Metschnikowia pulcherrima and Related Pulcherrimin-Producing Yeasts: Fuzzy Species Boundaries and Complex Antimicrobial Antagonism
Abstract
:1. Introduction
2. Taxonomy, Evolution and Taxonomic Identification
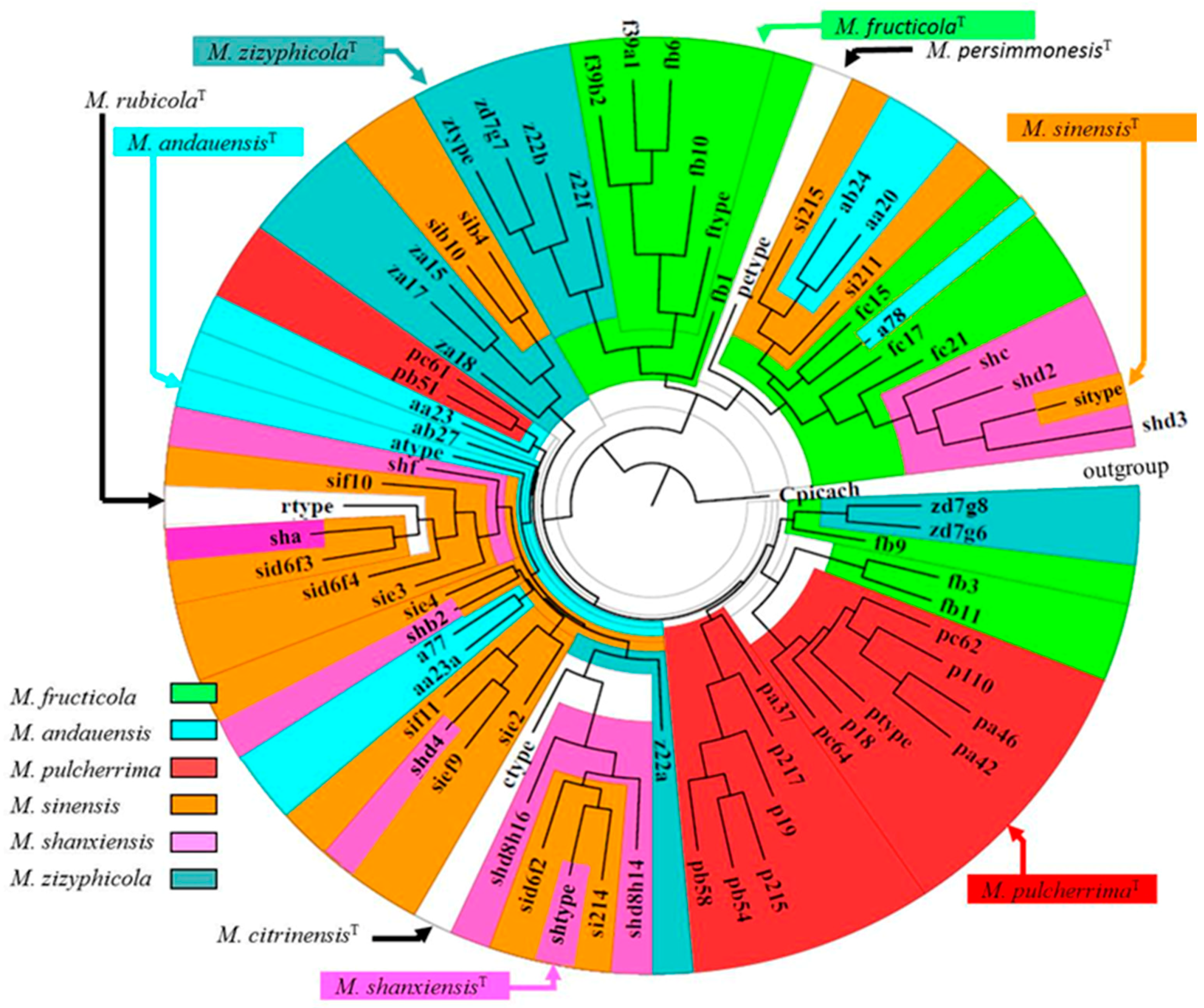
2.1. The M. pulcherrima Clade
2.2. Intragenomic rDNA Repeat Heterogeneity (Lack of rDNA Homogenisation): Limited Applicability of rDNA to Barcoding of Pulcherrimin-Producing Metschnikowia Species
2.3. The Possible Alternative to rDNA Homogenisation: Birth-and-Death Evolution of Repeats
2.4. Lack of Clear rDNA Barcode Gaps: Fuzzy Species Boundaries
2.5. Other Barcode Sequences
2.6. Hybridisation and Chimeric (“Hybrid”) Genomes
3. Antimicrobial Antagonism
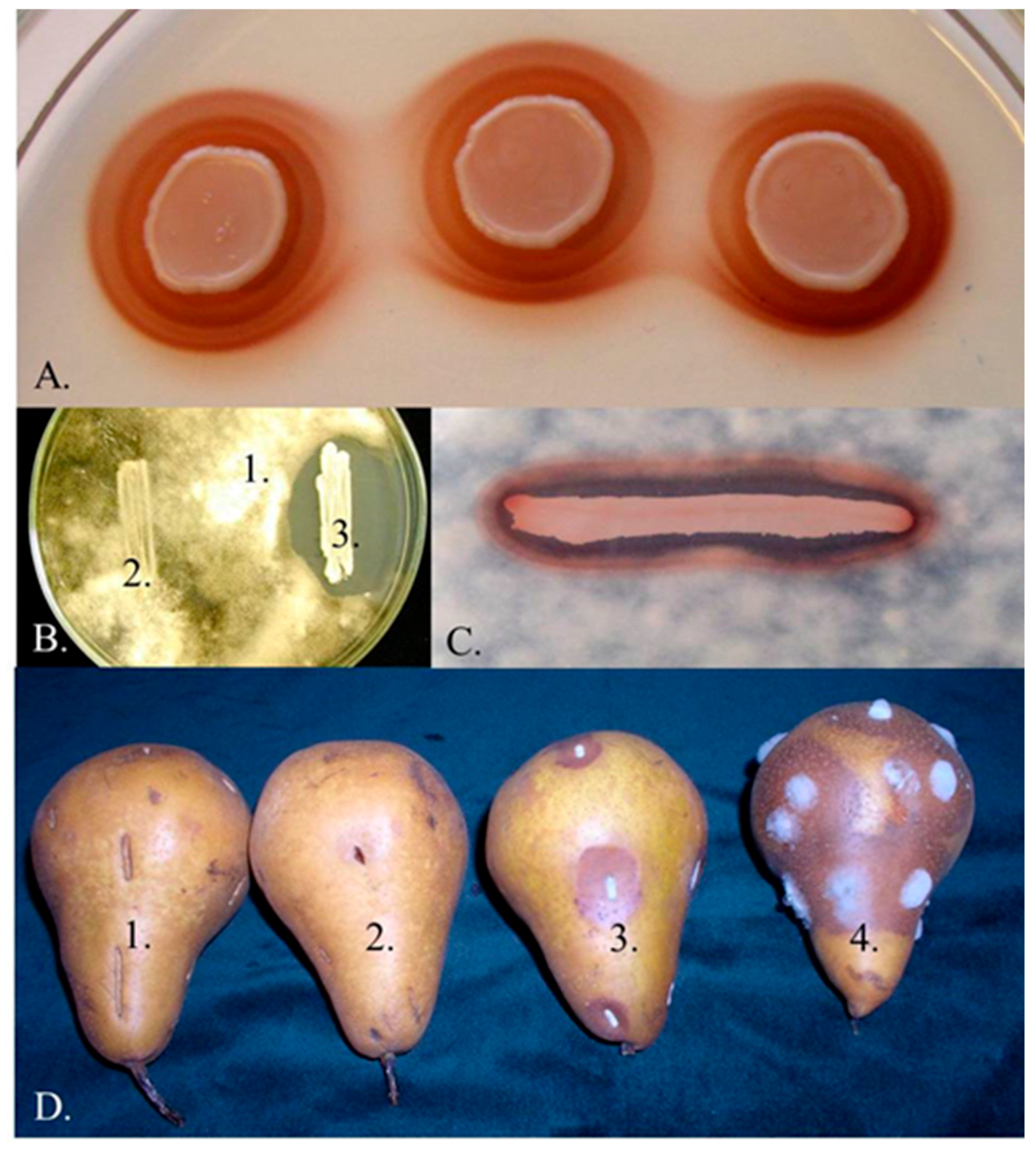
3.1. Iron Depletion by Pulcherrimin Production
3.2. Competition for Nutrients
3.3. Secretion of Enzymes
3.4. Release of Volatile Compounds
3.5. Biofilm Formation and Adhesion to the Plant Surface and Fungal Hyphae
3.6. Indirect Antagonism through Modulation of Plant Defence Response
4. Concluding Remarks and Perspectives
Supplementary Materials
Funding
Conflicts of Interest
References
- Lachance, M.A. Metschnikowia: Half tetrads, a regicide and the fountain of youth. Yeast 2016, 33, 563–574. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar, E.; Ward, T.J. Four new species of Metschnikowia and the transfer of seven Candida species to Metschnikowia and Clavispora as new combinations. Antonie Van Leeuwenhoek 2018, 111, 2017–2035. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Escott, C.; del Fresno, J.M.; Bañuelos, M.A.; Suárez-Lepe, J.A. Applications of Metschnikowia pulcherrima in wine biotechnology. Fermentation 2019, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Abeln, F.; Chuck, C.J. Achieving a high-density oleaginous yeast culture: Comparison of four processing strategies using Metschnikowia pulcherrima. Biotechnol. Bioeng. 2019, 116, 3200–3214. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Droby, S. Metschnikowia fructicola, a new ascosporic yeast with potential for biocontrol of postharvest fruit rots. Syst. Appl. Microbiol. 2001, 24, 395–399. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef] [Green Version]
- Saravanakumar, D.; Ciavorella, A.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Metschnikowia pulcherrima strain MACH1 outcompetes Botrytis cinerea, Alternaria alternata and Penicillium expansum in apples through iron depletion. Postharvest Biol. Technol. 2008, 49, 121–128. [Google Scholar] [CrossRef]
- Türkel, S.; Korukluoglu, M.; Yavuz, M. Biocontrol activity of the local strain of Metschnikowia pulcherrima on different postharvest pathogens. Biotechnol. Res. Int. 2014, 2014, 397167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kántor, A.; Hutková, J.; Petrová, J.; Hleba, L.; Kačániová, M. Antimicrobial activity of pulcherrimin pigment produced by Metschnikowia pulcherrima against various yeast species. J. Microbiol. Biotechnol. Food Sci. 2015, 5, 282–285. [Google Scholar] [CrossRef] [Green Version]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol ability and action mechanism of food-isolated yeast strains against Botrytis cinerea causing post-harvest bunch rot of table grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef]
