Involvement of the cbb3-Type Terminal Oxidase in Growth Competition of Bacteria, Biofilm Formation, and in Switching between Denitrification and Aerobic Respiration
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Media
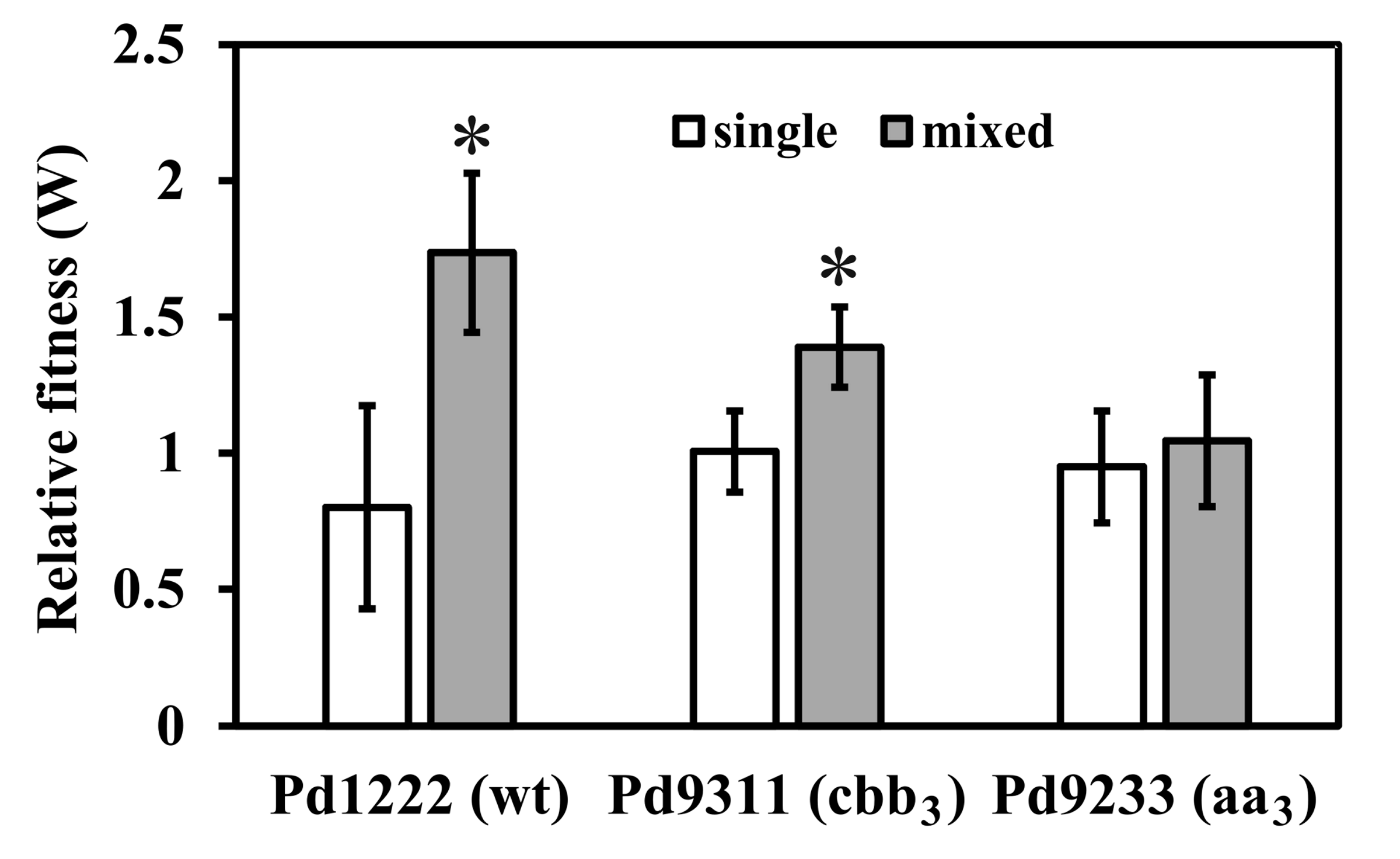
2.3. Growth Competition Assays
2.4. Aerobic Growth Study
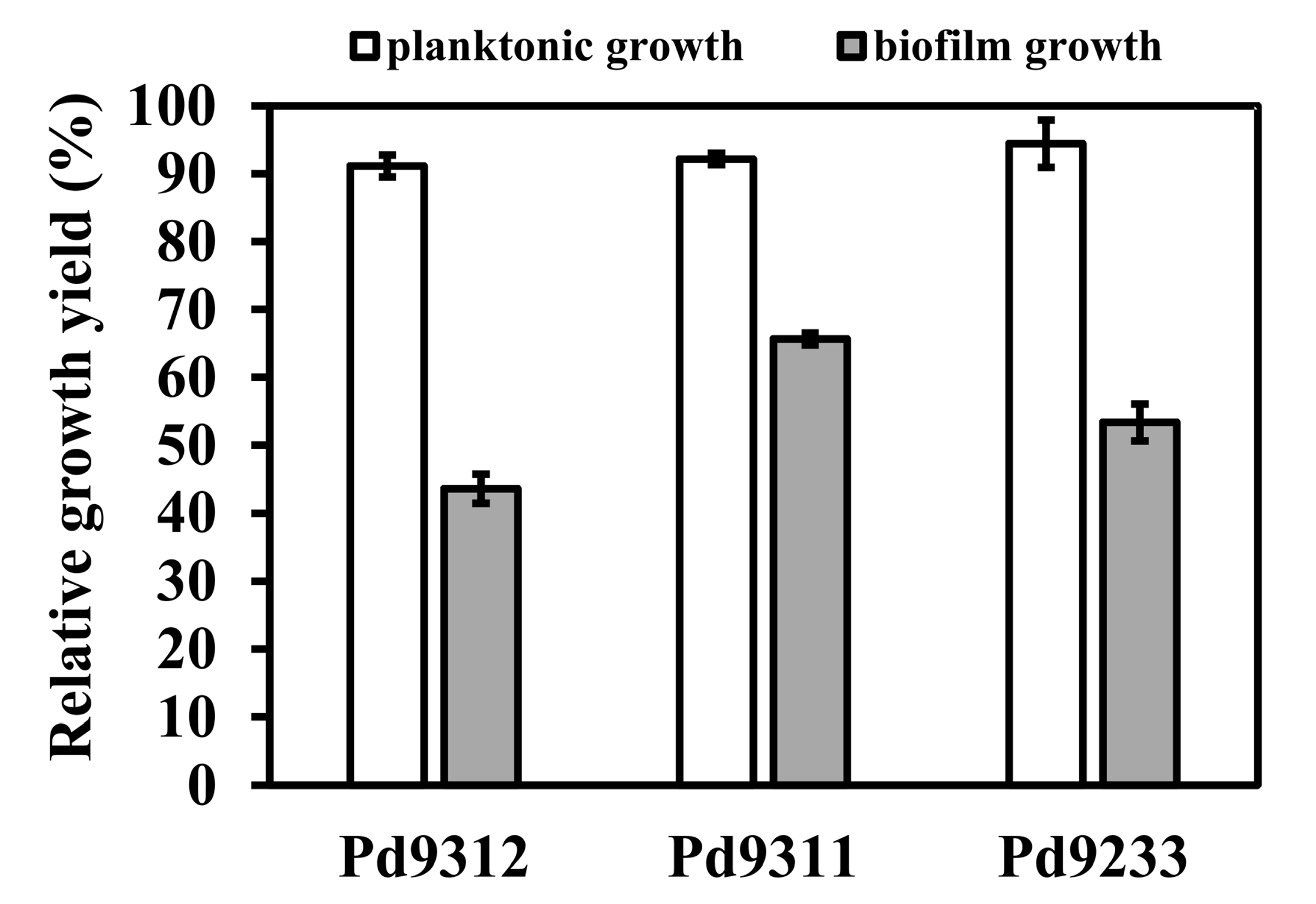
2.5. Biofilm Formation and Quantification
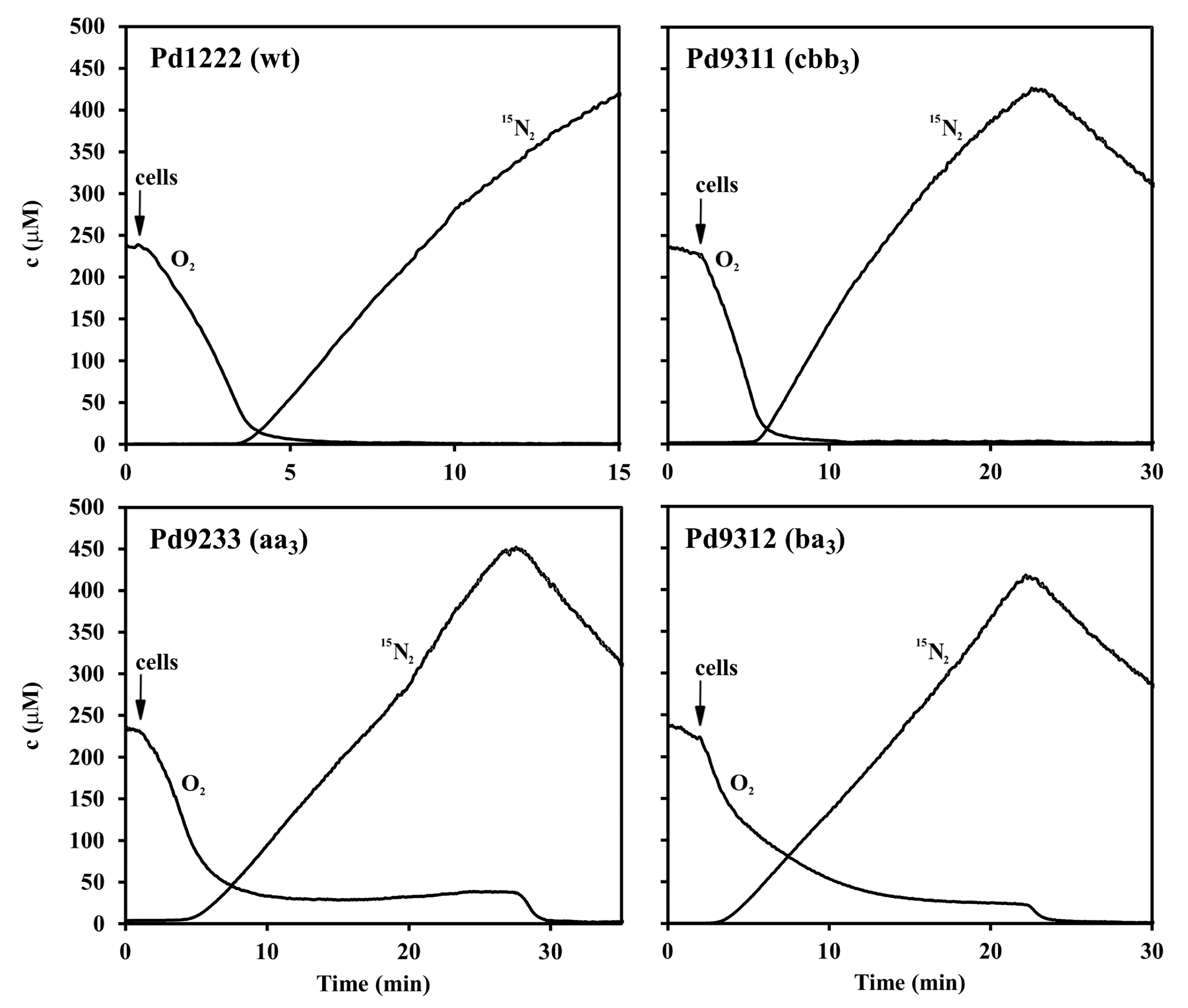
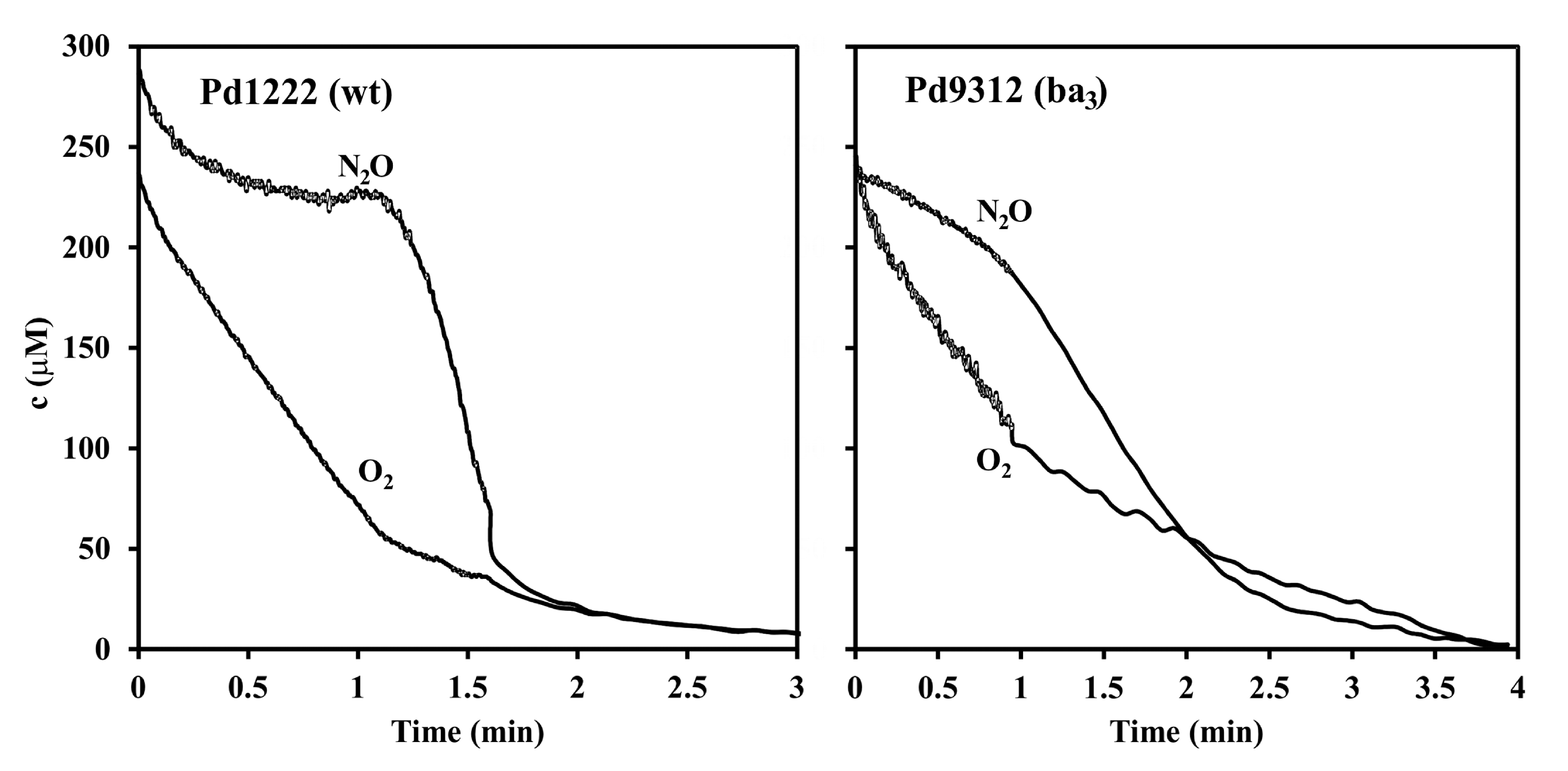
2.6. Membrane Inlet Mass Spectrometry (MIMS) Measurements
2.7. Cytochromes c Spectrophotometry
2.8. Statistical Analysis
3. Results
3.1. Comparative Growth Experiments
3.2. Biofilm-Forming Ability
3.3. Interaction between Aerobic Respiration and Denitrification
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [CrossRef]
- Coyne, M.S. Biological denitrification. In Nitrogen in Agricultural Systems; Schepers, J.S., Raun, W.R., Eds.; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2008; pp. 202–254. [Google Scholar]
- Kucera, I.; Dadak, V.; Dobry, R. The distribution of redox equivalents in the anaerobic respiratory chain of Paracoccus denitrificans. Eur. J. Biochem. 