Comparative Study of the Proteins Involved in the Fermentation-Derived Compounds in Two Strains of Saccharomyces cerevisiae during Sparkling Wine Second Fermentation
Abstract
:1. Introduction
2. Materials and Methods
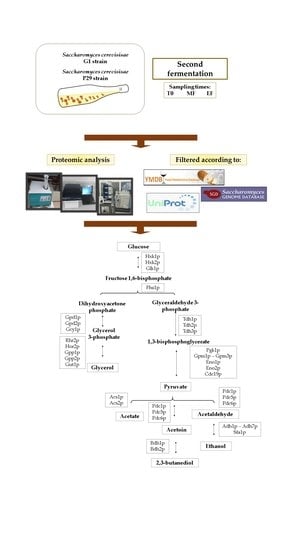
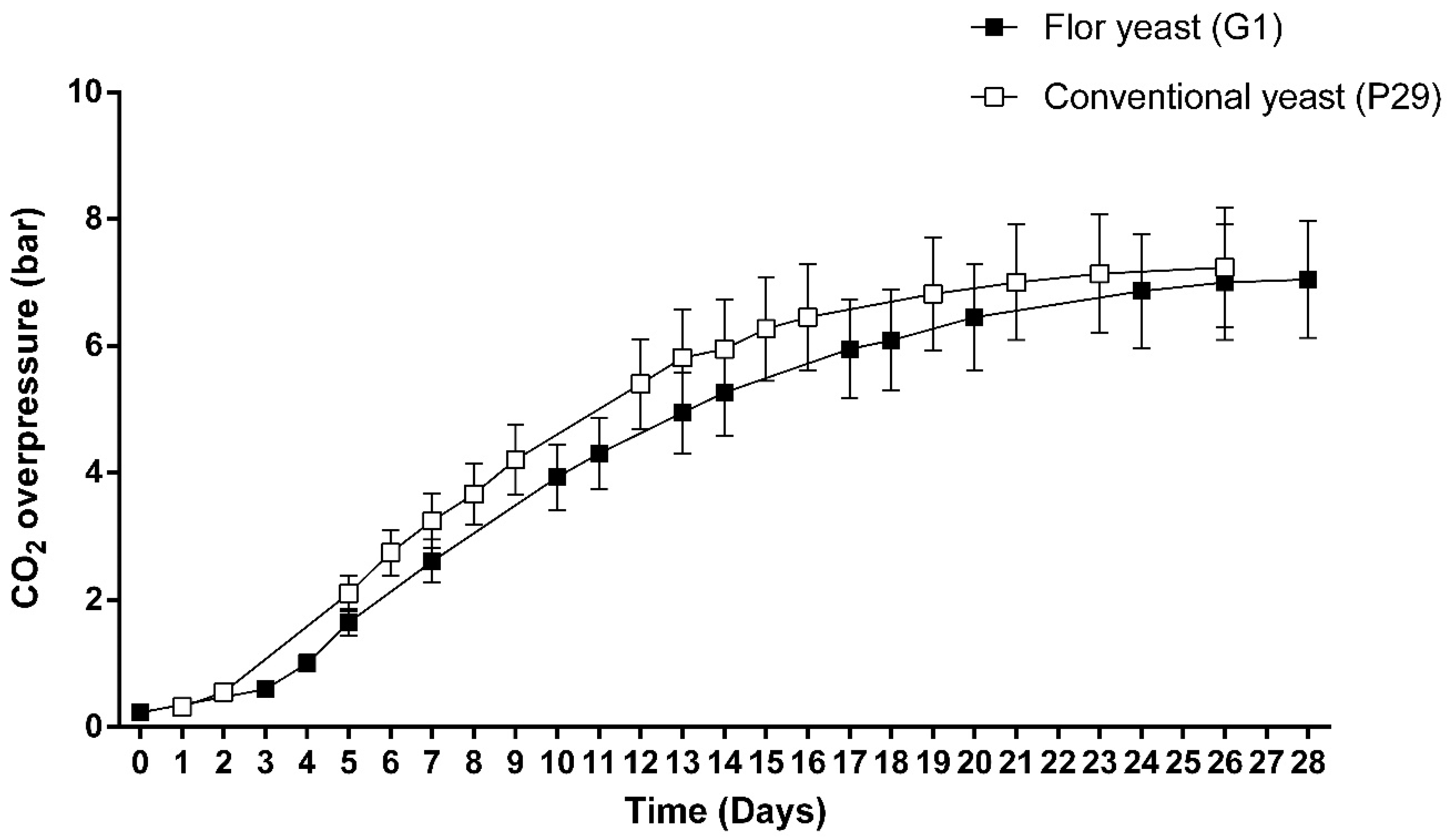
2.1. Microorganism and Experimental Conditions
2.2. Proteome Analysis
2.3. Statistical Analysis
3. Results and Discussion
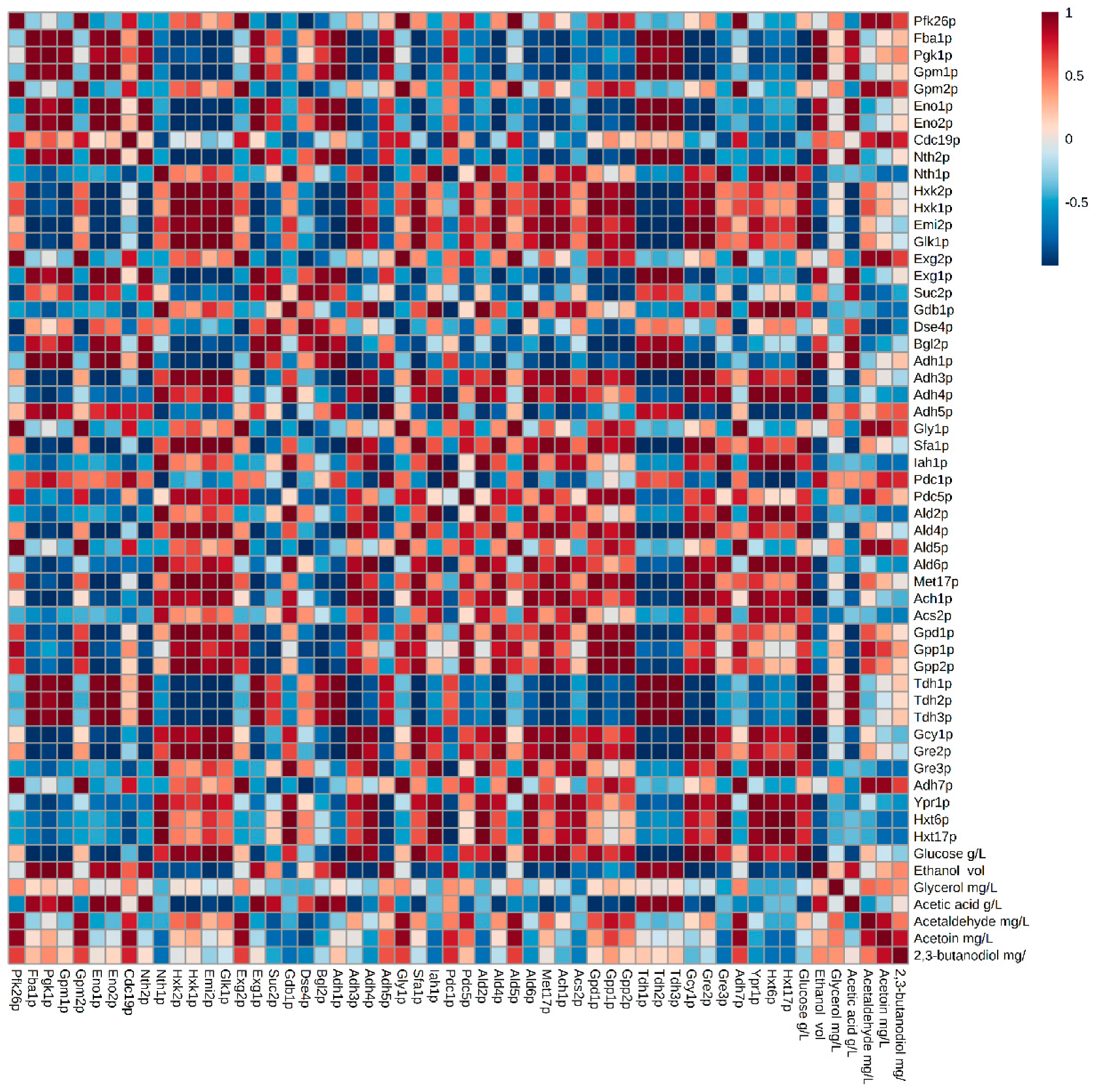
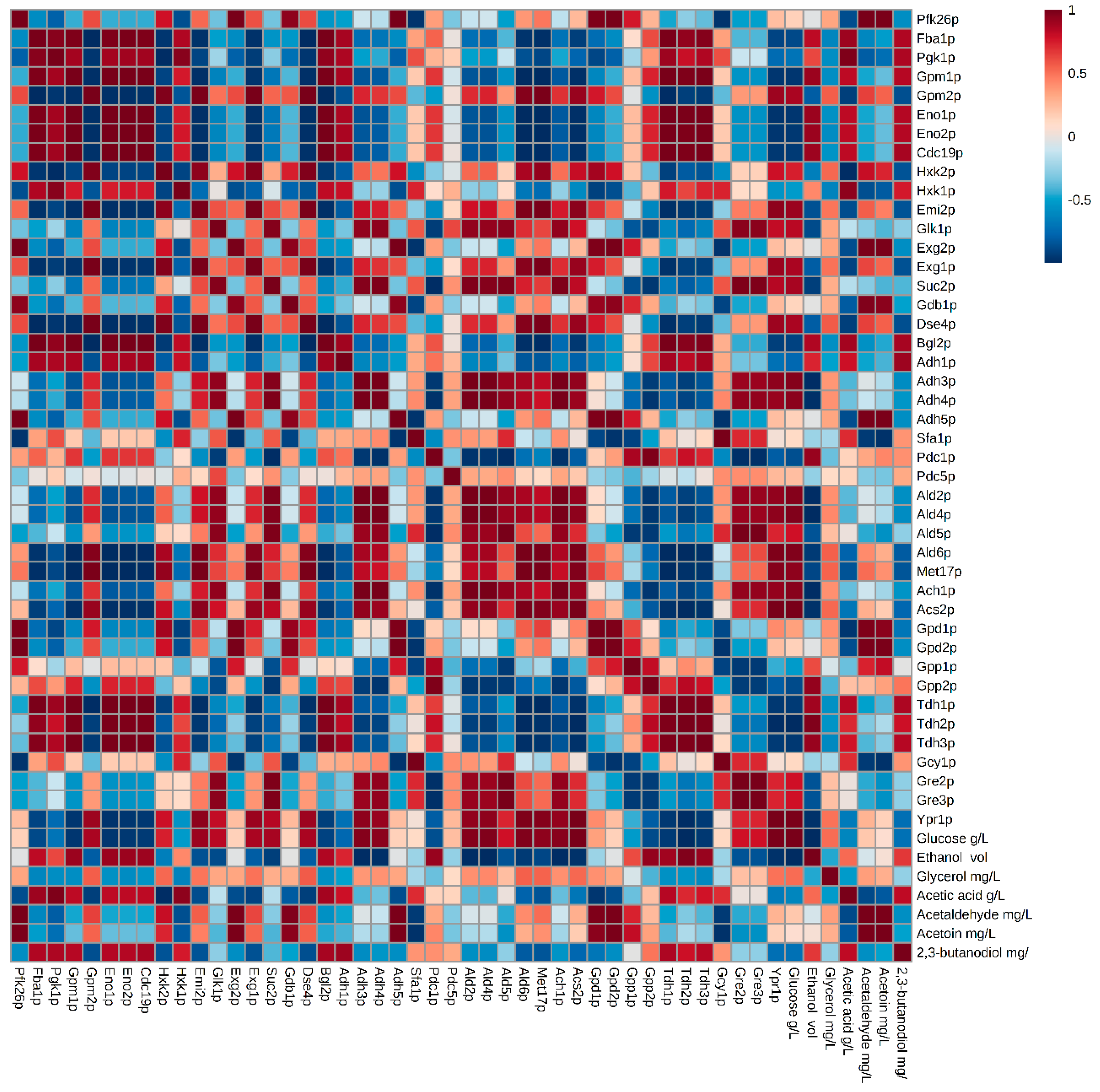
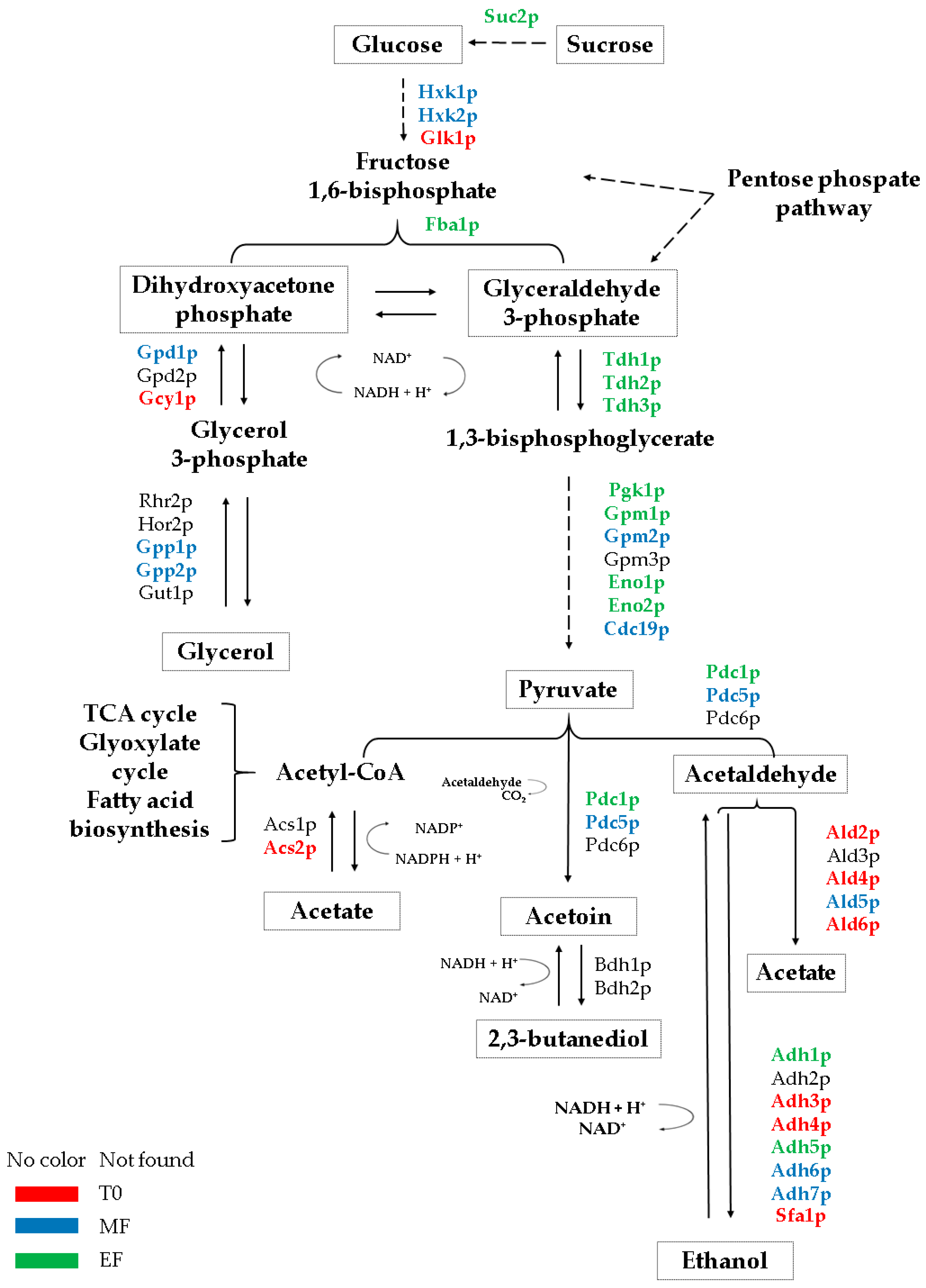
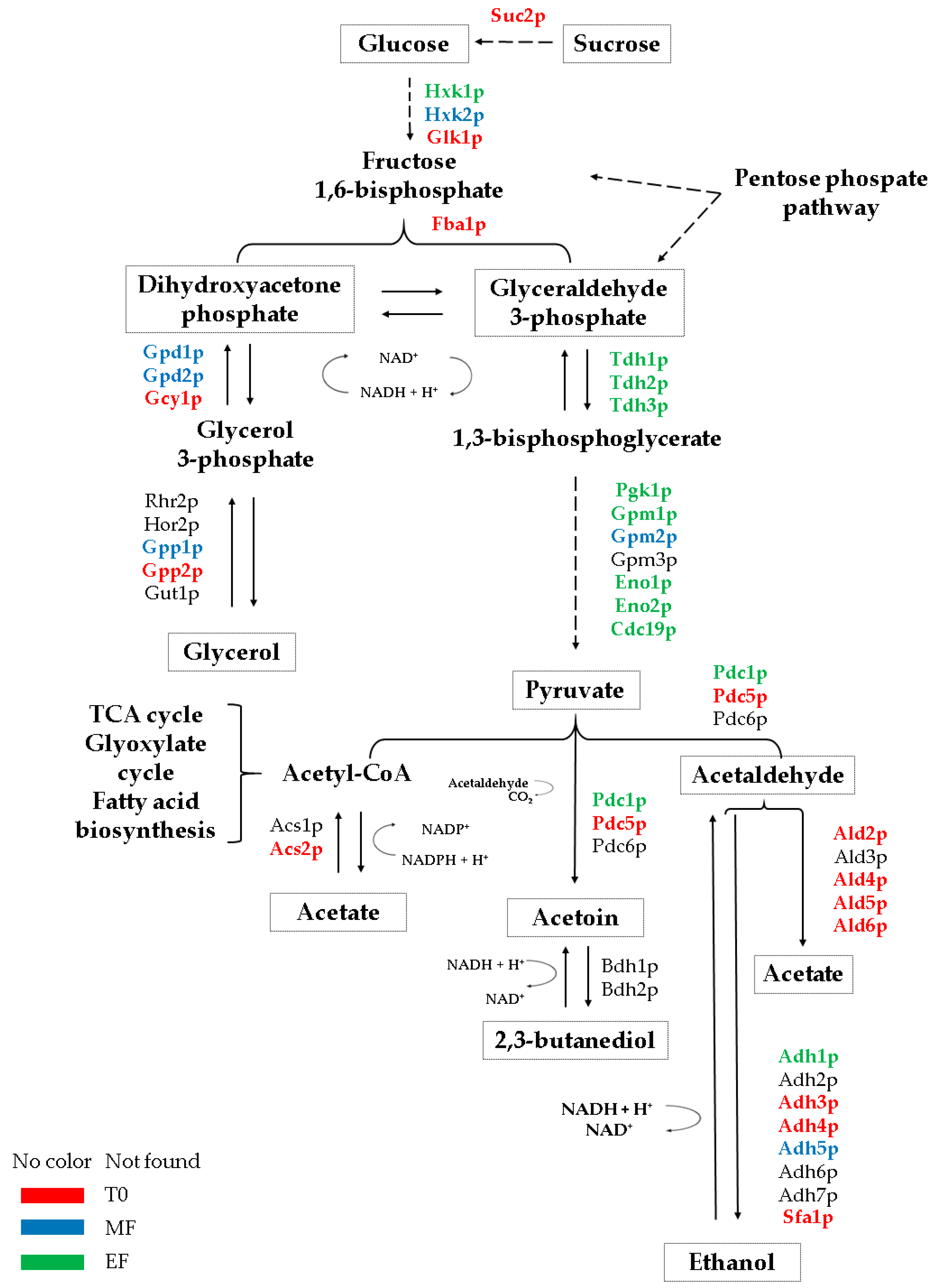
3.1. Glycolysis/Gluconeogenesis Proteome
3.2. Proteins Related to the Metabolism of Pyruvate to Ethanol and Acetic Acid
3.3. Proteins Related to the Metabolism of Pyruvate-Acetoin-2,3-Butanediol
3.4. Proteins Related to the Metabolism of Dihydroxyacetone Phosphate-Glycerol.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pozo-Bayón, M.Á.; Martínez-Rodríguez, A.; Pueyo, E.; Moreno-Arribas, M.V. Chemical and biochemical features involved in sparkling wine production: From a traditional to an improved winemaking technology. Trends Food Sci. Technol. 2009, 20, 289–299. [Google Scholar] [CrossRef]
- Buxaderas, S.; López-Tamames, E. Sparkling wines: Features and trends from tradition. In Advances in Food and Nutrition Research; Academic Press: Cambridge, MA, USA, 2012; Volume 66, pp. 1–45. [Google Scholar]
- Penacho, V.; Valero, E.; González, R. Transcription profiling of sparkling wine second fermentation. Int. J. Food Microbiol. 2012, 153, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Arena, P.M.; Laddomada, B.; Cappello, S.M.; Bleve, G.; Grieco, F.; Beneduce, L.; Berbegal, C.; Spano, G.; Capozzi, V. Starter cultures for sparkling wine. Fermentation 2016, 2, 21. [Google Scholar] [CrossRef]