- Liu, Y.; Yi, L.; Ruan, C.; Yao, S.; Deng, L.; Zeng, K. Proline increases pigment production to improve oxidative stress tolerance and biocontrol ability of Metschnikowia citriensis. Front. Microbiol. 2019, 10, 1273. [Google Scholar] [CrossRef] [PubMed]
- Pawlikowska, E.; James, S.A.; Breierova, E.; Antolak, H.; Kregiel, D. Biocontrol capability of local Metschnikowia sp. isolates. Antonie Van Leeuwenhoek 2019, 112, 1425–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sipiczki, M.; Pfliegler, W.P.; Holb, I.J. Metschnikowia species share a pool of diverse rRNA genes differing in regions that determine hairpin-loop structures and evolve by reticulation. PLoS ONE 2013, 8, e67384. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M.; Horvath, E.; Pfliegler, W.P. Birth-and-death evolution and reticulation of ITS segments of Metschnikowia andauensis and Metschnikowia fructicola rDNA repeats. Front. Microbiol. 2018, 9, 1193. [Google Scholar] [CrossRef] [Green Version]
- Piombo, E.; Sela, N.; Wisniewski, M.; Hoffmann, M.; Gullino, M.L.; Allard, M.W.; Levin, E.; Spadaro, D.; Droby, S. Genome sequence, assembly and characterization of two Metschnikowia fructicola strains used as biocontrol agents of postharvest diseases. Front. Microbiol. 2018, 9, 593. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, A.; Murray, A.L.; Boyle, A.B.; Quinn Farrington, L.; Maher, T.J.; Ó’Gaora, P.; Wolfe, K.H.; O’Brien, C.E.; Butler, G. Draft genome sequence of a highly heterozygous yeast strain from the Metschnikowia pulcherrima subclade, UCD127. Genome Announc. 2018, 6, e00550-18. [Google Scholar] [CrossRef] [Green Version]
- Gore-Lloyd, D.; Sumann, I.; Brachmann, A.O.; Schneeberger, K.; Ortiz-Merino, R.A.; Moreno-Beltrán, M.; Schläfli, M.; Kirner, P.; Santos Kron, A.; Rueda-Mejia, M.P.; et al. Snf2 controls pulcherriminic acid biosynthesis and antifungal activity of the biocontrol yeast Metschnikowia pulcherrima. Mol. Microbiol. 2019, 112, 317–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molnar, O.; Prillinger, H. Analysis of yeast isolates related to Metschnikowia pulcherrima using the partial sequences of the large subunit rDNA and the actin gene; description of Metschnikowia andauensis sp. nov. Syst. Appl. Microbiol. 2005, 28, 717–726. [Google Scholar] [CrossRef]
- Xue, M.L.; Zhang, L.Q.; Wang, Q.M.; Zhang, J.S.; Bai, F.Y. Metschnikowia sinensis sp. nov., Metschnikowia zizyphicola sp. nov. and Metschnikowia shanxiensis sp. nov., novel yeast species from jujube fruit. Int. J. Syst. Evol. Microbiol. 2006, 56, 2245–2250. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M. Overwintering of vineyard yeasts: Survival of interacting yeast communities in grapes mummified on vines. Front. Microbiol. 2016, 7, 212. [Google Scholar] [CrossRef]
- Türkel, S.; Ener, B. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z. Naturforsch. C J. Biosci. 2009, 64, 405–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Selection and evaluation of new antagonists for their efficacy against postharvest brown rot of peaches. Postharvest Biol. Technol. 2009, 55, 174–181. [Google Scholar] [CrossRef]
- Vadkertiova, R.; Molnarova, J.; Vranova, D.; Slavikova, E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can. J. Microbiol. 2012, 58, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Csutak, O.; Vassu, T.; Cornea, P.; Grebenisan, I. Genetic characterization of two new Metschnikowia strains with antifungal activity. Rom. Biotech. Lett. 2007, 12, 3175–3182. [Google Scholar]