1983, 130, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ni, B.J.; Lu, H.; Chandran, K.; Richardson, D.; Yuan, Z. Evaluating two concepts for the modelling of intermediates accumulation during biological denitrification in wastewater treatment. Water Res. 2015, 71, 21–31. [Google Scholar] [PubMed]
- Chen, J.; Strous, M. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys. Acta 2013, 1827, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.; Yang, K.; Zhu, L.; Jiang, Y.; Wang, H.Y.; Zhou, J.; Zhang, H.N. Aerobic denitrification: A review of important advances of the last 30 years. Biotechnol. Bioprocess. Eng. 2015, 20, 643–651. [Google Scholar] [CrossRef]
- Baker, S.C.; Ferguson, S.J.; Ludwig, B.; Page, M.D.; Richter, O.M.; van Spanning, R.J. Molecular genetics of the genus Paracoccus: Metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 1998, 62, 1046–1078. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.J. Paracoccus denitrificans Oxidative Phosphorylation: Retentions, Gains, Losses, and Lessons En Route to Mitochondria. IUBMB Life 2018, 70, 1214–1221. [Google Scholar] [CrossRef]
- Kaplan, P.; Kucera, I. Cytochromes c-dependent aerobic respiration of Paracoccus denitrificans. J. Basic Microbiol. 1993, 33, 397–404. [Google Scholar] [CrossRef]
