Pontiella desulfatans gen. nov., sp. nov., and Pontiella sulfatireligans sp. nov., Two Marine Anaerobes of the Pontiellaceae fam. nov. Producing Sulfated Glycosaminoglycan-like Exopolymers
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Growth Conditions and Substrates
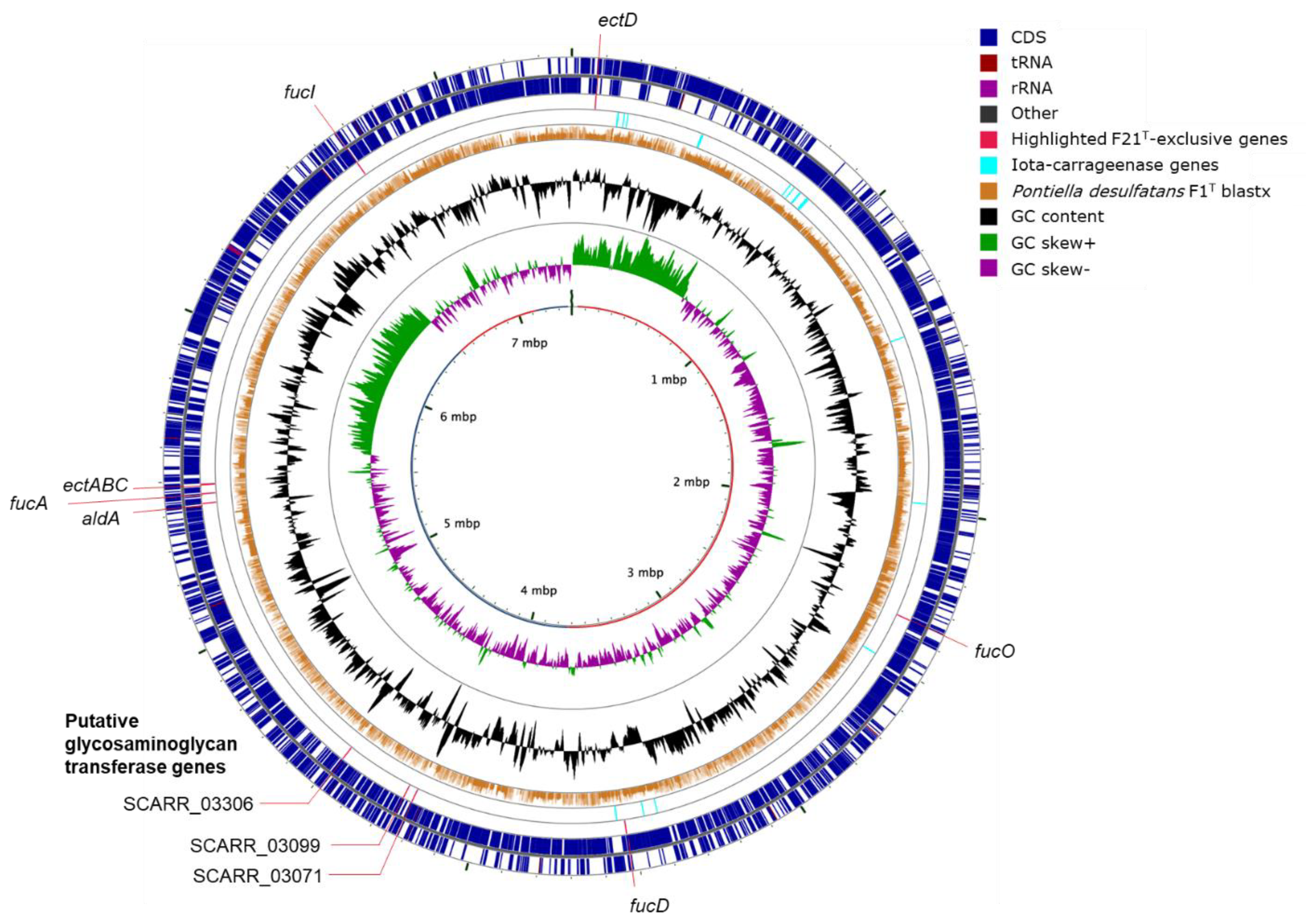
2.2. Genome Annotation and Visualization
2.3. Physiological Tests
2.4. Reduction of External Electron Acceptors
2.5. Oxygen Gradient Cultures
2.6. Energy Storage Compound Analysis
2.7. Extracellular Polymeric Substances Analysis
2.8. Phylogenetic Reconstruction
2.9. Lipid and Cellular Fatty Acid Analysis
3. Results
3.1. Phenotypic Characterization
3.2. Substrate Utilization and Genetic Capacity
3.3. Reduction of External Electron Acceptors during Anaerobic Growth on Sugars
3.4. Response to Different Redox Conditions and Oxygen
3.5. Formation of Energy Reserve Materials
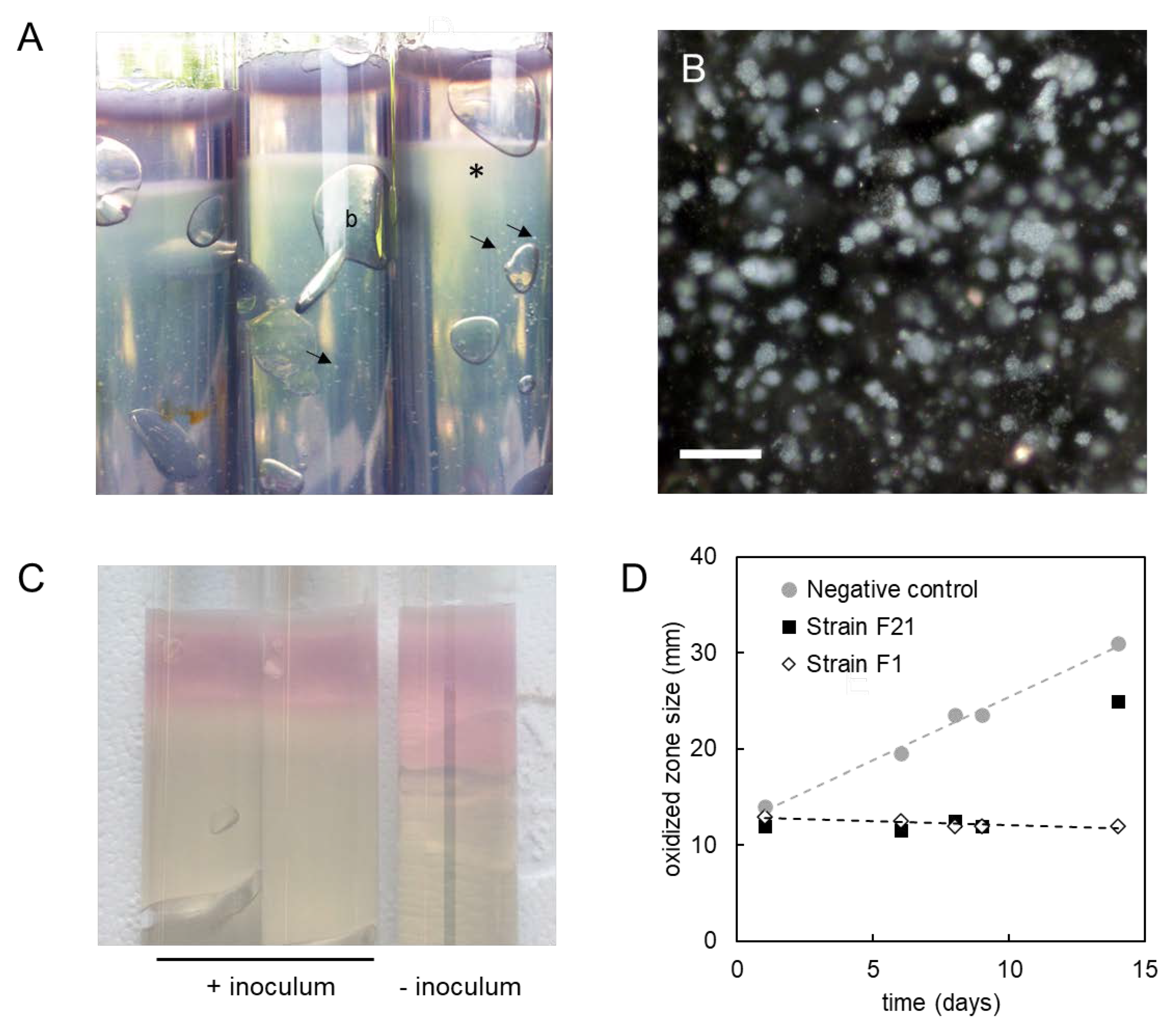
3.6. Production of Sulfated Glycosaminoglycan-like Exopolymers in Stationary Phase
3.7. Phylogenomics and Chemotaxonomy of the Class Kiritimatiellales
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jin, C.; Kenny, D.T.; Skoog, E.C.; Padra, M.; Adamczyk, B.; Vitizeva, V.; Thorell, A.; Venkatakrishnan, V.; Linden, S.K.; Karlsson, N.G. Structural diversity of human gastric mucin glycans. Mol. Cell. Proteomics 2017, 16, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Davidson, E.; Linker, A.; Hoffman, P. The acid mucopolysaccharides of connective tissue. Biochim. Biophys. Acta 1956, 21, 506–518. [Google Scholar] [CrossRef]
- Helbert, W. Marine polysaccharide sulfatases. Front. Mar. Sci. 2017, 4. [Google Scholar] [CrossRef]
- Barbeyron, T.; Brillet-Gueguen, L.; Carre, W.; Carriere, C.; Caron, C.; Czjzek, M.; Hoebeke, M.; Michel, G. Matching the diversity of sulfated biomolecules: creation of a classification database for sulfatases reflecting their substrate specificity. PLoS ONE 2016, 11, e0164846. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Kube, M.; Bauer, M.; Teeling, H.; Lombardot, T.; Ludwig, W.; Gade, D.; Beck, A.; Borzym, K.; Heitmann, K.; et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 2003, 100, 8298–8303. [Google Scholar] [CrossRef]
- Thrash, J.C.; Cho, J.C.; Vergin, K.L.; Morris, R.M.; Giovannoni, S.J. Genome sequence of Lentisphaera araneosa HTCC2155T, the type species of the order Lentisphaerales in the phylum Lentisphaerae. J. Bacteriol. 2010, 192, 2938–2939. [Google Scholar] [CrossRef][Green Version]
- Arndt, S.; Jørgensen, B.B.; LaRowe, D.E.; Middelburg, J.J.; Pancost, R.D.; Regnier, P. Quantifying the degradation of organic matter in marine sediments: A review and synthesis. Earth-Sci. Rev. 2013, 123, 53–86. [Google Scholar] [CrossRef]