- Csutak, O.; Vassu, T.; Sarbu, I.; Stoica, I.; Cornea, P. Antagonistic activity of three newly isolated yeast strains from the surface of fruits. Food Technol. Biotechnol. 2013, 51, 70–77. [Google Scholar]
- Hadwiger, L.A.; McDonel, H.; Glawe, D. Wild yeast strains as prospective candidates to induce resistance against potato late blight (Phytophthora infestans). Am. J. Potato Res. 2015, 92, 379–386. [Google Scholar] [CrossRef]
- Kang, Y.M.; Choi, J.E.; Komakech, R.; Park, J.H.; Kim, D.W.; Cho, K.M.; Kang, S.M.; Choi, S.H.; Song, K.C.; Ryu, C.M.; et al. Characterization of a novel yeast species Metschnikowia persimmonesis KCTC 12991BP (KIOM G15050 type strain) isolated from a medicinal plant, Korean persimmon calyx (Diospyros kaki Thumb). AMB Express 2017, 7, 199. [Google Scholar] [CrossRef] [Green Version]
- Pawlikowska, E.; Kręgiel, D. Enzymatic profiles and antimicrobial activity of the yeast Metschnikowia pulcherrima. Acta Innov. 2017, 23, 17–24. [Google Scholar]
- Liu, Y.; Wang, W.; Zhou, Y.; Yaoa, S.; Denga, L.; Zeng, K. Isolation, identification and in vitro screening of Chongqing orangery yeasts for the biocontrol of Penicillium digitatum on citrus fruit. Biol. Control 2017, 110, 18–24. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, S.; Deng, L.; Ming, J.; Zeng, K. Metschnikowia citriensis sp. nov., a novel yeast species isolated from leaves with potential for biocontrol of postharvest fruit rot. Biol. Control 2018, 125, 15–19. [Google Scholar] [CrossRef]
- Tian, Y.Q.; Li, W.; Jiang, Z.T.; Jing, M.M.; Shao, Y.Z. The preservation effect of Metschnikowia pulcherrima yeast on anthracnose of postharvest mango fruits and the possible mechanism. Food Sci. Biotechnol. 2017, 27, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Lage, P.; Esteves, M.; Chambel, L.; Mendes-Faia, A.; Mendes-Ferreira, A. Molecular and phenotypic characterization of Metschnikowia pulcherrima strains from Douro wine region. Fermentation 2018, 4, 8. [Google Scholar] [CrossRef] [Green Version]
- Lachance, M.A. Metschnikowia Kamienski (1899). In The Yeasts. A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 575–620. [Google Scholar]
- Genç, T.T. Effects of various environmental conditions on pulcherrimin production and extracellular enzyme profiles of Metschnikowia pulcherrima. SAR J. 2020, 3, 10–16. [Google Scholar]
- Naumov, G.I. Molecular and genetic differentiation of small_spored species of the yeast genus Metschnikowia Kamienski. Microbiology 2011, 80, 135–142. [Google Scholar] [CrossRef]
- Schlotterer, C.; Hauser, M.-T.; von Haeseler, A.; Tautz, D. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol. Biol. Evol. 1994, 11, 513–522. [Google Scholar]
- Hillis, D.M.; Dixon, M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991, 66, 411–453. [Google Scholar] [CrossRef]
- Lofgren, L.A.; Uehling, J.K.; Branco, S.; Bruns, T.D.; Martin, F.; Kennedy, P.G. Genome-based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol. Ecol. 2019, 28, 721–730. [Google Scholar] [CrossRef]
- Torres-Machorro, A.L.; Hernandez, R.; Cevallos, A.M.; Lopez-Villasenor, I. Ribosomal RNA genes in eukaryotic microorganisms: Witnesses of phylogeny? FEMS Microbiol. Rev. 2010, 34, 59–86. [Google Scholar] [CrossRef] [Green Version]
