Antibiogram Signatures of Some Enterobacteria Recovered from Irrigation Water and Agricultural Soil in two District Municipalities of South Africa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Collection of Samples and Isolation of Target Pathogens
2.3. Characterization of Target Pathogens
2.3.1. DNA Extraction
2.3.2. PCR Delineation of the Presumptive Enterobacterial Isolates
2.4. Antibiotic Susceptibility Testing of Confirmed Members of Enterobacteriales
2.5. Evaluation of Multiple Antibiotic Resistance Phenotypes (MARPs) and Multiple Antibiotic Resistance Indices (MARIs)
2.6. Screening for Antimicrobial Resistance Genes
2.7. Evaluation of the Patterns of Multiple Antibiotic Resistance Genotypes (MARGs)
2.8. Data Analysis
3. Results and Discussion
3.1. Antimicrobial Susceptibility Profiles of Members of Enterobacteriales
3.2. The Patterns of Multiple Antibiotic Resistance Phenotypes (MARPs) and Multiple Antibiotic Resistance Indices (MARI) in the Isolates
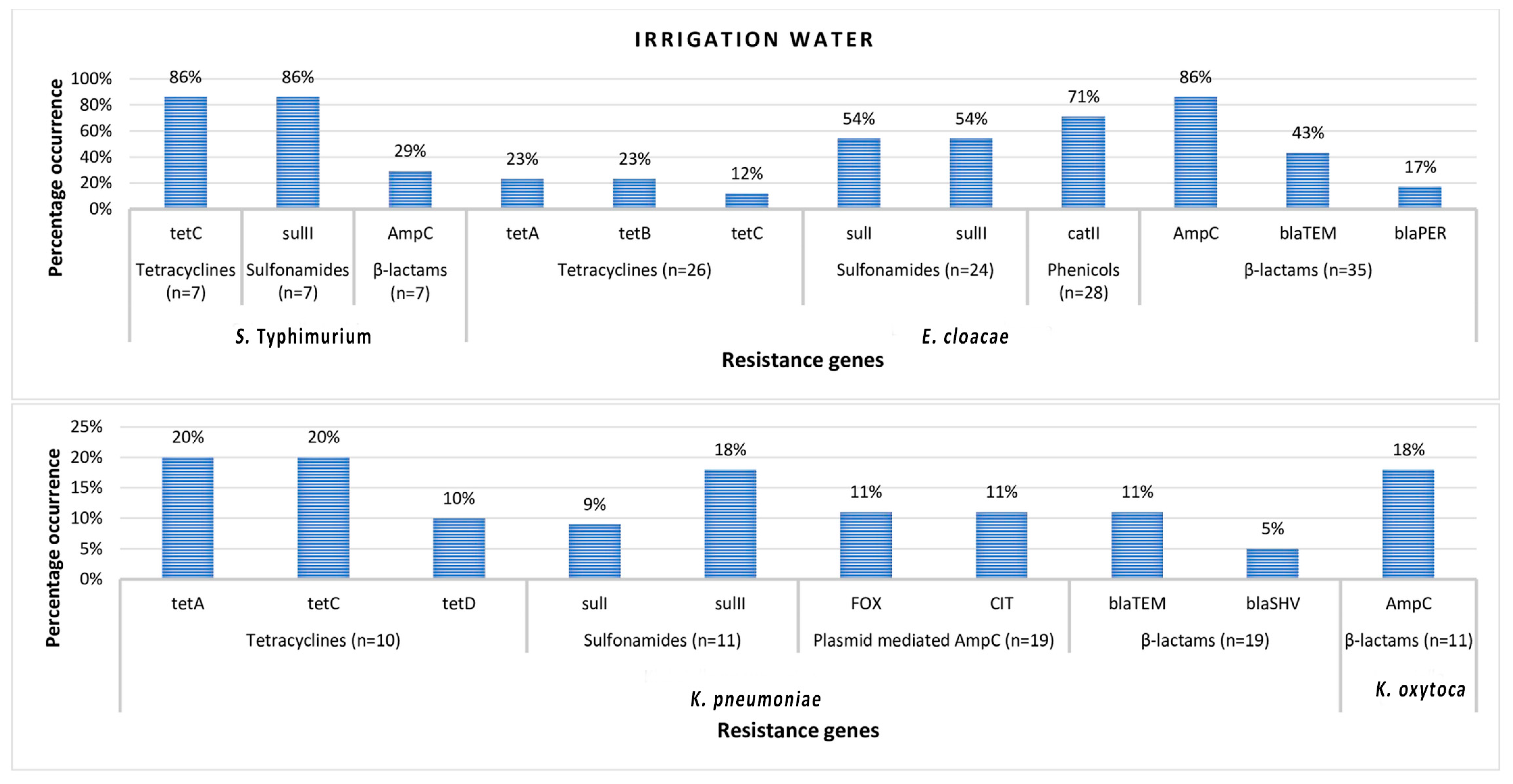
3.3. Antimicrobial Resistance Genes in Phenotypically Resistant Members of Enterobacteriales
3.4. The Patterns of Multiple Antibiotic Resistance Genotypes (MARGs) in the Isolates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Rajwar, A.; Srivastava, P.; Sahgal, M. Microbiology of Fresh Produce: Route of Contamination, Detection Methods, and Remedy. Crit. Rev. Food Sci. Nutr. 2016, 56, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Sharma, S.; Nara, S. Evaluation of gold nanoparticle based lateral flow assays for diagnosis of enterobacteriaceae members in food and water. Food Chem. 2015, 170, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Urbán, E. Resistance Trends and Epidemiology of Citrobacter-Enterobacter-Serratia in Urinary Tract Infections of Inpatients and Outpatients (RECESUTI): A 10-Year Survey. Medicina 2019, 55, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajdács, M.; Ábrók, M.; Lázár, A.; Burián, K. Comparative Epidemiology and Resistance Trends of Common Urinary Pathogens in a Tertiary-Care Hospital: A 10-Year Surveillance Study. Medicina 2019, 55, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramana, K.V.; Rao, R.; Sharada, C.V.; Kareem, M.; Reddy, L.R.; Ratna, M.M. Modified Hodge test: A useful and the low-cost phenotypic method for detection of carbapenemase producers in Enterobacteriaceae members. J. Nat. Sci. Biol. Med. 2013, 4, 346–348. [Google Scholar] [CrossRef] [Green Version]
- Paterson, D.L. Resistance in Gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control. 2006, 34, S20–S28. [Google Scholar] [CrossRef]
- Tosun, Ş.Y.; Alakavuk, D.Ü.; Mol, S. Isolation of Salmonella spp. and other members of Enterobacteriaceae from horse mackerel (Trachurus trachurus), sold in public markets of Istanbul, Turkey. J. Food Health Sci. 2016, 2, 82–89. [Google Scholar] [CrossRef] [Green Version]
- Pagadala, S.; Marine, S.C.; Micallef, S.A.; Wang, F.; Pahl, D.M.; Melendez, M.V.; Kline, W.L.; Oni, R.A.; Walsh, C.S.; Everts, K.L.; et al. Assessment of region, farming system, irrigation source and sampling time as food safety risk factors for tomatoes. Int. J. Food Microbiol. 2015, 196, 98–108. [Google Scholar] [CrossRef]