- De Gier, J.W.L.; Lubben, M.; Reijnders, W.N.M.; Tipker, C.A.; Slotboom, D.J.; van Spanning, R.J.M.; Stouthamer, A.H.; Vanderoost, J. The terminal oxidases of Paracoccus denitrificans. Mol. Microbiol. 1994, 13, 183–196. [Google Scholar] [CrossRef]
- Richter, O.M.; Tao, J.S.; Turba, A.; Ludwig, B. A cytochrome ba3 functions as a quinol oxidase in Paracoccus denitrificans. Purification, cloning, and sequence comparison. J. Biol. Chem. 1994, 269, 23079–23086. [Google Scholar]
- Pitcher, R.S.; Watmough, N.J. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 2004, 1655, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Arai, H.; Kawakami, T.; Osamura, T.; Hirai, T.; Sakai, Y.; Ishii, M. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 4206–4215. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Sexstone, A.J.; Parkin, T.B.; Revsbech, N.P.; Shelton, D.R. Anaerobic processes in soil. Plant Soil 1984, 76, 197–212. [Google Scholar] [CrossRef]
- Morris, R.L.; Schmidt, T.M. Shallow breathing: Bacterial life at low O2. Nat. Rev. Microbiol. 2013, 11, 205–212. [Google Scholar] [CrossRef] [PubMed]
- De Vries, G.E.; Harms, N.; Hoogendijk, J.; Stouthamer, A.H. Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a (NGATCN) DNA-modifying property. Arch. Microbiol. 1989, 152, 52–57. [Google Scholar] [CrossRef]