- Van Vliet, D.M.; Palakawong Na Ayudthaya, S.; Diop, S.; Villanueva, L.; Stams, A.J.M.; Sánchez-Andrea, I. Anaerobic degradation of sulfated polysaccharides by two novel Kiritimatiellales strains isolated from Black Sea sediment. Front. Microbiol. 2019, 10, 253. [Google Scholar] [CrossRef]
- Spring, S.; Bunk, B.; Spröer, C.; Schumann, P.; Rohde, M.; Tindall, B.J.; Klenk, H.-P. Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 2016, 10, 2801. [Google Scholar] [CrossRef]
- Frey, J.C.; Rothman, J.M.; Pell, A.N.; Nizeyi, J.B.; Cranfield, M.R.; Angert, E.R. Fecal bacterial diversity in a wild gorilla. Appl. Environ. Microbiol. 2006, 72, 3788–3792. [Google Scholar] [CrossRef]
- Steelman, S.M.; Chowdhary, B.P.; Dowd, S.; Suchodolski, J.; Janečka, J.E. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 2012, 8, 231. [Google Scholar] [CrossRef] [PubMed]
- Cardman, Z.; Arnosti, C.; Durbin, A.; Ziervogel, K.; Cox, C.; Steen, A.D.; Teske, A. Verrucomicrobia are candidates for polysaccharide-degrading bacterioplankton in an arctic fjord of Svalbard. Appl. Environ. Microbiol. 2014, 80, 3749–3756. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Yarza, P.; Rapp, J.Z.; Glöckner, F.O. Expanding the world of marine bacterial and archaeal clades. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Wegner, C.-E.; Richter-Heitmann, T.; Klindworth, A.; Klockow, C.; Richter, M.; Achstetter, T.; Glöckner, F.O.; Harder, J. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genom. 2013, 9, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Paley, S.M.; Krummenacker, M.; Latendresse, M.; Dale, J.M.; Lee, T.J.; Kaipa, P.; Gilham, F.; Spaulding, A.; Popescu, L.; et al. Pathway Tools version 23.0: integrated software for pathway/genome informatics and systems biology. Brief. Bioinform. 2010, 11, 40–79. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Burdige, D.J.; Nealson, K.H. Microbial manganese reduction by enrichment cultures from coastal marine sediments. Appl. Environ. Microbiol. 1985, 50, 491–497. [Google Scholar] [CrossRef]
- Stookey, L.L. Ferrozine---a new spectrophotometric reagent for iron. Anal. Chem. 1970, 42, 779–781. [Google Scholar] [CrossRef]
- Brewer, P.G.; Spencer, D.W. Colorimetric determination of manganese in anoxic waters. Limnol. Oceanogr. 1971, 16, 107–110. [Google Scholar] [CrossRef]
- Armstrong, P.B.; Lyons, W.B.; Gaudette, H.E. Application of formaldoxime colorimetric method for the determination of manganese in the pore water of anoxic estuarine sediments. Estuaries 1979, 2, 198–201. [Google Scholar] [CrossRef]
- Tanaka, T.; Kawasaki, K.; Daimon, S.; Kitagawa, W.; Yamamoto, K.; Tamaki, H.; Tanaka, M.; Nakatsu, C.H.; Kamagata, Y. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl. Environ. Microbiol. 2014, 80, 7659–7666. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, J.; Dreiseikelmann, B.; Rohde, M.; Meier-Kolthoff, J.P.; Bunk, B.; Rohde, C. First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol. J. 2014, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Spurr, A.R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969, 26, 31–43. [Google Scholar] [CrossRef]