- Miller, B. Fruit Recall Expanded by Another 5 Days to Ensure Safety for the Public. DUMBOUT. 2014. Available online: http://food-nc.blogspot.com/2014/08/fruit-recall-expanded-by-another-5-days.html (accessed on 1 May 2020).
- Warriner, K.; Huber, A.; Namvar, A.; Fan, W.; Dunfield, K. Recent Advances in the Microbial Safety of Fresh Fruits and Vegetables. Adv. Food Nutr. Res. 2009, 57, 155–208. [Google Scholar] [CrossRef] [PubMed]
- Micallef, S.A.; Rosenberg Goldstein, R.E.; George, A.; Kleinfelter, L.; Boyer, M.S.; McLaughlin, C.R.; Estrin, A.; Ewing, L.; Jean-Gilles Beaubrun, J.; Hanes, D.E.; et al. Occurrence and antibiotic resistance of multiple Salmonella serotypes recovered from water, sediment and soil on mid-Atlantic tomato farms. Environ. Res. 2012, 114, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Strawn, L.K.; Fortes, E.D.; Bihn, E.A.; Nightingale, K.K.; Gröhn, Y.T.; Worobo, R.W.; Wiedmann, M.; Bergholz, P.W. Landscape and Meteorological Factors Affecting Prevalence of Three Food-Borne Pathogens in Fruit and Vegetable Farms. Appl. Environ. Microbiol. 2013, 79, 588–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorski, L.; Parker, C.T.; Liang, A.; Cooley, M.B.; Jay-Russell, M.T.; Gordus, A.G.; Atwill, E.R.; Mandrell, R.E. Prevalence, Distribution, and Diversity of Salmonella enterica in a Major Produce Region of California. Appl. Environ. Microbiol. 2011, 77, 2734–2748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gemmell, M.E.; Schmidt, S. Microbiological assessment of river water used for the irrigation of fresh produce in a sub-urban community in Sobantu, South Africa. Food Res. Int. 2012, 47, 300–305. [Google Scholar] [CrossRef]
- Nontongana, N.; Sibanda, T.; Ngwenya, E.; Okoh, A. Prevalence and Antibiogram Profiling of Escherichia coli Pathotypes Isolated from the Kat River and the Fort Beaufort Abstraction Water. Int. J. Environ. Res. Public Health 2014, 11, 8213–8227. [Google Scholar] [CrossRef] [Green Version]
- Cooley, M.; Carychao, D.; Crawford-Miksza, L.; Jay, M.T.; Myers, C.; Rose, C.; Keys, C.; Farrar, J.; Mandrell, R.E. Incidence and Tracking of Escherichia coli O157:H7 in a Major Produce Production Region in California. PLoS ONE 2007, 2, e1159. [Google Scholar] [CrossRef]
- Tope, A.M.; Hitter, A.C.; Patel, S.V. Evaluation of Antimicrobial Resistance in Enterobacteriaceae and Coliforms Isolated on Farm, Packaged and Loose Vegetables in Kentucky. J. Food Microbiol. Safety Hyg. 2016, 1, 1–7. [Google Scholar] [CrossRef] [Green Version]
- FAO/WHO Food and Agricultural Organization of the United Nations/World Health Organization. Microbiological hazards in fresh leafy vegetables and herbs; Food & Agriculture Organization of the United: Rome, Italy, 2008. [Google Scholar]
- Jung, Y.; Jang, H.; Matthews, K.R. Effect of the food production chain from farm practices to vegetable processing on outbreak incidence. Microb. Biotechnol. 2014, 7, 517–527. [Google Scholar] [CrossRef]
- Gajdács, M. Epidemiology and antibiotic resistance trends of Pantoea species in a tertiary-care teaching hospital: A 12-year retrospective study. Dev. Health Sci. 2019, 2, 72–75. [Google Scholar] [CrossRef]
- Falomir, M.P.; Gozalbo, D.; Rico, H. Coliform bacteria in fresh vegetables: From cultivated lands to consumers. Formatex 2010, 2, 1175–1181. [Google Scholar]
