A Differential Proteomic Approach to Characterize the Cell Wall Adaptive Response to CO2 Overpressure during Sparkling Wine-Making Process
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Conditions
2.2. Proteomic Analysis
2.3. Confidence Parameters and Statistics
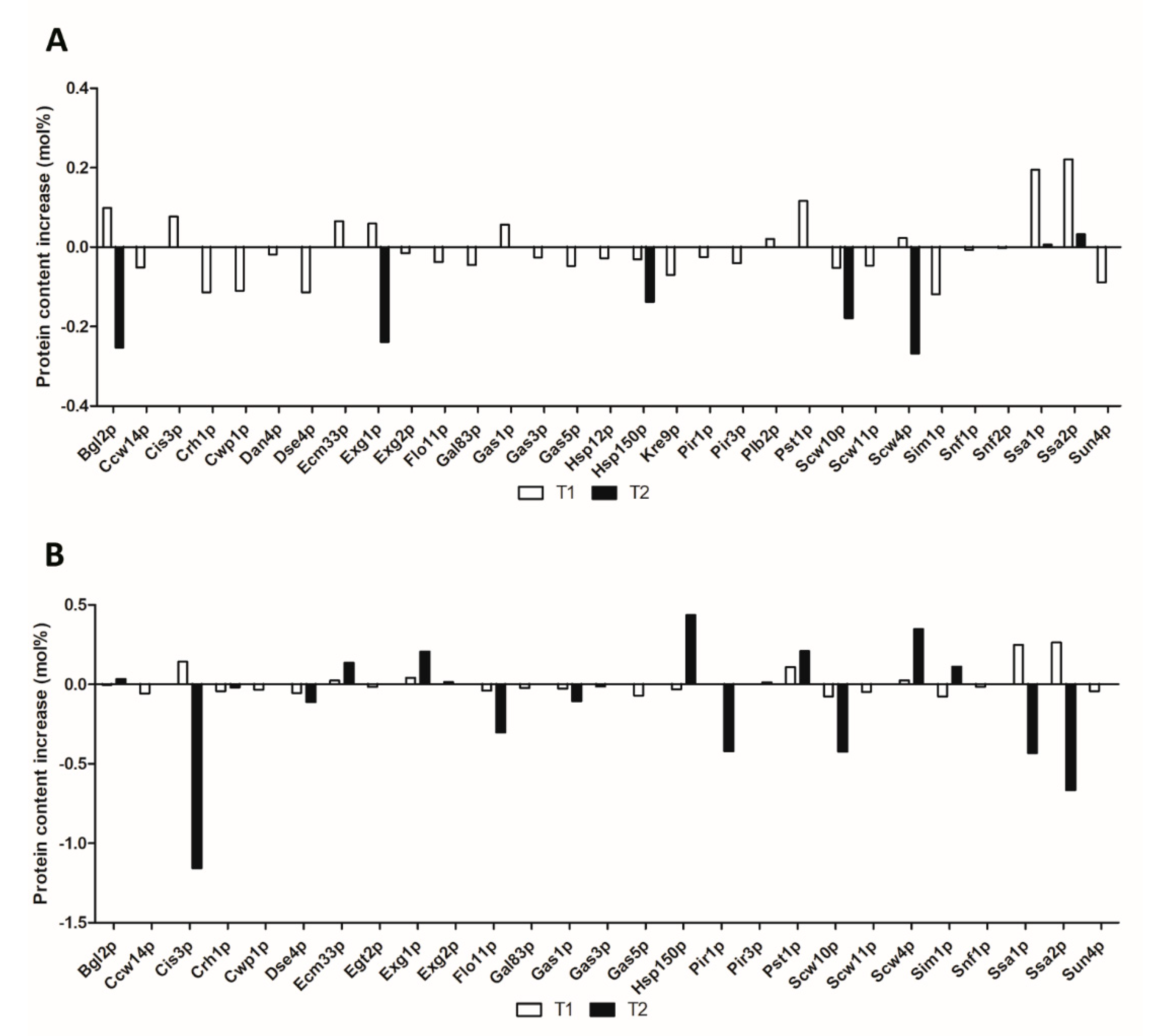
3. Results and Discussion
3.1. Glucan Processing and Remodeling
3.2. Glucan and Chitin Assembly
3.3. Mannoproteins
3.4. Cell Separation
3.5. Proteins Related to Flocculation, Cell Adhesion, and Biofilm Formation
3.6. Other Cell Wall Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alexandre, H.; Guilloux-Benatier, M. Yeast autolysis in sparkling wine-a review. Aust. J. Grape Wine Res. 2006, 12, 119–127. [Google Scholar] [CrossRef]
- Kemp, B.; Alexandre, H.; Robillard, B.; Marchal, R. Effect of production phase on bottle-fermented sparkling wine quality. J. Agric. Food Chem. 2015, 63, 19–38. [Google Scholar] [CrossRef]
- Penacho, V.; Valero, E.; Gonzalez, R. Transcription profiling of sparkling wine second fermentation. Int. J. Food Microbiol. 2012, 153, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Torresi, S.; Frangipane, M.T.; Anelli, G. Biotechnologies in sparkling wine production. Interesting approaches for quality improvement: A review. Food Chem. 2011, 129, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Kalebina, T.S.; Rekstina, V.V. Molecular organization of yeast cell envelope. Mol. Biol. 2019, 53, 850–861. [Google Scholar] [CrossRef]
- Charpentier, C.; Van Long, T.N.; Bonaly, R.; Feuillat, M. Alteration of cell wall structure in Saccharomyces cerevisiae and Saccharomyces bayanus during autolysis. Appl. Microbiol. Biotechnol. 1986, 24, 405–413. [Google Scholar] [CrossRef]
- Martínez-Rodríguez, A.J.; Polo, M.C.; Carrascosa, A.V. Structural and ultrastructural changes in yeast cells during autolysis in a model wine system and in sparkling wines. Int. J. Food Microbiol. 2001, 71, 45–51. [Google Scholar] [CrossRef]
- Piton, F.; Charpentier, M.; Troton, D. Cell Wall and Lipid Changes in Saccharomyces cerevisiae during Aging of Champagne Wine. Am. J. Enol. Vitic. 1988, 39, 221–226. [Google Scholar]
- Tudela, R.; Gallardo-Chacón, J.J.; Rius, N.; López-Tamames, E.; Buxaderas, S. Ultrastructural changes of sparkling wine lees during long-terms aging in real enological conditions. FEMS Yeast Res. 2012, 12, 466–476. [Google Scholar] [CrossRef] [Green Version]
- Klis, F.M.; Mol, P.; Hellingwerf, K.; Brul, S. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2002, 26, 239–256. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI pathway: Regulation of the transcriptional adaptative response to cell wall stress in yeast. J. Fungi 2018, 4, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durán, A.; Nombela, C. Fungal cell wall biogenesis: Building a dynamic interface with the environment. Microbiology 2004, 150, 3099–3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juega, M.; Núñez, Y.P.; Carrascosa, A.V.; Martínez-Rodríguez, A.J. Influence of yeast mannoproteins in the aroma improvement of white wines. J. Food Sci. 2012, 77, 499–504. [Google Scholar] [CrossRef] [Green Version]
- Guadalupe, Z.; Martínez, L.; Ayestarán, B. Yeast Mannoproteins in Red Winemaking: Effect on Polysaccharide, Polyphenolic, and Color Composition. Am. J. Enol. Vitic. 2010, 61, 191–200. [Google Scholar]