- Havemeyer, S. Polyphosphate Storage in the Family Beggiatoaceae with a Focus on the Species Beggiatoa Alba; Universität Bremen: Bremen, Germany, 2013. [Google Scholar]
- Bienkowski, M.J.; Conrad, H.E. Structural characterization of the oligosaccharides formed by depolymerization of heparin with nitrous acid. J. Biol. Chem. 1985, 260, 356–365. [Google Scholar] [PubMed]
- Rodriguez-R, L.M.; Gunturu, S.; Harvey, W.T.; Rossello-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Delmont, T.O.; Quince, C.; Shaiber, A.; Esen, Ö.C.; Lee, S.T.M.; Rappé, M.S.; McLellan, S.L.; Lücker, S.; Eren, A.M. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 2018, 3, 804–813. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1901. [Google Scholar] [CrossRef]
- Bale, N.J.; Rijpstra, W.I.C.; Sahonero-Canavesi, D.X.; Oshkin, I.Y.; Belova, S.E.; Dedysh, S.N.; Sinninghe Damste, J.S. Fatty acid and hopanoid adaption to cold in the methanotroph Methylovulum psychrotolerans. Front. Microbiol. 2019, 10, 589. [Google Scholar] [CrossRef]
- Ollivier, B.; Caumette, P.; Garcia, J.L.; Mah, R.A. Anaerobic bacteria from hypersaline environments. Microbiol. Mol. Biol. Rev. 1994, 58, 27–38. [Google Scholar] [CrossRef]
- Ndeh, D.; Rogowski, A.; Cartmell, A.; Luis, A.S.; Baslé, A.; Gray, J.; Venditto, I.; Briggs, J.; Zhang, X.; Labourel, A.; et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 2017, 544, 65–70. [Google Scholar] [CrossRef]
- Damrow, R.; Maldener, I.; Zilliges, Y. The multiple functions of common microbial carbon polymers, glycogen and PHB, during stress responses in the non-diazotrophic cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 2016, 7, 966. [Google Scholar] [CrossRef]
- Khadem, A.F.; van Teeseling, M.C.; van Niftrik, L.; Jetten, M.S.; Op den Camp, H.J.; Pol, A. Genomic and physiological analysis of carbon storage in the verrucomicrobial methanotroph "Ca. Methylacidiphilum Fumariolicum" SolV. Front. Microbiol. 2012, 3, 345. [Google Scholar] [CrossRef]
- Rubin-Blum, M.; Antony, C.P.; Sayavedra, L.; Martinez-Perez, C.; Birgel, D.; Peckmann, J.; Wu, Y.C.; Cardenas, P.; MacDonald, I.; Marcon, Y.; et al. Fueled by methane: deep-sea sponges from asphalt seeps gain their nutrition from methane-oxidizing symbionts. ISME J. 2019, 13, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, P.L. Microbial glycosaminoglycan glycosyltransferases. Glycobiology 2002, 12, 9R–16R. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 11, 2399–2406. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Bale, N.J.; Sorokin, D.Y.; Hopmans, E.C.; Koenen, M.; Rijpstra, W.I.C.; Villanueva, L.; Wienk, H.; Sinninghe Damsté, J.S. New insights into the polar lipid composition of extremely halo(alkali)philic Euryarchaea from hypersaline lakes. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Giordano, A.; Vella, F.M.; Romano, I.; Gambacorta, A. Structural elucidation of a novel phosphoglycolipid isolated from six species of Halomonas. J. Lipid Res. 2007, 48, 1825–1831. [Google Scholar] [CrossRef]
- Ale, M.T.; Meyer, A.S. Fucoidans from brown seaweeds: an update on structures, extraction techniques and use of enzymes as tools for structural elucidation. RSC Adv. 2013, 3, 8131–8141. [Google Scholar] [CrossRef]
- Dong, S.; Chang, Y.; Shen, J.; Xue, C.; Chen, F. Purification, expression and characterization of a novel α-L-fucosidase from a marine bacteria Wenyingzhuangia fucanilytica. Protein Expr. Purif. 2017, 129, 9–17. [Google Scholar] [CrossRef]
- Colin, S.; Deniaud, E.; Jam, M.; Descamps, V.; Chevolot, Y.; Kervarec, N.; Yvin, J.C.; Barbeyron, T.; Michel, G.; Kloareg, B. Cloning and biochemical characterization of the fucanase FcnA: definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology 2006, 16, 1021–1032. [Google Scholar] [CrossRef]
- Descamps, V.; Colin, S.; Lahaye, M.; Jam, M.; Richard, C.; Potin, P.; Barbeyron, T.; Yvin, J.C.; Kloareg, B. Isolation and culture of a marine bacterium degrading the sulfated fucans from marine brown algae. Mar. Biotechnol. 2006, 8, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Silchenko, A.S.; Kusaykin, M.I.; Kurilenko, V.V.; Zakharenko, A.M.; Isakov, V.V.; Zaporozhets, T.S.; Gazha, A.K.; Zvyagintseva, T.N. Hydrolysis of fucoidan by fucoidanase isolated from the marine bacterium, Formosa algae. Mar. Drugs 2013, 11, 2413–2430. [Google Scholar] [CrossRef] [PubMed]
- Ohshiro, T.; Harada, N.; Kobayashi, Y.; Miki, Y.; Kawamoto, H. Microbial fucoidan degradation by Luteolibacter algae H18 with deacetylation. Biosci. Biotechnol. Biochem. 2012, 76, 620–623. [Google Scholar] [CrossRef] [PubMed]
- Silchenko, A.S.; Rasin, A.B.; Zueva, A.O.; Kusaykin, M.I.; Zvyagintseva, T.N.; Kalinovsky, A.I.; Kurilenko, V.V.; Ermakova, S.P. Fucoidan sulfatases from marine bacterium Wenyingzhuangia fucanilytica CZ1127T. Biomolecules 2018, 8, 98. [Google Scholar] [CrossRef]
- Fernández-Gomez, B.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.G.; González, J.M.; Pedros-Alio, C. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 2013, 7, 1026. [Google Scholar] [CrossRef]