- Alexandre, H.; Guilloux–Nematier, M. Yeast autolysis in sparkling wine—A review. Aust. J. Grape Wine Res. 2006, 12, 119–127. [Google Scholar] [CrossRef]
- Sartor, S.; Toaldo, I.M.; Panceri, C.P.; Caliari, V.; Luna, A.S.; de Gois, J.S.; Bordignon-Luiz, M.T. Changes in organic acids, polyphenolic and elemental composition of rosé sparkling wines treated with mannoproteins during over-lees aging. Food Res. Int. 2019, 124, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Ortega, N.; Martín-Santamaría, M.; Acedo, A.; Marquina, D.; Pascual, O.; Rozès, N.; Zamora, F.; Santos, A.; Belda, I. Occurrence and enological properties of two new non-conventional yeasts (Nakazawaea ishiwadae and Lodderomyces elongisporus) in wine fermentations. Int. J. Food Microbiol. 2019, 305, 108255. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.; Moreno-García, J.; López-Muñoz, B.; Mauricio, J.C.; García-Martínez, T. Use of a flor velum yeast for modulating colour, ethanol and major aroma compound contents in red wine. Food Chem. 2016, 213, 90–97. [Google Scholar] [CrossRef]
- Quincozes, L.; Santos, P.; Vieira, L.; Gabbardo, M.; Eckhardt, D.P.; Cunha, W.; Costa, V.; Zigiotto, L.; Schumacher, R. Influence of yeasts of the genus Saccharomyces and not Saccharomyces in elaboration of white wines. BIO Web Conf. 2019, 12, 02014. [Google Scholar] [CrossRef]
- Alonso-del-Real, J.; Lairón-Peris, M.; Barrio, E.; Querol, A. Effect of temperature on the prevalence of Saccharomyces non cerevisiae species against a S. cerevisiae wine strain in wine fermentation: Competition, physiological fitness, and influence in final wine composition. Front. Microbiol. 2017, 8, 150. [Google Scholar] [CrossRef] [Green Version]