- Blasco, L.; Viñas, M.; Villa, T.G. Proteins influencing foam formation in wine and beer: The role of yeast. Int. Microbiol. 2011, 14, 61–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pozo-Bayón, M.A.; Martínez-Rodríguez, A.; Pueyo, E.; Moreno-Arribas, M.V. Chemical and biochemical features involved in sparkling wine production: From a traditional to an improved winemaking technology. Trends Food Sci. Technol. 2009, 20, 289–299. [Google Scholar] [CrossRef]
- Soares, E.V. Flocculation in Saccharomyces cerevisiae: A review. J. Appl. Microbiol. 2010, 110, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Zheng, C.; Chen, Y.; Ying, H. FLO genes family and transcription factor MIG1 regulate Saccharomyces cerevisiae biofilm formation during immobilized fermentation. Front. Microbiol. 2018, 9, 1860. [Google Scholar] [CrossRef]
- Barrales, R.R.; Jimenez, J.; Ibeas, J.I. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics 2008, 178, 145–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dyk, D.; Pretorius, I.S.; Bauer, F.F. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccahromyces cerevisiae. Genetics 2005, 169, 91–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zara, G.; Zeidan, M.B.; Fancello, F.; Sanna, M.S.; Mannazzu, I.; Budroni, M.; Zara, S. The administration of L-cysteine and L-arginine inhibits biofilm formation in wild-type biofilm-forming yeast by modulating FLO11 gene expression. Appl. Microbiol. Biotechnol. 2019, 103, 7675–7685. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.L.; Moreno-García, J.; Zara, S.; Zara, G.; García-Martínez, T.; Mauricio, J.C.; Mannazzu, I.; Coi, A.L.; Zeidan, M.B.; Dequin, S.; et al. Flor yeast: New perspectives beyond wine aging. Front. Microbiol. 2016, 7, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porras-Agüera, J.A.; Moreno-García, J.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. First Proteomic Approach to Identify Cell Death Biomarkers in Wine Yeasts during Sparkling Wine Production. Microorganisms 2019, 7, 542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Alexandre, H. Flor yeasts of Saccharomyces cerevisiae-Their ecology, genetics and metabolism. Int. J. Food Microbiol. 2013, 167, 269–275. [Google Scholar] [CrossRef]
- Teparić, R.; Mrša, V. Proteins involved in building, maintaining and remodeling of yeast cell walls. Curr. Genet. 2013, 59, 171–185. [Google Scholar] [CrossRef]
- Caro, L.H.; Tettelin, H.; Vossen, J.H.; Ram, A.F.; van de Ende, H.; Klis, F.M. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 1997, 13, 1477–1489. [Google Scholar] [CrossRef]
- Capellaro, C.; Mrša, V.; Tanner, W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 1998, 180, 5030–5037. [Google Scholar] [CrossRef] [Green Version]
- Cao, Q.; Zheng, F.; Zhao, J.; Zhang, M.; Li, J.; Wang, J.; Li, Q. The isolation, screening and identification of yeasts for mulberry wine. Food Ferment. Ind. 2017, 43, 94–98. [Google Scholar]
- Spellman, P.T.; Sherlock, G.; Zhang, M.Q.; Iyer, V.R.; Anders, K.; Eisen, M.B.; Brown, P.O.; Botstein, D.; Futcher, B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 1998, 9, 3273–3297. [Google Scholar] [CrossRef] [PubMed]
- Sestak, S.; Hagen, I.; Tanner, W.; Strahl, S. Scw10p, a cell-wall glucanase/transglucosidase important for cell-wall stability in Saccharomyces cerevisiae. Microbiology 2004, 150, 3197–3208. [Google Scholar] [CrossRef] [Green Version]