- Barbeyron, T.; Thomas, F.; Barbe, V.; Teeling, H.; Schenowitz, C.; Dossat, C.; Goesmann, A.; Leblanc, C.; Oliver Glöckner, F.; Czjzek, M.; et al. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans DsijT. Environ. Microbiol. 2016, 18, 4610–4627. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.; Brazel, D.M.; Swan, B.K.; Arnosti, C.; Chain, P.S.; Reitenga, K.G.; Xie, G.; Poulton, N.J.; Lluesma Gomez, M.; Masland, D.E.; et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS ONE 2012, 7, e35314. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef]
- He, S.; Stevens, S.L.R.; Chan, L.K.; Bertilsson, S.; Glavina Del Rio, T.; Tringe, S.G.; Malmstrom, R.R.; McMahon, K.D. Ecophysiology of freshwater Verrucomicrobia inferred from metagenome-assembled genomes. mSphere 2017, 2, e00277-00217. [Google Scholar] [CrossRef] [PubMed]
- Rabus, R.; Hansen, T.A.; Widdel, F. Dissimilatory sulfate-and sulfur-reducing prokaryotes. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2013; pp. 309–404. [Google Scholar]
- Elshahed, M.S.; Youssef, N.H.; Luo, Q.; Najar, F.Z.; Roe, B.A.; Sisk, T.M.; Buhring, S.I.; Hinrichs, K.U.; Krumholz, L.R. Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide- and sulfur-rich Zodletone Spring, Oklahoma. Appl. Environ. Microbiol. 2007, 73, 4707–4716. [Google Scholar] [CrossRef] [PubMed]
- Slobodkina, G.B.; Kovaleva, O.L.; Miroshnichenko, M.L.; Slobodkin, A.I.; Kolganova, T.V.; Novikov, A.A.; van Heerden, E.; Bonch-Osmolovskaya, E.A. Thermogutta terrifontis gen. nov., sp. nov. and Thermogutta hypogea sp. nov., thermophilic anaerobic representatives of the phylum Planctomycetes. Int. J. Syst. Evol. Microbiol. 2015, 65, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Slobodkina, G.B.; Panteleeva, A.N.; Beskorovaynaya, D.A.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Thermostilla marina gen. nov., sp. nov., a thermophilic, facultatively anaerobic planctomycete isolated from a shallow submarine hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2016, 66, 633–638. [Google Scholar] [CrossRef]
- Sánchez-Andrea, I.; Florentino, A.P.; Semerel, J.; Strepis, N.; Sousa, D.Z.; Stams, A.J.M. Co-culture of a novel fermentative bacterium, Lucifera butyrica gen. nov. sp. nov., with the sulfur reducer Desulfurella amilsii for enhanced sulfidogenesis. Front. Microbiol. 2018, 9, 3108. [Google Scholar] [CrossRef]
- Duval, S.; Ducluzeau, A.L.; Nitschke, W.; Schoepp-Cothenet, B. Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol. Biol. 2008, 8, 206. [Google Scholar] [CrossRef]
- Wasmund, K.; Mussmann, M.; Loy, A. The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 2017, 9, 323–344. [Google Scholar] [CrossRef]
- Benz, M.; Schink, B.; Brune, A. Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl. Environ. Microbiol. 1998, 64, 4507–4512. [Google Scholar] [CrossRef]