- Ciani, M.; Comitini, F. Non-Saccharomyces wine yeasts have a promising role in biotechnological approaches to winemaking. Ann. Microbiol. 2011, 61, 25–32. [Google Scholar] [CrossRef]
- Nuñez-Guerrero, M.A.; Paez-Lerma, J.B.; Rutiaga-Quiñones, O.M.; González-Herrera, S.M.; Soto-Cruz, N.O. Performance of mixtures of Saccharomyces and non-Saccharomyces native yeasts during alcoholic fermentation of Agave duranguensis juice. Food Microbiol. 2016, 54, 91–97. [Google Scholar] [CrossRef]
- Suárez-Lepe, J.A.; Morata, A. New trends in yeast selection for winemaking. Trends Food Sci. Technol. 2012, 23, 39–50. [Google Scholar] [CrossRef]
- Ciani, M.; Morales, P.; Comitini, F.; Tronchoni, J.; Canonico, L.; Curiel, J.A.; Oro, L.; Rodrigues, A.J.; Gonzalez, R. Non-conventional yeast species for lowering ethanol content of wines. Front. Microbiol. 2016, 7, 642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina-Trujillo, L.; González-Royo, E.; Sieczkowski, N.; Heras, J.; Canals, J.M.; Zamora, F. Effect of sequential inoculation (Torulaspora delbrueckii/Saccharomyces cerevisiae) in the first fermentation on the foaming properties of sparkling wine. Eur. Food Res. Technol. 2017, 243, 681–688. [Google Scholar] [CrossRef]
- Alexandre, H. Flor yeasts of Saccharomyces cerevisiae-their ecology, genetics and metabolism. Int. J. Food Microbiol. 2013, 167, 269–275. [Google Scholar] [CrossRef]
- Martínez-García, R.; Roldán-Romero, Y.; Moreno, J.; Puig-Pujol, A.; Mauricio, J.C.; García-Martínez, T. Use of a flor yeast strain for the second fermentation of sparkling wines: Effect of endogenous CO2 over-pressure on the volatilome. Food Chem. 2020, 308, 125555. [Google Scholar] [CrossRef]