- Nei, M.; Rooney, A.P. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005, 39, 121–152. [Google Scholar] [CrossRef] [Green Version]
- Eickbush, T.H.; Eickbush, D.G. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics 2007, 175, 477–485. [Google Scholar] [CrossRef] [Green Version]
- Naidoo, K.; Steenkamp, E.T.; Coetzee, M.P.A.; Wingfield, M.J.; Wingfield, B.D. Concerted evolution in the ribosomal RNA cistron. PLoS ONE 2013, 8, e59355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtzman, C.P. Yeast systematics-from phenotype to genotype. Food Technol. Biotechnol. 1998, 36, 261–266. [Google Scholar]
- Kurtzman, C.P. Recognition of yeast species from gene sequence comparisons. Open Appl. Inform. J. 2011, 5, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Wurzbacher, C.; Larsson, E.; Bengtsson-Palme, J.; Van den Wyngaert, S.; Svantesson, S.; Kristiansson, E.; Kagami, M.; Nilsson, R.H. Introducing ribosomal tandem repeat barcoding for fungi. Mol. Ecol. Resour. 2019, 19, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Bourret, T.B.; Grove, G.G.; Vandemark, G.J.; Henick-Kling, T.; Glawe, D.A. Diversity and molecular determination of wild yeasts in a central Washington State vineyard. N. Am. Fungi 2013, 8, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Guo, J.; Liu, F.; Liu, Y. Identification of indigenous yeast flora isolated from the five winegrape varieties harvested in Xiangning, China. Antonie Van Leeuwenhoek 2014, 105, 533–540. [Google Scholar] [CrossRef]
- Brysch-Herzberg, M.; Seidel, M. Yeast diversity on grapes in two German wine growing regions. Int. J. Food Microbiol. 2015, 214, 137–144. [Google Scholar] [CrossRef]
- Savini, V.; Hendrickx, M.; Sisti, M.; Masciarelli, G.; Favaro, L.; Arzeni, D.; Astolfi, D.; Catavitello, C.; Polilli, E.; Farina, C.; et al. An atypical, pigment-producing Metschnikowia strain from a leukaemia patient. Med. Mycol. 2013, 51, 438–443. [Google Scholar] [CrossRef] [Green Version]
- Rooney, A.P.; Ward, T.J. Evolution of a large ribosomal RNA multigene family in filamentous fungi: Birth and death of a concerted evolution paradigm. Proc. Natl. Acad. Sci. USA 2005, 102, 5084–5089. [Google Scholar] [CrossRef] [Green Version]
- Vu, D.; Groenewald, M.; Szoke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vries, M.; Verkleij, G.J.M.; Crous, P.W.; Boelhout, T.; et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Lachance, M.A.; Daniel, H.M.; Meyer, W.; Prasad, G.S.; Gautam, S.P.; Boundy-Mills, K. The D1/D2 domain of the large subunit rDNA of the yeast species Clavispora lusitaniae is unusually polymorphic. FEMS Yeast Res. 2003, 4, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Ganley, A.R.; Kobayashi, T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007, 17, 184–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alper, I.; Frenette, M.; Labrie, S. Ribosomal DNA polymorphisms in the yeast Geotrichum candidum. Fungal. Biol. 2011, 115, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Lindner, D.L.; Banik, M.T. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia 2011, 103, 731–740. [Google Scholar] [CrossRef] [Green Version]