- Smits, G.J.; Kapteyn, J.C.; van den Ende, H.; Klis, F.M. Cell wall dynamics in yeast. Curr. Opin. Microbiol. 1999, 2, 348–352. [Google Scholar] [CrossRef]
- Mrša, V.; Klebl, F.; Tanner, W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J. Bacteriol. 1993, 175, 2102–2106. [Google Scholar] [CrossRef] [Green Version]
- Nebreda, A.R.; Villa, T.G.; Villanueva, J.R.; del Rey, F. Cloning of genes related to exo-beta-glucanase production in Saccharomyces cerevisiae: Characterization of an exo-beta-glucanase structural gene. Gene 1986, 47, 245–259. [Google Scholar] [CrossRef]
- Jiang, B.; Ram, A.F.; Sheraton, J.; Klis, F.M.; Bussey, H. Regulation of cell wall beta-glucan assembly: PTC1 negatively affects PBS2 action in a pathway that includes modulation of EXG1 transcription. Mol. Gen. Genet. 1995, 248, 260–269. [Google Scholar] [CrossRef] [Green Version]
- Kalebina, T.S.; Farkas, V.; Laurinavichiute, D.K.; Gorlovoy, P.M.; Fominov, G.V.; Bartek, P.; Kulaev, I.S. Deletion of BGL2 results in an increased chitin level in the cell wall of Saccharomyces cerevisiae. Anton. Leeuw. 2003, 84, 179–184. [Google Scholar] [CrossRef]
- Plotnikova, T.A.; Selyakh, I.O.; Kalebina, T.S.; Kulaev, I.S. Bgl2p and Gas1p are the major glucan transferases forming the molecular ensemble of yeast cell wall. Dokl. Biochem. Biophys. 2006, 409, 244–247. [Google Scholar] [CrossRef]
- Kalebina, T.S.; Plotnikova, T.A.; Karpova, E.V.; Kulaev, I.S. A new phenotypic manifestation of deletion of the BGL2 gene encoding the cell-wall glucanotransferase Bgl2p in the yeast Saccharomyces cerevisiae. Microbiology 2006, 75, 625–626. [Google Scholar] [CrossRef]
- Brown, J.L.; Bussey, H. The yeast KRE9 gene encodes an O glycoprotein involved in cell surface beta-glucan assembly. Mol. Cell Biol. 1993, 13, 6346–6356. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, A.; Suzuki, T.; Goshima, T.; Hoshino, T. Evaluation of Saccharomyces cerevisiae GAS1 with respect to its involvement in tolerance to low pH and salt stress. J. Biosci. Bioeng. 2017, 124, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Mazáň, M.; Ragni, E.; Popolo, L.; Farkaš, V. Catalytic properties of the Gas family β-(1,3)-glucanosyltransferases active in fungal cell-wall biogenesis as determined by a novel fluorescent assay. Biochem. J. 2011, 438, 275–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouyna, I.; Fontaine, T.; Vai, M.; Monod, M.; Fonzi, W.A.; Diaquin, M.; Popolo, L.; Hartland, R.P.; Latgé, J.P. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 2000, 275, 14882–14889. [Google Scholar] [CrossRef] [Green Version]
- Ragni, E.; Coluccio, A.; Rolli, E.; Rodriguez-Peña, J.M.; Colasante, G.; Arroyo, J.; Neiman, A.M.; Popolo, L. GAS2 and GAS4, a pair of developmentally regulated genes required for spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 2007, 6, 302–316. [Google Scholar] [CrossRef] [Green Version]
- Cabib, E.; Blanco, N.; Grau, C.; Rodríguez-Peña, J.M.; Arroyo, J. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 2007, 63, 921–935. [Google Scholar] [CrossRef]