- Zekar, F.M.; Granier, S.A.; Marault, M.; Yaici, L.; Gassilloud, B.; Manceau, C.; Touati, A.; Millemann, Y. From Farms to Markets: Gram-Negative Bacteria Resistant to Third-Generation Cephalosporins in Fruits and Vegetables in a Region of North Africa. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Saldinger, S.S.; Manulis-Sasson, S. What else can we do to mitigate contamination of fresh produce by foodborne pathogens? Microb. Biotechnol. 2015, 8, 29–31. [Google Scholar] [CrossRef] [PubMed]
- Falomir, M.P.; Rico, H.; Gozalbo, D. Enterobacter and Klebsiella species isolated from fresh vegetables marketed in Valencia (spain) and their clinically relevant resistances to chemotherapeutic agents. Foodborne Pathog. Dis. 2013, 10, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Pezzoli, L.; Elson, R.; Little, C.L.; Yip, H.; Fisher, I.; Yishai, R.; Anis, E.; Valinsky, L.; Biggerstaff, M.; Patel, N.; et al. Packed with Salmonella—Investigation of an International Outbreak of Salmonella Senftenberg Infection Linked to Contamination of Prepacked Basil in 2007. Foodborne Pathog. Dis. 2008, 5, 661–668. [Google Scholar] [CrossRef]
- Bartz, F.E.; Lickness, J.S.; Heredia, N.; Fabiszewski de Aceituno, A.; Newman, K.L.; Hodge, D.W.; Jaykus, L.-A.; García, S.; Leon, J.S. Contamination of Fresh Produce by Microbial Indicators on Farms and in Packing Facilities: Elucidation of Environmental Routes. Appl. Environ. Microbiol. 2017, 83, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Aarestrup, F.M.; Wegener, H.C.; Collignon, P. Resistance in bacteria of the food chain: Epidemiology and control strategies. Expert Rev. Anti Infect. Ther. 2008, 6, 733–750. [Google Scholar] [CrossRef]
- Walsh, C.; Fanning, S. Antimicrobial Resistance in Foodborne Pathogens—A Cause for Concern? Curr. Drug Targets 2008, 9, 808–815. [Google Scholar] [CrossRef]
- Iwu, C.J.; Iweriebor, B.C.; Obi, L.C.; Basson, A.K.; Okoh, A.I. Multidrug-Resistant Salmonella Isolates from Swine in the Eastern Cape Province, South Africa. J. Food Prot. 2016, 79, 1234–1239. [Google Scholar] [CrossRef]
- Iwu, C.D.; Okoh, A.I. Preharvest Transmission Routes of Fresh Produce Associated Bacterial Pathogens with Outbreak Potentials: A Review. Int. J. Environ. Res. Public Health 2019, 16, 4407. [Google Scholar] [CrossRef] [Green Version]
- Iwu, C.D.; Korsten, L.; Okoh, A.I. The incidence of antibiotic resistance within and beyond the agricultural ecosystem: A concern for public health. MicrobiologyOpen 2020, e1035. [Google Scholar] [CrossRef]
- Gajdács, M. The concept of an ideal antibiotic: Implications for drug design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajdács, M.; Bátori, Z.; Ábrók, M.; Lázár, A.; Burián, K. Characterization of Resistance in Gram-Negative Urinary Isolates Using Existing and Novel Indicators of Clinical Relevance: A 10-Year Data Analysis. Life 2020, 10, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, J.P.; Clark, N.M.; Zhanel, G.G. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum β-lactamases and carbapenemases). Expert Opin. Pharmacother. 2013, 14, 199–210. [Google Scholar] [CrossRef]
- Amathole District Municipality (DC12)- Overview Municipalities of South Africa. Available online: https://municipalities.co.za/overview/102/amathole-district-municipality (accessed on 19 March 2020).