- Chand Dakal, T.; Giudici, P.; Solieri, L. Contrasting patterns of rDNA homogenization within the Zygosaccharomyces rouxii species complex. PLoS ONE 2016, 11, e0160744. [Google Scholar] [CrossRef] [Green Version]
- Sternes, P.R.; Lee, D.; Kutyna, D.R.; Borneman, A.R. A combined meta-barcoding and shotgun metagenomic analysis of spontaneous wine fermentation. Gigascience 2017, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Guzmán, B.; Lachance, M.A.; Herrera, C.M. Phylogenetic analysis of the angiosperm-floricolous insect-yeast association: Have yeast and angiosperm lineages co-diversified? Mol. Phylogenet. Evol. 2013, 68, 161–175. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, K.H. Origin of the yeast whole-genome duplication. PLoS Biol. 2015, 13, e1002221. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M. Interspecies hybridisation and genome chimerisation in Saccharomyces: Combining of gene pools of species and its biotechnological perspectives. Front. Microbiol. 2018, 9, 3071. [Google Scholar] [CrossRef]
- Hershkovitz, V.; Sela, N.; Taha-Salaime, L.; Liu, J.; Rafael, G.; Kessler, C.; Aly, R.; Levy, M.; Wisniewski, M.; Droby, S. De-novo assembly and characterization of the transcriptome of Metschnikowia fructicola reveals differences in gene expression following interaction with Penicillium digitatum and grapefruit peel. BMC Genom. 2013, 14, 168. [Google Scholar] [CrossRef] [Green Version]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol yeasts: Mechanisms and applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Sui, Y.; Wisniewski, M.; Droby, S.; Liu, Y. Review: Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int. J. Food Microbiol. 2013, 167, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yao, S.; Deng, L.; Ming, J.; Zeng, K. Different mechanisms of action of isolated epiphytic yeasts against Penicillium digitatum and Penicillium italicum on citrus fruit. Postharvest Biol. Technol. 2019, 152, 100–110. [Google Scholar] [CrossRef]
- Muccilli, S.; Restuccia, C. Bioprotective role of yeasts. Microorganisms 2015, 3, 588–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlikowska, E.; Kręgiel, D. Niekonwencjonalne drozdze Metschnikowia pulcherrima i ich zastosowanie w biotechnologii. Post. Mikrobiol. 2017, 56, 405–415. [Google Scholar]
- Dukare, A.S.; Paul, S.; Nambi, V.E.; Gupta, R.K.; Singh, R.; Sharma, K.; Vishwakarma, R.R. Exploitation of microbial antagonists for the control of postharvest diseases of fruits: A review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1498–1513. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Nardi, T. Microbial resources as a tool for enhancing sustainability in winemaking. Microorganisms 2020, 8, 507. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Verma, J.P.; Gaurav, A.K.; Chouhan, G.K.; Patel, J.S.; El-Lstif Hesham, A. Yeast a potential bio-agent: Future for plant growth and postharvest disease management for sustainable agriculture. Appl. Microbiol. Biotechnol. 2020, 104, 1497–1510. [Google Scholar] [CrossRef]
- Oro, L.; Ciani, M.; Comitini, F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 2014, 116, 1209–1217. [Google Scholar] [CrossRef]
