGTACCCGGGCGGGACGAAGAGAACCGTTACAAGAATAAAGGAGGTTTTTTATGGCACAGGTTATGGACTTC |
| pGANMT-sahH-rv | CTGCAGGTCGACTCTAGAGTTAGTAGCGGTAGTGCTCCGG |
| Plasmids | Description | Source |
|---|---|---|
| pK19mobsacB | KmR; E. coli/C. glutamicum shuttle vector for construction of insertion and deletion mutants in C. glutamicum (pK19 oriVEc sacB lacZα) | [50] |
| pK19-Δvdh::PilvC-aroGD146N | pK19mobsacB with a construct for replacement of vdh (cg2953) by aroGD146N from E. coli under control of C. glutamicum promoter PilvC | [36] |
| pK19-ΔldhA | pK19mobsacB with a construct for deletion of ldhA (cg3219) | [52] |
| pK19-ΔsugR | pK19mobsacB with a construct for deletion of sugR (cg2115) | [53] |
| pK19-ΔaroR::PilvC | pK19mobsacB with a construct for replacement of aroR and the native promoter of aroF by C. glutamicum promoter PilvC and an artificial RBS | This work |
| pK19-ΔqsuABCD::Ptuf-qsuC | pK19mobsacB with a construct for replacement of qsuABCD (cg0501-cg0504) by qsuC (cg0503) with an artificial RBS under control of C. glutamicum promoter Ptuf | This work |
| pK19-Δppc::Psod-aroB | pK19mobsacB with a construct for replacement of ppc (cg1787) by aroB (cg1827) with an artificial RBS under control of C. glutamicum promoter Psod | This work |
| pK19-ΔPtkt::Ptuf | pK19mobsacB with a construct for replacement of the tkt (cg1774) promoter by C. glutamicum promoter Ptuf and artificial RBS | This work |
| pK19-ΔiolR::Ptuf-aroE | pK19mobsacB with a construct for replacement of iolR (cg0196) by aroE (cg1835) with an artificial RBS under control of C. glutamicum promoter Ptuf | This work |
| pK19-ΔsugR::sugR | pK19mobsacB with a construct for reintegration of sugR (cg2115) into its native locus | This work |
| pEKEx3 | SpecR, PtaclacIq, pBL1 oriVCg, C. glutamicum/E. coli expression shuttle vector | [54] |
| pEKEx3-trpEFBR | SpecR, pEKEx3 overexpressing trpES40F from E. coli K12 containing an artificial RBS | This work |
| pEC-XK99E | KmR, PtrclacIq, pGA1 oriVEc, C. glutamicum/E. coli expression shuttle vector | [55] |
| pGold | KmR, PtrclacIq, pGA1 oriVEc, C. glutamicum/E. coli expression shuttle vector with BsaI recognition site for Golden Gate assembly | This work |
| pGold-anmt | KmR, pGold overexpressing codon harmonized anmt from Ruta graveolens with an artificial RBS | This work |
| pGold-anmt-sahH | KmR, pGold overexpressing a synthetic operon with codon harmonized anmt from R. graveolens with an artificial RBS and sahH from C. glutamicum with an artificial RBS | This work |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walter, T.; Al Medani, N.; Burgardt, A.; Cankar, K.; Ferrer, L.; Kerbs, A.; Lee, J.-H.; Mindt, M.; Risse, J.M.; Wendisch, V.F. Fermentative N-Methylanthranilate Production by Engineered Corynebacterium glutamicum. Microorganisms 2020, 8, 866. https://doi.org/10.3390/microorganisms8060866
Walter T, Al Medani N, Burgardt A, Cankar K, Ferrer L, Kerbs A, Lee J-H, Mindt M, Risse JM, Wendisch VF. Fermentative N-Methylanthranilate Production by Engineered Corynebacterium glutamicum. Microorganisms. 2020; 8(6):866. https://doi.org/10.3390/microorganisms8060866
Chicago/Turabian StyleWalter, Tatjana, Nour Al Medani, Arthur Burgardt, Katarina Cankar, Lenny Ferrer, Anastasia Kerbs, Jin-Ho Lee, Melanie Mindt, Joe Max Risse, and Volker F. Wendisch. 2020. "Fermentative N-Methylanthranilate Production by Engineered Corynebacterium glutamicum" Microorganisms 8, no. 6: 866. https://doi.org/10.3390/microorganisms8060866
APA StyleWalter, T., Al Medani, N., Burgardt, A., Cankar, K., Ferrer, L., Kerbs, A., Lee, J.-H., Mindt, M., Risse, J. M., & Wendisch, V. F. (2020). Fermentative N-Methylanthranilate Production by Engineered Corynebacterium glutamicum. Microorganisms, 8(6), 866. https://doi.org/10.3390/microorganisms8060866