- Chris Hani District Municipality (DC13)- Demographic Municipalities of South Africa. Available online: https://municipalities.co.za/demographic/104/chris-hani-district-municipality (accessed on 19 March 2020).
- Maugeri, T.L.; Carbone, M.; Fera, M.T.; Irrera, G.P.; Gugliandolo, C. Distribution of potentially pathogenic bacteria as free living and plankton associated in a marine coastal zone. J. Appl. Microbiol. 2004, 97, 354–361. [Google Scholar] [CrossRef]
- Lopez-Saucedo, C.; Cerna, J.F.; Villegas-Sepulveda, N.; Thompson, R.; Velazquez, F.R.; Torres, J.; Tarr, P.I.; Estrada-Garcia, T. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg. Infect. Dis. 2003, 9, 127–131. [Google Scholar] [CrossRef]
- CLSI Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Titilawo, Y.; Sibanda, T.; Obi, L.; Okoh, A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ. Sci. Pollut. Res. 2015, 22, 10969–10980. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Multiple Antibiotic Resistance Indexing of Escherichia coli to Identify High-Risk Sources of Fecal Contamination of Foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Titilawo, Y.; Obi, L.; Okoh, A. Antimicrobial resistance determinants of Escherichia coli isolates recovered from some rivers in Osun State, South-Western Nigeria: Implications for public health. Sci. Total. Environ. 2015, 523, 82–94. [Google Scholar] [CrossRef]
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Kocsis, B.; Szabó, D. Antibiotic resistance mechanisms in Enterobacteriaceae. Formatex 2013, 1, 251–257. [Google Scholar]
- Ye, Q.; Wu, Q.; Zhang, S.; Zhang, J.; Yang, G.; Wang, H.; Huang, J.; Chen, M.; Xue, L.; Wang, J. Antibiotic-resistant extended spectrum β-lactamase- and plasmid-mediated AmpC-producing Enterobacteriaceae isolated from retail food products and the pearl river in Guangzhou, China. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Iredell, J.; Brown, J.; Tagg, K. Antibiotic resistance in Enterobacteriaceae: Mechanisms and clinical implications. BMJ 2016, 352, h6420. [Google Scholar] [CrossRef] [PubMed]
- Hudson, C.M.; Bent, Z.W.; Meagher, R.J.; Williams, K.P. Resistance Determinants and Mobile Genetic Elements of an NDM-1-Encoding Klebsiella pneumoniae Strain. PLoS ONE 2014, 9, e99209. [Google Scholar] [CrossRef] [Green Version]
- Iweriebor, B.C.; Iwu, C.J.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in Eastern Cape of South Africa. BMC Microbiol. 2015, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Machado, E.; Coque, T.M.; Canton, R.; Sousa, J.C.; Silva, D.; Ramos, M.; Rocha, J.; Ferreira, H.; Peixe, L. Leakage into Portuguese aquatic environments of extended-spectrum- -lactamase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2009, 63, 616–618. [Google Scholar] [CrossRef] [Green Version]
- Tacão, M.; Correia, A.; Henriques, I. Resistance to Broad-Spectrum Antibiotics in Aquatic Systems: Anthropogenic Activities Modulate the Dissemination of blaCTX-M-Like Genes. Appl. Environ. Microbiol. 2012, 78, 4134–4140. [Google Scholar] [CrossRef] [Green Version]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [Green Version]
- Cantón, R.; Novais, A.; Valverde, A.; Machado, E.; Peixe, L.; Baquero, F.; Coque, T.M. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2008, 14, 144–153. [Google Scholar] [CrossRef] [Green Version]
- Yagi, T.; Kurokawa, H.; Shibata, N.; Shibayama, K.; Arakawa, Y. A preliminary survey of extended-spectrum beta-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol. Lett. 2000, 184, 53–56. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, J.C.; Penha Filho, R.A.C.; Andrade, L.N.; Berchieri Junior, A.; Darini, A.L.C. IncI1/ST113 and IncI1/ST114 conjugative plasmids carrying blaCTX-M-8 in Escherichia coli isolated from poultry in Brazil. Diagn. Microbiol. Infect. Dis. 2014, 80, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Zarfel, G.; Galler, H.; Luxner, J.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Kittinger, C.; Grisold, A.J.; Pless, P.; Feierl, G. Multiresistant bacteria isolated from chicken meat in Austria. Int. J. Environ. Res. Public Health 2014, 11, 12582–12593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitout, J.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- APUA Alliance for the Prudent Use of Antibiotics. About Bacteria & Antibiotics. Available online: https://apua.org/about-resistance (accessed on 17 February 2020).