- Pretscher, J.; Fischkal, T.; Branscheidt, S.; Jäger, L.; Kahl, S.; Schlander, M.; Thines, E.; Claus, H. Yeasts from different habitats and their potential as biocontrol agents. Fermentation 2018, 4, 31. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Ruan, C.; Yi, L.; Deng, L.; Yao, S.; Zeng, K. Biocontrol ability and action mechanism of Metschnikowia citriensis against Geotrichum citri-aurantii causing sour rot of postharvest citrus fruit. Food Microbiol. 2020, 87, 103375. [Google Scholar] [CrossRef] [PubMed]
- Piano, S.; Neyrotti, V.; Migheli, Q.; Gullino, M.L. Biocontrol capability of Metschnikowia pulcherrima against Botrytis postharvest rot of apple. Postharvest Biol. Technol. 1997, 11, 131–140. [Google Scholar] [CrossRef]
- Manso, T.; Vero, S.; González, M.E.; Nunes, C. Study of modes of action of the biocontrol agent Metschnikowia andauensis PBC-2. In Environmentally Friendly and Safe Technologies for Quality of Fruit and Vegetables; Nunes, C., Ed.; Universidade do Algarve: Faro, Portugal, 2011; pp. 140–146. [Google Scholar]
- Saravanakumar, D.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Detection of enzymatic activity and partial sequence of a chitinase gene in Metschnikowia pulcherrima strain MACH1 used as postharvest biocontrol agent. Eur. J. Plant Pathol. 2009, 123, 183–193. [Google Scholar] [CrossRef]
- Oro, L.; Feliziani, E.; Ciani, M.; Romanazzi, G.; Comitini, F. Volatile organic compounds from Wickerhamomyces anomalus, Metschnikowia pulcherrima and Saccharomyces cerevisiae inhibit growth of decay causing fungi and control postharvest diseases of strawberries. Int. J. Food Microbiol. 2018, 265, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Macarisin, D.; Droby, S.; Bauchan, G.; Wisniewski, M. Superoxide anion and hydrogen peroxide in the yeast antagonist-fruit interaction: A new role for reactive oxygen species in postharvest biocontrol? Postharvest Biol. Technol. 2010, 58, 194–202. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 52, 137–145. [Google Scholar] [CrossRef]
- Hershkovitz, V.; Ben-Dayan, C.; Raphael, G.; Pasmanik-Chor, M.; Liu, J.; Belusov, E.; Aly, R.; Wisniewski, M.; Droby, S. Global changes in gene expression of grapefruit peel tissue in response to the yeast biocontrol agent Metschnikowia fructicola. Mol. Plant Pathol. 2012, 13, 338–349. [Google Scholar] [CrossRef]
- Li, X.; Wang, D.; Cai, D.; Zhan, Y.; Wang, Q.; Chen, S. Identification and high-level production of pulcherrimin in Bacillus licheniformis DW2. Appl. Biochem. Biotechnol. 2017, 183, 1323–1335. [Google Scholar] [CrossRef]
- Randazzo, P.; Aubert-Frambourg, A.; Guillot, A.; Auger, S. The MarR-like protein PchR (YvmB) regulates expression of genes involved in pulcherriminic acid biosynthesis and in the initiation of sporulation in Bacillus subtilis. BMC Microbiol. 2016, 16, 190. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhan, Y.; Cai, D.; Li, X.; Wang, Q.; Chen, S. Regulation of the synthesis and secretion of the iron chelator cyclodipeptide pulcherriminic acid in Bacillus licheniformis. Appl. Environ. Microbiol. 2018, 84, e00262-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, D.J.; Kominek, J.; Opulente, D.A.; Shen, X.X.; Zhou, X.; Langdon, Q.K.; DeVirgilio, J.; Hulfachor, A.B.; Kurtzman, C.P.; Rokas, A.; et al. Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proc. Natl. Acad. Sci. USA 2018, 115, 11030–11035. [Google Scholar] [CrossRef] [Green Version]