- González-Jiménez, M.C.; Moreno-García, J.; García-Martínez, T.; Moreno, J.J.; Puig-Pujol, A.; Capdevilla, F.; Mauricio, J.C. Differential analysis of proteins involved in ester metabolism in two Saccharomyces cerevisiae strains during the second fermentation in sparkling wine elaboration. Microorganisms 2020, 8, 403. [Google Scholar] [CrossRef] [Green Version]
- Mauricio, J.C.; Moreno, J.J.; Ortega, J.M. In vitro specific activities of alcohol and aldehyde dehydrogenases from two flor yeast during controlled wine ageing. J. Agric. Food Chem. 1997, 45, 1967–1971. [Google Scholar] [CrossRef]
- Porras-Agüera, J.A.; Moreno-García, J.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. First proteomic approach to identify cell death biomarkers in wine yeasts during sparkling wine production. Microorganisms 2019, 7, 542. [Google Scholar] [CrossRef] [Green Version]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Moreno-García, J.; García-Martínez, T.; Moreno, J.; Mauricio, J.C. Proteins involved in flor yeast carbon metabolism under biofilm formation conditions. Food Microbiol. 2015, 46, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Porras-Agüera, J.A.; Moreno-García, J.; González-Jiménez, M.C.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. Autophagic proteome in two Saccharomyces cerevisiae strains during second fermentation for sparkling wine elaboration. Microorganisms 2020, 8, 523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seisonen, S.; Vene, K.; Koppel, K. The current practice in the application of chemometrics for correlation of sensory and gas chromatography data. Food Chem. 2016, 210, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, R.; García-Martínez, T.; Puig-Pujol, A.; Mauricio, J.C.; Moreno, J. Changes