- Iwu, C.D.; Okoh, A.I. Characterization of antibiogram fingerprints in Listeria monocytogenes recovered from irrigation water and agricultural soil samples. PLoS ONE 2020, 15, e0228956. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Antibiotics | Potency (µg) | S. Typhimurium n = 13 (Frequency/Percent) | E. cloacae n = 58 (Frequency/Percent) | K. pneumoniae n = 36 (Frequency/Percent) | K. oxytoca n = 21 (Frequency/Percent) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | I | S | R | I | S | R | I | S | R | I | S | ||
| Gentamicin | 10 | 0/0 | 0/0 | 13/100 | 1/1.7 | 0/0 | 57/98.3 | 1/2.8 | 0/0 | 35/97.2 | 0/0 | 0/0 | 21/100 |
| Amikacin | 30 | 0/0 | 0/0 | 13/100 | 0/0 | 0/0 | 58/100 | 0/0 | 0/0 | 36/100 | 0/0 | 0/0 | 21/100 |
| Amoxicillin/ Clavulanic acid | 20/10 | 5/38.5 | 2/15.4 | 6/46.2 | 45/77.6 | 2/3.4 | 11/19.0 | 29/80.6 | 3/8.3 | 4/11.1 | 13/61.9 | 0/0 | 8/38.1 |
| Ampicillin | 10 | 9/69.2 | 0/0 | 4/30.8 | 49/84.5 | 3/5.2 | 6/10.3 | 32/88.9 | 0/0 | 4/11.1 | 13/61.9 | 0/0 | 8/38.1 |
| Imipenem | 10 | 0/0 | 0/0 | 13/100 | 0/0 | 0/0 | 58/100 | 2/5.6 | 4/11.1 | 30/83.3 | 0/0 | 0/0 | 21/100 |
| Meropenem | 10 | 0/0 | 0/0 | 13/100 | 1/1.7 | 4/6.9 | 53/91.4 | 0/0 | 3/8.3 | 33/91.7 | 0/0 | 2/9.5 | 19/90.5 |
| Cefotaxime | 30 | 2/15.4 | 0/0 | 11/84.6 | 30/51.7 | 6/10.3 | 22/37.9 | 16/44.4 | 3/8.3 | 17/47.2 | 10/47.6 | 0/0 | 11/52.4 |
| Cefuroxime | 30 | 5/38.5 | 8/61.5 | 0/0 | 47/81.0 | 5/8.6 | 6/10.3 | 22/61.1 | 13/36.1 | 1/2.8 | 12/57.1 | 8/38.1 | 1/4.8 |
| Ciprofloxacin | 5 | 0/0 | 0/0 | 13/100 | 1/1.7 | 7/12.1 | 50/86.2 | 4/11.1 | 1/2.8 | 31/86.1 | 3/14.3 | 3/14.3 | 15/71.4 |
| Norfloxacin | 30 | 0/0 | 0/0 | 13/100 | 2/3.4 | 7/12.1 | 49/84.5 | 4/11.1 | 1/2.8 | 31/86.1 | 5/23.8 | 2/9.5 | 14/66.7 |
| Nitrofurantoin | 300 | 1/7.7 | 1/7.7 | 11/84.6 | 47/81.0 | 2/3.4 | 9/15.5 | 18/50.0 | 2/5.6 | 16/44.4 | 13/61.9 | 0/0 | 8/38.1 |
| Chloramphenicol | 30 | 0/0 | 0/0 | 13/100 | 32/55.2 | 10/17.2 | 16/27.6 | 15/41.7 | 2/5.6 | 19/52.8 | 11/52.4 | 2/9.5 | 8/38.1 |
| Nalidixic acid | 30 | 2/15.4 | 0/0 | 11/84.6 | 28/48.3 | 4/6.9 | 26/44.8 | 14/38.9 | 2/5.6 | 20/55.6 | 8/38.1 | 0/0 | 13/61.9 |
| Trimethoprim/ Sulfamethoxazole | 1.25/23.75 | 3/23.1 | 0/0 | 10/76.9 | 29/50.0 | 1/5.2 | 26/44.8 | 10/27.8 | 1/2.8 | 25/69.4 | 8/38.1 | 0/0 | 13/61.9 |