- De Gier, J.W.L.; Schepper, M.; Reijnders, W.N.M.; van Dyck, S.J.; Slotboom, D.J.; Warne, A.; Saraste, M.; Krab, K.; Finel, M.; Stouthamer, A.H.; et al. Structural and functional analysis of aa3-type and cbb3-type cytochrome c oxidases of Paracoccus denitrificans reveals significant differences in proton-pump design. Mol. Microbiol. 1996, 20, 1247–1260. [Google Scholar] [CrossRef]
- Otten, M.F.; Stork, D.R.; Reijnders, W.N.M.; Westerhoff, H.V.; van Spanning, R.J.M. Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur. J. Biochem. 2001, 268, 2486–2497. [Google Scholar] [CrossRef]
- Marrs, B.; Gest, H. Genetic mutations affecting respiratory electron-transport system of photosynthetic bacterium Rhodopseudomonas capsulata. J. Bacteriol. 1973, 114, 1045–1051. [Google Scholar] [CrossRef]
- Lenski, R.E.; Rose, M.R.; Simpson, S.C.; Tadler, S.C. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am. Nat. 1991, 138, 1315–1341. [Google Scholar] [CrossRef]
- Wijtzes, T.; de Wit, J.C.; In, H.; Van’t, R.; Zwietering, M.H. Modelling bacterial growth of Lactobacillus curvatus as a function of acidity and temperature. Appl. Environ. Microbiol. 1995, 61, 2533–2539. [Google Scholar] [CrossRef]
- Kumar, S.; Spiro, S. Environmental and genetic determinants of biofilm formation in Paracoccus denitrificans. mSphere 2017, 2, e00350. [Google Scholar] [CrossRef]