- Lovley, D. Dissimilatory Fe(III)- and Mn(IV)-reducing prokaryotes. In The Prokaryotes; Rosenberg, E., DeLong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Vandieken, V.; Marshall, I.P.G.; Niemann, H.; Engelen, B.; Cypionka, H. Labilibaculum manganireducens gen. nov., sp. nov. and Labilibaculum filiforme sp. nov., novel Bacteroidetes isolated from subsurface sediments of the Baltic Sea. Front. Microbiol. 2017, 8, 2614. [Google Scholar] [CrossRef]
- Mehta, T.; Coppi, M.V.; Childers, S.E.; Lovley, D.R. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 2005, 71, 8634–8641. [Google Scholar] [CrossRef]
- Voordeckers, J.W.; Kim, B.C.; Izallalen, M.; Lovley, D.R. Role of Geobacter sulfurreducens outer surface c-type cytochromes in reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl. Environ. Microbiol. 2010, 76, 2371–2375. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, H. Chapter V: Enrichment cultures of prokaryotic organisms. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 1970; Volume 3, pp. 305–361. [Google Scholar]
- Khan, M.T.; Duncan, S.H.; Stams, A.J.M.; van Dijl, J.M.; Flint, H.J.; Harmsen, H.J.M. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012, 6, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; van der Ark, K.C.; Davids, M.; Claassens, N.J.; Robert Finestra, T.; de Vos, W.M.; Belzer, C. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl. Environ. Microbiol. 2016, 82, 6983–6993. [Google Scholar] [CrossRef] [PubMed]
- Pitcher, R.S.; Watmough, N.J. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta (BBA)-Bioenerg. 2004, 1655, 388–399. [Google Scholar] [CrossRef]
- Borisov, V.B.; Gennis, R.B.; Hemp, J.; Verkhovsky, M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 1398–1413. [Google Scholar] [CrossRef]
- Cho, J.C.; Vergin, K.L.; Morris, R.M.; Giovannoni, S.J. Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environ. Microbiol. 2004, 6, 611–621. [Google Scholar] [CrossRef]
- Delbarre-Ladrat, C.; Sinquin, C.; Lebellenger, L.; Zykwinska, A.; Colliec-Jouault, S. Exopolysaccharides produced by marine bacteria and their applications as glycosaminoglycan-like molecules. Front. Chem. 2014, 2, 85. [Google Scholar] [CrossRef]
- Aquino, R.S.; Landeira-Fernandez, A.M.; Valente, A.P.; Andrade, L.R.; Mourão, P.A.S. Occurrence of sulfated galactans in marine angiosperms: evolutionary implications. Glycobiology 2005, 15, 11–20. [Google Scholar] [CrossRef]
- Troeberg, L.; Mulloy, B.; Ghosh, P.; Lee, M.H.; Murphy, G.; Nagase, H. Pentosan polysulfate increases affinity between ADAMTS-5 and TIMP-3 through formation of an electrostatically driven trimolecular complex. Biochem. J. 2012, 443, 307–315. [Google Scholar] [CrossRef][Green Version]
- DeAngelis, P.L. Evolution of glycosaminoglycans and their glycosyltransferases: Implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 2002, 268, 317–326. [Google Scholar] [CrossRef]