| Tetracycline | 30 | 12/92.3 | 0/0 | 1/7.7 | 35/60.3 | 5/8.6 | 18/31.0 | 13/36.1 | 2/5.6 | 21/58.3 | 8/38.1 | 1/4.8 | 12/57.1 |
| Doxycycline | 30 | 2/15.4 | 7/53.8 | 4/30.8 | 32/55.2 | 10/17.2 | 16/27.6 | 7/19.4 | 12/33.3 | 17/47.2 | 2/9.5 | 6/28.6 | 13/61.9 |
| SN | MAR Phenotypes | No. of Antibiotics | No. of Isolates | MARI |
|---|---|---|---|---|
| S. Typhimurium | ||||
| 1 | AUG-AP-CTX-CXM-NI-T | 6 | 1 | 0.4 |
| 2 | CTX-CXM-T-DXT | 4 | 1 | 0.3 |
| 3 | AP-CXM-T | 3 | 1 | 0.2 |
| 4 | AUG-AP-NA-T-DXT | 5 | 1 | 0.3 |
| 5 | AUG-AP-CXM-T | 4 | 1 | 0.3 |
| E. cloacae | ||||
| 1 | AUG-AP-CXM-NI-C-NA-T-DXT | 8 | 3 | 0.5 |
| 2 | AUG-AP-CXM-NI-NA-T-DXT | 7 | 1 | 0.4 |
| 3 | AUG-AP-CTX-CXM-CIP-NI-NA-T-DXT | 9 | 1 | 0.6 |
| 4 | AUG-CXM-NI-NA-T-DXT | 6 | 1 | 0.4 |
| 5 | AUG-AP-CXM-NI-T-DXT | 6 | 1 | 0.4 |
| 6 | AUG-AP-CTX-CXM-NI-C-T-DXT | 8 | 1 | 0.5 |
| 7 | AUG-AP-CTX-CXM-NI-C-T-DXT | 8 | 1 | 0.5 |
| 8 | AUG-CXM-NI-C | 4 | 1 | 0.3 |
| 9 | AUG-AP-CXM-NI-C | 5 | 2 | 0.3 |
| 10 | AUG-AP-CTX-CXM-NI-C | 6 | 1 | 0.4 |
| 11 | AP-CXM-NI-C-T-DXT | 6 | 1 | 0.4 |
| 12 | AUG-AP-CXM-NOR-NI-C-NA-T-DXT | 9 | 1 | 0.6 |
| 13 | AP-CXM-NI-C-T-DXT | 6 | 1 | 0.4 |
| 14 | AP-NI-C-T-DXT | 5 | 1 | 0.3 |
| 15 | AP-CXM-NI-C-T-DXT | 6 | 1 | 0.4 |
| 16 | AP-CTX-CXM-NI-C | 5 | 1 | 0.3 |
| 17 | AP-CTX-CXM-NI-C-NA-T-DXT | 8 | 2 | 0.5 |
| 18 | AUG-AP-CTX-CXM-NI-C-NA-T-DXT | 9 | 1 | 0.6 |
| 19 | AUG-AP-CTX-CXM-NI-C-NA-T | 8 | 1 | 0.5 |
| 20 | AUG-AP-CTX-CXM-NI-NA-TS-T-DXT | 9 | 1 | 0.6 |
| 21 | AUG-AP-CTX-CXM-NI-NA-T-DXT | 8 | 1 | 0.5 |
| 22 | AUG-AP-CTX-CXM-NI-C-T-DXT | 8 | 1 | 0.5 |
| 23 | AUG-AP-CTX-CXM-NOR-NI-C-T-DXT | 9 | 1 | 0.6 |
| 24 | AUG-AP-CTX-CXM-NI-C-NA-T-DXT | 9 | 3 | 0.6 |
| 25 | AUG-AP-CTX-CXM-NI-C-NA | 7 | 2 | 0.4 |
| 26 | GM-AUG-AP-MEM-CTX-CXM-NI-C-NA | 9 | 1 | 0.6 |
| 27 | AUG-NI-C-T-DXT | 5 | 1 | 0.3 |
| K. pneumoniae | ||||
| 1 | AUG-AP-CTX-CXM-NI-C-NA | 7 | 1 | 0.4 |
| 2 | AUG-AP-IMI-CTX-CXM-NI C-NA | 8 | 1 | 0.5 |
| 3 | AUG-AP-CTX-CXM-NI-C | 6 | 1 | 0.4 |
| 4 | AUG-AP-CTX-CXM-CIP-NOR-NI-C-NA-TS-T | 11 | 2 | 0.7 |
| 5 | AUG-AP-IMI-CXM-NI-C-NA-TS-T | 9 | 1 | 0.6 |
| 6 | AUG-AP-CTX-CXM-NI-C-NA-TS-T | 9 | 1 | 0.6 |
| 7 | AP-CTX-CXM-NI-C-NA-TS-T | 8 | 1 | 0.5 |
| 8 | GM-AUG-AP-MEM-CTX-CXM-NI-C-NA-TS-DXT | 11 | 1 | 0.7 |
| 9 | AUG-AP-MEM-CTX-CXM-NOR-NI-NA-TS | 9 | 1 | 0.6 |