- Bermejo, C.; Rodríguez, E.; García, R.; Rodríguez-Peña, J.M.; Rodríguez de la Concepción, M.L.; Rivas, C.; Arias, P.; Nombela, C.; Posas, F.; Arroyo, J. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol. Biol. Cell 2008, 19, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Ndlovu, T.; Divol, B.; Bauer, F.F. Yeast cell wall chitin reduces wine haze formation. Appl. Environ. Microbiol. 2018, 84, e00668-18. [Google Scholar] [CrossRef] [Green Version]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [Green Version]
- Popolo, L.; Gualtieri, T.; Ragni, E. The yeast cell wall salvage pathway. Med. Mycol. 2001, 39, 111–121. [Google Scholar] [CrossRef]
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toh-e, A.; Yasunaga, S.; Nisogi, H.; Tanaka, K.; Oguchi, T.; Matsui, Y. Three yeast genes, PIR1, PIR2 and PIR3, containing internal tandem repeats, are related to each other, and PIR1 and PIR2 are required for tolerance to heat shock. Yeast 1993, 9, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Yu, Z.; Jia, D.; Xie, Z.; Zhang, K.; Xia, Z.; Lei, L.; Qiao, M. The contribution of Pir protein family to yeast cell surface display. Appl. Microbiol. Biotechnol. 2014, 98, 2897–2905. [Google Scholar] [CrossRef] [PubMed]
- Moukadiri, I.; Jaafar, L.; Zueco, J. Identification of two mannoproteins released from cell walls of a Saccharomyces cerevisiae mnn1 mnn9 double mutant by reducing agents. J. Bacteriol. 1999, 181, 4741–4745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumita, T.; Yoko-o, T.; Shimma, Y.; Jigami, Y. Comparison of Cell Wall Localization among Pir Family Proteins and Functional Dissection of the Region Required for Cell Wall Binding and Bud Scar Recruitment of Pir1p. Eukaryot. Cell 2005, 4, 1872–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapteyn, J.C.; Van Egmond, P.; Sievi, E.; Van Den Ende, H.; Makarow, M.; Klis, F.M. The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and beta 1,6-glucan-deficient mutants. Mol. Microbiol. 1999, 31, 1835–1844. [Google Scholar] [CrossRef]
- Mrša, V.; Tanner, W. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of the Pir protein family) in stability of the Saccharomyces cerevisiae cell wall. Yeast 1999, 15, 813–820. [Google Scholar] [CrossRef]
- Russo, P.; Simonen, M.; Uimari, A.; Teesalu, T.; Makarow, M. Dual regulation by heat and nutrient stress of the yeast HSP150 gene encoding a secretory glycoprotein. Mol. Gen. Genet. 1993, 239, 273–280. [Google Scholar] [CrossRef]
- Moukadiri, I.; Armero, J.; Abad, A.; Sentandreu, R.; Zueco, J. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 1997, 179, 2154–2162. [Google Scholar] [CrossRef] [Green Version]
- Mrša, V.; Ecker, M.; Strahl-Bolsinger, S.; Nimtz, M.; Lehle, L.; Tanner, W. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 1999, 181, 3076–3086. [Google Scholar] [CrossRef] [Green Version]
- Moreno-García, J.; Coi, A.L.; Zara, G.; García-Martínez, T.; Mauricio, J.C.; Budroni, M. Study of the role of the covalently linked cell wall protein (Ccw14p) and yeast glycoprotein (Ygp1p) within biofilm formation in a flor yeast strain. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Van der Vaart, J.M.; Caro, L.H.; Chapman, J.W.; Klis, F.M.; Verrips, C.T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 1995, 177, 3104–3110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abramova, N.; Sertil, O.; Mehta, S.; Lowry, C.V. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 2001, 183, 2881–2887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardo, M.; Monteoliva, L.; Pla, J.; Sánchez, M.; Gil, C.; Nombela, C. Two-dimensional analysis of proteins secreted by Saccharomyces cerevisiae regenerating protoplasts: A novel approach to study the cell wall. Yeast 1999, 15, 459–472. [Google Scholar] [CrossRef]