- Widner, B.; Behr, R.; Von Dollen, S.; Tang, M.; Heu, T.; Sloma, A.; Sternberg, D.; Deangelis, P.L.; Weigel, P.H.; Brown, S. Hyaluronic acid production in Bacillus subtilis. Appl. Environ. Microbiol. 2005, 71, 3747–3752. [Google Scholar] [CrossRef] [PubMed]
- Felz, S.; Neu, T.R.; van Loosdrecht, M.C.M.; Lin, Y. Aerobic granular sludge contains Hyaluronic acid-like and sulfated glycosaminoglycans-like polymers. Water Res. 2020, 169, 115291. [Google Scholar] [CrossRef] [PubMed]
- Bourven, I.; Bachellerie, G.; Costa, G.; Guibaud, G. Evidence of glycoproteins and sulphated proteoglycan-like presence in extracellular polymeric substance from anaerobic granular sludge. Environ. Technol. 2015, 36, 2428–2435. [Google Scholar] [CrossRef] [PubMed]
- Boleij, M.; Kleikamp, H.; Pabst, M.; Neu, T.R.; Van Loosdrecht, M.C.M.; Lin, Y. Decorating the anammox house: sialic acids and sulfated glycosaminoglycans in the extracellular polymeric substances of anammox granular sludge. Environ. Sci. Technol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kusche-Gullberg, M.; Kjellén, L. Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 2003, 13, 605–611. [Google Scholar] [CrossRef] [PubMed]




| Polysaccharide | Source/Type | Distributor | Lot Number |
|---|---|---|---|
| alginic acid | NR | Thermo Fisher Scientific (Waltham, MA, US) | NR |
| arabinan | sugar beet | Megazyme (Bray, Ireland) | 80902b |
| cellulose | microgranular, CC41 | Whatman (Maidstone, UK) | 1441024 |
| chitin | shrimp shells | Sigma-Aldrich (St. Louis, MO, US) | SLBL2694V |
| chitosan | shrimp shells | Sigma-Aldrich (St. Louis, MO, US) | BCBQ3414V |
| chondroitin sulfate | bovine trachea | Sigma-Aldrich (St. Louis, MO, US) | NR |
| laminarin | Eisenia bicyclis | abcr (Karlsruhe, Baden-Württemberg, Germany) | 1025869 |
| pectin | apple | Sigma-Aldrich (St. Louis, MO, US) | BCBK7271V |
| pullulan | Aureobasidium pullulans | Sigma-Aldrich (St. Louis, MO, US) | NR |
| starch | soluble | Sigma-Aldrich (St. Louis, MO, US) | SLBL2691V |
| xanthan gum | Xanthomonas campestris | Sigma-Aldrich (St. Louis, MO, US) | 100M0218V |
| xylan | beechwood | Sigma-Aldrich (St. Louis, MO, US) | 107H1209 |
| κ-carrageenan | NR | Sigma-Aldrich (St. Louis, MO, US) | BCBR6980V |
| ι-carrageenan | NR | Sigma-Aldrich (St. Louis, MO, US) | SLBJ7874V |
| Species | P. desulfatans | P. sulfatireligans | K. glycovorans |
|---|---|---|---|
| Type Strain | F1T | F21T | L21-Fru-ABT |
| Isolation source | Anoxic marine sediment * | Anoxic marine sediment * | Hypersaline microbial mat |
| Cell diameter (μm) | 0.5–1.2 * | 0.5–1.0 * | 1.0–2.0 |
| Genome size (Mbp) | 8.6 * | 7.4 * | 3.0 |
| DNA G+C content (mol%) | 56.0 | 54.6 | 63.3 |
| Quinones | MK-7, MK-6, MK-8 | MK-9, MK-8, MK-6, MK-7 | none |
| Major CFAs (>5% of total) | C18:0, i-C12:0, i-C14:0, i-C18:0 | C18:0, i-C12:0, i-C18:0, i-C14:0, i-C16:0 | i-C14:0, C18:0, i-C18:0 |