| 10 | AUG-AP-MEM-CTX-CXM-CIP-NOR-C-NA-TS-T-DXT | 12 | 1 | 0.8 |
| 11 | AUG-AP-CXM-NI-C-NA-TS-T-DXT | 9 | 1 | 0.6 |
| 12 | AUG-AP-CTX-CXM-NI-C-NA-TS-T-DXT | 10 | 1 | 0.6 |
| 13 | AUG-AP-NI | 3 | 2 | 0.2 |
| 14 | AUG-AP-CTX-CXM-NI-T | 6 | 1 | 0.4 |
| 15 | AUG-AP-CTX-CXM-NA | 5 | 1 | 0.3 |
| 16 | AUG-AP-CXM | 3 | 1 | 0.2 |
| K. oxytoca | ||||
| 1 | AUG-AP-CTX-CXM-CIP-NI -C-NA-TS | 9 | 1 | 0.6 |
| 2 | AUG-AP-CTX-CXM-NI-C-NA-TS-T-DXT | 10 | 1 | 0.6 |
| 3 | AUG-AP-CTX-CXM-NOR-NI-C-T | 8 | 1 | 0.5 |
| 4 | AUG-AP-CTX-CXM-NOR-NI-C-NA-TS-T | 10 | 1 | 0.6 |
| 5 | AUG-AP-CTX-CXM-CIP-NOR-NI-C-NA-TS-T | 11 | 1 | 0.7 |
| 6 | AUG-AP-CTX-CXM-CIP-NOR-NI-C-NA-TS-T | 11 | 1 | 0.7 |
| 7 | AUG-AP-CTX-CXM-NI-C-T | 7 | 1 | 0.4 |
| 8 | AUG-AP-CXM-NI-T | 5 | 1 | 0.3 |
| 9 | AUG-AP-CTX-CXM-NOR-NI-C-NA-TS-T | 10 | 1 | 0.6 |
| 10 | AUG-AP-CTX-CXM-NI-C-NA-TS | 8 | 1 | 0.5 |
| SN | MAR Phenotypes | No. of Antibiotics | No. of Isolates | MARI |
|---|---|---|---|---|
| S. Typhimurium | ||||
| 1 | AUG-AP-T | 3 | 2 | 0.2 |
| 2 | AP-NA-T | 3 | 1 | 0.2 |
| E. cloacae | ||||
| 1 | AUG-NA-T-DXT | 4 | 1 | 0.3 |
| 2 | AUG-AP-T | 3 | 1 | 0.2 |
| 3 | AUG-AP-CTX-CXM-NI-C-NA-T-DXT | 9 | 1 | 0.6 |
| 4 | AUG-AP-NI | 3 | 1 | 0.2 |
| 5 | AUG-AP-CXM | 3 | 2 | 0.2 |
| 6 | AUG-AP-CTX-CXM-NI-C | 6 | 1 | 0.4 |
| 7 | AUG-AP-CTX-CXM-NI | 5 | 1 | 0.3 |
| 8 | AP-CXM-NI-T | 4 | 1 | 0.3 |
| 9 | PB-T-DXT | 3 | 1 | 0.2 |
| 10 | AUG-AP-CXM-C-T-DXT | 6 | 1 | 0.4 |
| 11 | AUG-AP-CTX | 3 | 1 | 0.2 |
| 12 | AUG-AP-CTX-CXM-NI-NA-T-DXT | 8 | 2 | 0.5 |
| 13 | AUG-AP-CTX-CXM-NI-NA | 6 | 2 | 0.4 |
| 14 | AUG-AP-CTX-CXM-NI-NA-TS-T-DXT | 9 | 1 | 0.6 |
| 15 | AUG-AP-CTX-CXM-NI-NA-TS | 7 | 1 | 0.4 |
| 16 | CXM-NI-C | 3 | 1 | 0.2 |
| K. pneumoniae | ||||
| 1 | AUG-AP-T-DXT | 4 | 1 | 0.3 |
| 2 | AUG-AP-CTX-CXM-NI-C-T-DXT | 8 | 2 | 0.5 |
| 3 | AUG-AP-CTX-CXM-NI-C-NA-T | 8 | 1 | 0.5 |
| 4 | AUG-AP-CIP | 3 | 1 | 0.2 |
| 5 | AUG-AP-CXM | 3 | 2 | 0.2 |
| K. oxytoca | ||||
| 1 | AUG-AP-CTX-CXM-NI-NA-TS-DXT | 8 | 1 | 0.5 |
| 2 | AUG-AP-CXM | 3 | 1 | 0.2 |
| SN | MAR Genotypes | No. of Resistance Genes other than β-lactamases | No. of β-lactamases | No. of Observed Pattern |
|---|---|---|---|---|
| S. Typhimurium | ||||
| 1 | tetC- sulII | 2 | 0 | 3 |
| 2 | tetC- blaAmpC | 1 | 1 | 1 |
| 3 | tetC -sulII- blaAmpC | 2 | 1 | 1 |
| E. cloacae | ||||