- Pardo, M.; Monteoliva, L.; Vázquez, P.; Martínez, R.; Molero, G.; Nombela, C.; Gil, C. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 2004, 150, 4157–4170. [Google Scholar] [CrossRef] [Green Version]
- Terashima, H.; Yabuki, N.; Arisawa, M.; Hamada, K.; Kitada, K. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000, 264, 64–74. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Phornsanthia, S.; Yin, X.; Li, Q. Strengthening of Cell Wall Structure Enhances Stress Resistance and Fermentation Performance in Lager. J. Am. Soc. Brew. Chem. 2014, 72, 88–94. [Google Scholar] [CrossRef]
- Kovacech, B.; Nasmyth, K.; Schuster, T. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol. Cell. Biol. 1996, 16, 3264–3274. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Heitman, J. Sok2 Regulates Yeast Pseudohyphal Differentiation via a Transcription Factor Cascade That Regulates Cell-Cell Adhesion. Mol. Cell Biol. 2000, 20, 8364–8372. [Google Scholar] [CrossRef] [Green Version]
- Baladrón, V.; Ufano, S.; Dueñas, E.; Martín-Cuadrado, A.B.; del Rey, F.; Vázquez de Aldana, C.R. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 774–786. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsov, E.; Kucerova, H.; Vachova, L.; Palkova, Z. SUN family proteins Sun4p, Uth1p and Sim1p are secreted from Saccharomyces cerevisiae and produced dependently on oxygen level. PLoS ONE 2013, 8, e73882. [Google Scholar] [CrossRef] [PubMed]
- Mouassite, M.; Camougrand, N.; Schwob, E.; Demaison, G.; Laclau, M.; Guérin, M. The ‘SUN’ family: Yeast SUN4/SCW3 is involved in cell septation. Yeast 2000, 16, 905–919. [Google Scholar] [CrossRef]
- Tofalo, R.; Perpetuini, G.; Di Gianvito, P.; Arfelli, G.; Shirone, M.; Corsetti, A.; Suzzi, G. Characterization of specialized flocculent yeasts to improve sparkling wine fermentation. J. Appl. Microbiol. 2016, 120, 1574–1584. [Google Scholar] [CrossRef]
- Barrales, R.R.; Korber, P.; Jimenez, J.; Ibeas, J.I. Chromatin modulation at the FLO11 promoter of Saccharomyces cerevisiae by HDAC and Swi/Snf complexes. Genetics 2012, 191, 791–803. [Google Scholar] [CrossRef] [Green Version]
- Kuchin, S.; Vyas, V.K.; Carlson, M. Snf1 Protein Kinase and the Repressors Nrg1 and Nrg2 Regulate FLO11, Haploid Invasive Growth, and Diploid Pseudohyphal Differentiation. Mol. Cell. Biol. 2002, 22, 3994–4000. [Google Scholar] [CrossRef] [Green Version]
- Vyas, V.K.; Kuchin, S.; Berkey, C.D.; Carlson, M. Snf1 Kinases with Different β-Subunit Isoforms Play Distinct Roles in Regulating Haploid Invasive Growth. Mol. Cell Biol. 2003, 23, 1341–1348. [Google Scholar] [CrossRef] [Green Version]
- Welker, S.; Rudolph, B.; Frenzel, E.; Hagn, F.; Liebisch, G.; Schmitz, G.; Scheuring, J.; Kerth, A.; Blume, A.; Weinkauf, S.; et al. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell 2010, 39, 507–520. [Google Scholar] [CrossRef]
- Zara, S.; Antonio Farris, G.; Budroni, M.; Bakalinsky, A.T. HSP12 is essential for biofilm formation by a Sardinian wine strain of S. cerevisiae. Yeast 2002, 19, 269–276. [Google Scholar] [CrossRef]