in sparkling wine aroma during the second fermentation under CO2 pressure in sealed bottle. Food Chem. 2017, 237, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Valadi, Å.; Ansell, R.; Gustafsson, L.; Adler, L.; Norbeck, J.; Blomberg, A. NADH-reductive stress in Saccharomyces cerevisiae induces the expression of the minor isoform of glyceraldehyde-3-phosphate dehydrogenase (TDH1). Curr. Genet. 2004, 45, 90–95. [Google Scholar] [PubMed]
- Esteve-Zarzoso, B.; Peris-Torán, M.J.; Garcıa-Maiquez, E.; Uruburu, F.; Querol, A. Yeast population dynamics during the fermentation and biological aging of sherry wines. Appl. Environ. Microb. 2001, 67, 2056–2061. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M.; Botstein, D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell 1982, 28, 145–154. [Google Scholar] [CrossRef]
- Byrne, S.; Howell, G. Acetaldehyde: How to Limit its Formation during Fermentation; Aust, N.Z., Ed.; Grapegrower Winemaker: Sydney, NSW, Australia, 2017; pp. 68–69. [Google Scholar]
- Mojzita, D.; Hohmann, S. Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae. Mol. Genet. Genom. 2006, 276, 147–161. [Google Scholar] [CrossRef]
- Moreno-García, J.; García-Martínez, T.; Millán, M.C.; Mauricio, J.C.; Moreno, J. Proteins involved in wine aroma compounds metabolism by a Saccharomyces cerevisiae flor-velum yeast strain grown in two conditions. Food Microbiol. 2015, 51, 1–9. [Google Scholar] [CrossRef]
- Ugliano, M.; Henschke, P.A. Yeasts and Wine Flavour. In Wine Chemistry and Biochemistry; Springer: Berlin/Heidelberg, Germany, 2009; pp. 313–392. [Google Scholar]
- Remize, F.; Andrieu, E.; Dequin, S. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: Role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl. Environ. Microbiol. 2000, 66, 3151–3159. [Google Scholar] [CrossRef] [Green Version]