| 1 | tetA- tetB- sulI- sulII- catII- blaTEM | 5 | 1 | 1 |
| 2 | tetA- tetB- sulI- sulII | 4 | 0 | 1 |
| 3 | tetA- tetB- sulI- sulII- blaTEM | 4 | 1 | 2 |
| 4 | tetA- sulI- sulII- blaTEM | 3 | 1 | 1 |
| 5 | tetA- tetB- blaTEM | 2 | 1 | 1 |
| 6 | tetC- sulI- sulII- catII- blaPER | 4 | 1 | 1 |
| 7 | tetC- catII- blaTEM- blaPER | 2 | 2 | 1 |
| 8 | blaTEM- blaPER | 0 | 2 | 1 |
| 9 | catII- blaTEM- blaPER | 1 | 2 | 1 |
| 10 | tetC- sulI- sulII- catII- blaTEM | 4 | 1 | 1 |
| 11 | sulI- sulII- catII- blaTEM | 3 | 1 | 1 |
| 12 | tetB-sulI-sulII-catII- blaAmpC- blaTEM | 4 | 2 | 1 |
| 13 | sulI- catII- blaTEM | 2 | 1 | 1 |
| 14 | sulI- sulII- catII- blaTEM | 3 | 1 | 1 |
| 15 | catII- blaTEM- blaPER | 1 | 2 | 1 |
| 16 | sulI- sulII- catII | 3 | 0 | 2 |
| K. pneumoniae | ||||
| 1 | blaFOX- blaCIT | 0 | 2 | 2 |
| 2 | tetD- sulI- blaTEM | 2 | 1 | 1 |
| 3 | sulII- blaTEM | 1 | 1 | 1 |
| 4 | tetA- blaSHV | 1 | 1 | 1 |
| 5 | tetA- tetC | 2 | 0 | 1 |
| SN | MAR Genotypes | No. of Resistance Genes other than β-lactamases | No. of β-lactamases | No. Observed |
|---|---|---|---|---|
| S. Typhimurium | ||||
| 1 | tetC- sulII | 2 | 0 | 6 |
| E. cloacae | ||||
| 1 | blaAmpC- blaPER | 0 | 2 | 1 |
| 2 | tetA- blaAmpC | 1 | 1 | 1 |
| K. pneumoniae | ||||
| 1 | blaFOX- blaCIT- blaCTX-Ma | 0 | 3 | 1 |
| 2 | blaFOX- blaCIT | 0 | 2 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwu, C.D.; du Plessis, E.M.; Korsten, L.; Nontongana, N.; Okoh, A.I. Antibiogram Signatures of Some Enterobacteria Recovered from Irrigation Water and Agricultural Soil in two District Municipalities of South Africa. Microorganisms 2020, 8, 1206. https://doi.org/10.3390/microorganisms8081206
Iwu CD, du Plessis EM, Korsten L, Nontongana N, Okoh AI. Antibiogram Signatures of Some Enterobacteria Recovered from Irrigation Water and Agricultural Soil in two District Municipalities of South Africa. Microorganisms. 2020; 8(8):1206. https://doi.org/10.3390/microorganisms8081206
Chicago/Turabian StyleIwu, Chidozie Declan, Erika M du Plessis, Lise Korsten, Nolonwabo Nontongana, and Anthony Ifeanyi Okoh. 2020. "Antibiogram Signatures of Some Enterobacteria Recovered from Irrigation Water and Agricultural Soil in two District Municipalities of South Africa" Microorganisms 8, no. 8: 1206. https://doi.org/10.3390/microorganisms8081206