| Major IPLs | PG, LCL, CL, MGDG | PG, CL, MGDG, LCL | PG, CL, MGDG, PG-Gly, LCL |
| Oxidase activity | − | + | − |
| Temp. for growth (°C) | |||
| Range | 10–30 * | 0–25 | 20–40 |
| Optimum | 25 * | 25 * | 28 |
| NaCl conc. for growth (g L−1) | |||
| Range | 10–31 | 10–50 | 20–180 |
| Optimum | 23 | 23 | 60–70 |
| pH for growth | |||
| Range | 6.5–8.5 | 6.0–8.5 | 6.5–8.0 |
| Optimum | 7.5 | 7.5 | 7.5 |
| Substrate utilization | |||
| Chondroitin sulfate | + * | + * | - |
| Fucoidan | + * | + * | +/- |
| Iota-carrageenan | − * | + * | +/- |
| Arabinose | + * | − * | − |
| Cellobiose | + * | + * | − |
| Fructose | + * | + * | − |
| Fucose | + * | + * | − |
| Galactose | + * | + * | +/− |
| Galacturonate | − * | + * | NDA |
| Lactose | + * | + * | − |
| Maltose | + * | + * | − |
| Mannitol | − * | + * | − |
| Mannose | − * | + * | + |
| Rhamnose | + * | + * | +/− |
| Sucrose | + * | + * | − |
| Tagatose | + * | - * | NDA |
| Trehalose | + * | + * | − |
| Major fermentation productsfrom L-fucose | Acetate, H2, ethanol, lactate * | Acetate, ethanol, H2, 1,2-propanediol * | − |
| Major non-gaseous fermentation products from D-glucose | Acetate, ethanol, lactate | Acetate, ethanol, lactate | Ethanol, acetate |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Vliet, D.M.; Lin, Y.; Bale, N.J.; Koenen, M.; Villanueva, L.; Stams, A.J.M.; Sánchez-Andrea, I. Pontiella desulfatans gen. nov., sp. nov., and Pontiella sulfatireligans sp. nov., Two Marine Anaerobes of the Pontiellaceae fam. nov. Producing Sulfated Glycosaminoglycan-like Exopolymers. Microorganisms 2020, 8, 920. https://doi.org/10.3390/microorganisms8060920
van Vliet DM, Lin Y, Bale NJ, Koenen M, Villanueva L, Stams AJM, Sánchez-Andrea I. Pontiella desulfatans gen. nov., sp. nov., and Pontiella sulfatireligans sp. nov., Two Marine Anaerobes of the Pontiellaceae fam. nov. Producing Sulfated Glycosaminoglycan-like Exopolymers. Microorganisms. 2020; 8(6):920. https://doi.org/10.3390/microorganisms8060920
Chicago/Turabian Stylevan Vliet, Daan M., Yuemei Lin, Nicole J. Bale, Michel Koenen, Laura Villanueva, Alfons J. M. Stams, and Irene Sánchez-Andrea. 2020. "Pontiella desulfatans gen. nov., sp. nov., and Pontiella sulfatireligans sp. nov., Two Marine Anaerobes of the Pontiellaceae fam. nov. Producing Sulfated Glycosaminoglycan-like Exopolymers" Microorganisms 8, no. 6: 920. https://doi.org/10.3390/microorganisms8060920
APA Stylevan Vliet, D. M., Lin, Y., Bale, N. J., Koenen, M., Villanueva, L., Stams, A. J. M., & Sánchez-Andrea, I. (2020). Pontiella desulfatans gen. nov., sp. nov., and Pontiella sulfatireligans sp. nov., Two Marine Anaerobes of the Pontiellaceae fam. nov. Producing Sulfated Glycosaminoglycan-like Exopolymers. Microorganisms, 8(6), 920. https://doi.org/10.3390/microorganisms8060920