- Lopez-Ribot, J.L.; Chaffin, W.L. Members of the Hsp70 family of proteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 1996, 178, 4724–4726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bukau, B.; Horwich, A.L. The Hsp70 and Hsp60 chaperone machines. Cell 1998, 92, 351–366. [Google Scholar] [CrossRef] [Green Version]
- Merkel, O.; Fido, M.; Mayr, J.A.; Prüger, H.; Raab, F.; Zandonella, G.; Kohlwein, S.D.; Paltauf, F. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 28121–28127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilera, F.; Peinado, R.A.; Millán, C.; Ortega, J.M.; Mauricio, J.C. Relationship between ethanol tolerance, H+ -ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains. Int. J. Food Microbiol. 2006, 110, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Terashima, H.; Hamada, K.; Kitada, K. The localization change of Ybr078w/Ecm33, a yeast GPI-associated protein, from the plasma membrane to the cell wall, affecting the cellular function. FEMS Microbiol. Lett. 2003, 218, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Astorga, M.A.; Gardner, J.M.; Walker, M.E.; Grbin, P.R.; Jiranek, V. Disruption of the cell wall integrity gene ECM33 results in improved fermentation by wine yeast. Metab. Eng. 2018, 45, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Mouassite, M.; Guérin, M.G.; Camougrand, N.M. The SUN family of Saccharomyces cerevisiae: The double knock-out of UTH1 and SIM1 promotes defects in nucleus migration and increased drug sensitivity. FEMS Microbiol. Lett. 2000, 182, 137–141. [Google Scholar] [CrossRef] [PubMed]

| Yeast Strains | S. cerevisiae P29 | S. cerevisiae G1 | ||||||
|---|---|---|---|---|---|---|---|---|
| Conditions | T1 | Funcion | T2 | Function | T1 | Function | T2 | Function |
| Protein | Ecm33p (2.1) | GPI-anchored protein | - | - | Cis3p (1.8) | Mannoprotein | Hsp150p (1.8) | O-mannosylated heat shock protein |
| Gas1p (1.8) | β-1,3-glucanosyltransferase | - | - | Exg2p (2.4) | β-glucan assembly | Pst1p (1.8) | Cell wall protein with GPI (Glycosylphosphatidylinositol)-attachment site | |
| Pst1p (2.3) | Cell wall protein with GPI-attachment site | - | - | Pst1p (1.9) | Cell wall protein with GPI-attachment site | Scw4p (3) | Cell wall protein with similarity to glucanases | |
| Ssa1p (2) | Protein folding | - | - | Ssa1p (2.1) | Protein folding | - | - | |
| Ssa2p (1.9) | Protein folding | - | - | Ssa2p (2.1) | Protein folding | - | - | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porras-Agüera, J.A.; Mauricio, J.C.; Moreno-García, J.; Moreno, J.; García-Martínez, T. A Differential Proteomic Approach to Characterize the Cell Wall Adaptive Response to CO2 Overpressure during Sparkling Wine-Making Process. Microorganisms 2020, 8, 1188. https://doi.org/10.3390/microorganisms8081188
Porras-Agüera JA, Mauricio JC, Moreno-García J, Moreno J, García-Martínez T. A Differential Proteomic Approach to Characterize the Cell Wall Adaptive Response to CO2 Overpressure during Sparkling Wine-Making Process. Microorganisms. 2020; 8(8):1188. https://doi.org/10.3390/microorganisms8081188
Chicago/Turabian StylePorras-Agüera, Juan Antonio, Juan Carlos Mauricio, Jaime Moreno-García, Juan Moreno, and Teresa García-Martínez. 2020. "A Differential Proteomic Approach to Characterize the Cell Wall Adaptive Response to CO2 Overpressure during Sparkling Wine-Making Process" Microorganisms 8, no. 8: 1188. https://doi.org/10.3390/microorganisms8081188