- Pigeau, G.M.; Inglis, D.L. Response of wine yeast (Saccharomyces cerevisiae) aldehyde dehydrogenases to acetaldehyde stress during Icewine fermentation. J. Appl. Microbiol. 2007, 103, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- González, V.F. Aroma, aromas y olores del vino. In Análisis Sensorial y Cata de los Vinos de España; Dialnet: Logroño, Spain, 2001; pp. 207–233. [Google Scholar]
- Vilela-Moura, A.; Schuller, D.; Mendes-Faia, A.; Silva, R.D.; Chaves, S.R.; Sousa, M.J.; Corte-Real, M. The impact of acetate metabolism on yeast fermentative performance and wine quality: Reduction of volatile acidity of grape musts and wines. Appl. Microbiol. Biotechnol. 2011, 89, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boubekeur, S.; Camougrand, N.; Bunoust, O.; Rigoulet, M.; Guérin, B. Participation of acetaldehyde dehydrogenases in ethanol and pyruvate metabolism of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 2001, 268, 5057–5065. [Google Scholar] [CrossRef] [PubMed]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; Van Dijken, J.P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 1990, 136, 395–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, P.; Suzzi, G. Origin and Production of Acetoin during Wine Yeast Fermentation. Appl. Environ. Microbiol. 1996, 62, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Michnick, S.; Roustan, J.L.; Remize, F.; Barre, P.; Dequin, S. Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol 3-phosphate dehydrogenase. Yeast 1997, 13, 783–793. [Google Scholar] [CrossRef]