- Nigro, F.; Finetti Sialer, M.M.; Gallitelli, D. Transformation of Metschnikowia pulcherrima 320, biocontrol agent of storage rot, with the green fuorescent protein gene. J. Plant Pathol. 1999, 81, 205–208. [Google Scholar]
- Liu, X.; Li, M.; Xia, X.; Li, X.; Chen, Z. Mechanism of chromatin remodelling revealed by the Snf2–nucleosome structure. Nature 2017, 544, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Talibi, I.; Boubaker, H.; Boudyach, E.H.; Ait Ben Aoumar, A. Alternative methods for the control of postharvest citrus diseases. J. Appl. Microbiol. 2014, 117, 1–17. [Google Scholar] [CrossRef]
- Ferraz, P.; Cássio, F.; Lucas, C. Potential of yeasts as biocontrol agents of the phytopathogen causing cacao witches’ broom disease: Is microbial warfare a solution? Front. Microbiol. 2019, 10, 1766. [Google Scholar] [CrossRef] [Green Version]
- Sanz Ferramola, M.I.; Benuzzi, D.; Calvente, V.; Calvo, J.; Sansone, G.; Cerutti, S.; Raba, J. The use of siderophores for improving the control of postharvest diseases in stored fruits and vegetables. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2013; pp. 1385–1394. [Google Scholar]
- Albelda-Berenguer, M.; Monachon, M.; Joseph, E. Siderophores: From natural roles to potential applications. Adv. Appl. Microbiol. 2019, 106, 193–225. [Google Scholar]
- Saha, M.; Sarkar, S.; Sarkar, B.; Sharma, B.K.; Bhattacharjee, S.; Tribedi, P. Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. Int. 2016, 23, 3984–3999. [Google Scholar] [CrossRef]
- Fatima, N.; Javaid, K.; Lahmo, K.; Banday, S.; Sharma, P.; Masoodi, L. Siderophores in fungal physiology and virulence. J. Pharmacog. Phytochem. 2017, 6, 1073–1080. [Google Scholar]
- Zajc, J.; Gostincar, C.; Cernosa, A.; Gunde-Cimerman, N. Stress tolerant yeasts: Opportunistic pathogenicity versus biocontrol potential. Genes (Basel) 2019, 10, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vadkertiová, R.; Sláviková, E. Killer activity of yeasts isolated from natural environments against some medically important Candida species. Pol. J. Microbiol. 2007, 56, 39–43. [Google Scholar] [PubMed]
- Sangorrín, M.P.; Lopes, C.A.; Jofré, V.; Querol, A.; Caballero, A.C. Spoilage yeasts from Patagonian cellars: Characterization and potential biocontrol based on killer interactions. World J. Microbiol. Biotechnol. 2008, 24, 945–953. [Google Scholar] [CrossRef]
- Lopes, C.A.; Sangorrin, M.P. Optimization of killer assays for yeast selection protocols. Rev. Argent. Microbiol. 2010, 42, 298–306. [Google Scholar]
- Robledo-Leal, E.; Rivera-Morales, L.G.; Sangorrín, M.P.; González, G.M.; Ramos-Alfano, G.; Adame-Rodriguez, J.M.; Alcocer-Gonzalez, J.M.; Arechiga-Carvajal, E.T.; Rodriguez-Padilla, C. Identification and susceptibility of clinical isolates of Candida spp. to killer toxins. Braz. J. Biol. 2018, 78, 742–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buzzini, P.; Turchetti, B.; Vaughan-Martini, A.E. The use of killer sensitivity patterns for biotyping yeast strains: The state of the art, potentialities and limitations. FEMS Yeast Res. 2007, 7, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Fia, G.; Giovani, G.; Rosi, I. Study of β-glucosidase production by wine-related yeasts during alcoholic fermentation. A new rapid fluorimetric method to determine enzymatic activity. J. Appl. Microbiol. 2005, 99, 509–517. [Google Scholar] [CrossRef]