- Romano, P.; Brandolini, V.; Ansaloni, C.; Menziani, E. The production of 2,3-butanediol as a differentiating character in wine yeasts. World J. Microbiol. Biotechnol. 1998, 14, 649–653. [Google Scholar] [CrossRef]
- Borrull, A.; Poblet, M.; Rozès, N. New insights into the capacity of commercial wine yeasts to grow on sparkling wine media. Factor screening for improving wine yeast selection. Food Microbiol. 2015, 48, 41–48. [Google Scholar] [CrossRef]
- Kemp, B.; Alexandre, H.; Robillard, B.; Marchal, R. Effect of production phase on bottle-fermented sparkling wine quality. J. Agric. Food Chem. 2015, 63, 19–38. [Google Scholar] [CrossRef]
- Goold, H.D.; Kroukamp, H.; Williams, T.C.; Paulsen, I.T.; Varela, C.; Pretorius, I.S. Yeast′s balancing act between ethanol and glycerol production in low-alcohol wines. Microb. Biotechnol. 2017, 10, 264–278. [Google Scholar] [CrossRef]
- Hohmann, S. An integrated view on a eukaryotic osmoregulation system. Curr. Genet. 2015, 61, 373–382. [Google Scholar] [CrossRef] [PubMed]
- García-Mauricio, J.C.; García-Martínez, T. Role of Yeasts in Sweet Wines. In Book Sweet, Reinforced and Fortified Wines: Grape Biochemistry, Technology and Vinification; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Remize, F.; Cambon, B.; Barnavon, L.; Dequin, S. Glycerol formation during wine fermentation is mainly linked to Gpd1p and is only partially controlled by the HOG pathway. Yeast 2003, 20, 1243–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nissen, T.L.; Kielland Brandt, M.C.; Nielsen, J.; Villadsen, J. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab. Eng. 2000, 2, 69–77. [Google Scholar] [CrossRef] [PubMed]
| Flor Yeast | Conventional Yeast | |||||
|---|---|---|---|---|---|---|
| T0 | MF | EF | T0 | MF | EF | |