- Banani, H.; Spadaro, D.; Zhang, D.; Matic, S.; Garibaldi, A.; Gullino, M.L. Postharvest application of a novel chitinase cloned from Metschnikowia fructicola and overexpressed in Pichia pastoris to control brown rot of peaches. Int. J. Food Microbiol. 2015, 199, 54–61. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Ali, M.; Cheng, C.; Ahmad, H.; Hayat, S. Reactive oxygen species (ROS) as defences against a broad range of plant fungal infections and case study on ROS employed by crops against Verticillium dahliae wilts. J. Plant Interact. 2018, 13, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Breitenbach, M.; Weber, M.; Rinnerthaler, M.; Karl, T.; Breitenbach-Koller, L. Oxidative stress in fungi: Its function in signal transduction, interaction with plant hosts, and lignocellulose degradation. Biomolecules 2015, 5, 318–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, S.; Singh, S.P.; Upadhyay, S.K. Role of superoxide dismutases (SODs) in stress tolerance in plants. In Molecular Approaches in Plant Biology and Environmental Challenges; Singh, S.P., Upadhyay, S.K., Pandey, A., Kumar, S., Eds.; Springer Nature: Singapore, 2019; pp. 51–77. [Google Scholar]



| Mechanism 1,2 | Strains | References |
|---|---|---|
| Iron immobilisation by pulcherrimin production | 02.4.3.38, 02.11.1.21, 7.3.37, 17.1.IV, 17.3.1, 152, 160, 192, 2305, 446, 523, 648, APC 1.2, CBS 610NT, FL01, MACH1, MPR3, Msp50, Msp51, UMY12, UMY14, UMY15 | [6,7,8,9,10,11,17,21,71,72,73] |
| Competition for nitrogen source | 2.33, 4.4 | [74] |
| Competition for carbon source | Anonymous | [31] |
| Chitinase secretion | AP47, FL01, MACH1, NCYC 3728 (PBC-2) | [11,75,76] |
| Chitinase gene up-regulation | 277 (CBS 8853) | [61] |
| DNAse secretion | PO1C004 | [26] |
| Release of volatile compound(s) | FL01, MPR3 | [10,64] |
| Release of ethyl acetate | Disva 267 | [77] |
| Biofilm formation | FL01, MPR3, six anonymous strains | [10,11] |
| Adherence to plant surface | FL01 | [73] |
| Adherence to fungal hyphae | FL01, FL02 | [64] |
| Increased ROS production | 277 (CBS 8853) | [78] |
| Induction of increased ROS production in plant tissue | 277 (CBS 8853) | [78,79,80] |
| Unknown, independent of pulcherrimin production | NCYC 3728 (PBC-2), snf2 | [17,75] |
| Unknown | KIOM_G15050 | [27] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sipiczki, M. Metschnikowia pulcherrima and Related Pulcherrimin-Producing Yeasts: Fuzzy Species Boundaries and Complex Antimicrobial Antagonism. Microorganisms 2020, 8, 1029. https://doi.org/10.3390/microorganisms8071029
Sipiczki M. Metschnikowia pulcherrima and Related Pulcherrimin-Producing Yeasts: Fuzzy Species Boundaries and Complex Antimicrobial Antagonism. Microorganisms. 2020; 8(7):1029. https://doi.org/10.3390/microorganisms8071029
Chicago/Turabian StyleSipiczki, Matthias. 2020. "Metschnikowia pulcherrima and Related Pulcherrimin-Producing Yeasts: Fuzzy Species Boundaries and Complex Antimicrobial Antagonism" Microorganisms 8, no. 7: 1029. https://doi.org/10.3390/microorganisms8071029
APA StyleSipiczki, M. (2020). Metschnikowia pulcherrima and Related Pulcherrimin-Producing Yeasts: Fuzzy Species Boundaries and Complex Antimicrobial Antagonism. Microorganisms, 8(7), 1029. https://doi.org/10.3390/microorganisms8071029