| Ethanol (% v/v) | 10.23 ± 0.02 | 10.76 ± 0.04 | 11.4 ± 0.1 | 10.23 ± 0.02 | 10.85 ± 0.04 | 11.60 ± 0.03 |
| Acetaldehyde (mg/L) | 87 ± 1 | 132 ± 1 | 87 ± 16 | 87.2 ± 1.1 | 133.9 ± 7.3 | 85.2 ± 0.2 |
| Acetoin (mg/L) | 19 ± 1 | 61 ± 1 | 31 ± 2 | 19.3 ± 1.3 | 127.5 ± 11.8 | 24.5 ± 6.3 |
| 2,3-butanediol (mg/L) | 171 ± 7 | 221 ± 4 | 200 ± 33 | 171 ± 6 | 166 ± 4 | 192 ± 12 |
| Acetic acid (g/L) | 0.23 ± 0.02 | 0.20 ± 0.00 | 0.28 ± 0.02 | 0.23 ± 0.02 | 0.20 ± 0.00 | 0.22 ± 0.00 |
| Glycerol (mg/L) | 4020 ± 656 | 4493 ± 164 | 4227 ± 297 | 4019 ± 655 | 4557 ± 212 | 3513 ± 163 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Jiménez, M.d.C.; García-Martínez, T.; Mauricio, J.C.; Sánchez-León, I.; Puig-Pujol, A.; Moreno, J.; Moreno-García, J. Comparative Study of the Proteins Involved in the Fermentation-Derived Compounds in Two Strains of Saccharomyces cerevisiae during Sparkling Wine Second Fermentation. Microorganisms 2020, 8, 1209. https://doi.org/10.3390/microorganisms8081209
González-Jiménez MdC, García-Martínez T, Mauricio JC, Sánchez-León I, Puig-Pujol A, Moreno J, Moreno-García J. Comparative Study of the Proteins Involved in the Fermentation-Derived Compounds in Two Strains of Saccharomyces cerevisiae during Sparkling Wine Second Fermentation. Microorganisms. 2020; 8(8):1209. https://doi.org/10.3390/microorganisms8081209
Chicago/Turabian StyleGonzález-Jiménez, María del Carmen, Teresa García-Martínez, Juan Carlos Mauricio, Irene Sánchez-León, Anna Puig-Pujol, Juan Moreno, and Jaime Moreno-García. 2020. "Comparative Study of the Proteins Involved in the Fermentation-Derived Compounds in Two Strains of Saccharomyces cerevisiae during Sparkling Wine Second Fermentation" Microorganisms 8, no. 8: 1209. https://doi.org/10.3390/microorganisms8081209