- Thomsen, J.K.; Geest, T.; Cox, R.P. Mass spectrometric studies of the effect of pH on the accumulation of intermediates in denitrification by Paracoccus denitrificans. Appl. Environ. Microbiol. 1994, 60, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Gevantman, L.H. Solubility of selected gases in water. In CRC Handbook of Chemistry and Physics; Lide, D.R., Ed.; CRC Press: Boca Raton, FL, USA, 1992; pp. 82–83. [Google Scholar]
- Kim, I.S.; Jang, A.; Ivanov, V.; Stabnikova, O.; Ulanov, M. Denitrification of drinking water using biofilms formed by Paracoccus denitrificans and microbial adhesion. Environ. Eng. Sci. 2004, 21, 283–290. [Google Scholar] [CrossRef]
- Yoshida, K.; Toyofuku, M.; Obana, N.; Nomura, N. Biofilm formation by Paracoccus denitrificans requires a type I secretion system-dependent adhesin BapA. FEMS Microbiol. Lett. 2017, 364, fnx029. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.L.; Alves, R.J.; Ribeiro, M.A.; Pereira-Leal, J.B.; Teixeira, M.; Pereira, M.M. The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim. Biophys. Acta 2012, 1817, 629–637. [Google Scholar] [CrossRef]
- Rauhamaki, V.; Wikstrom, M. The causes of reduced proton-pumping efficiency in type B and C respiratory heme-copper oxidases, and in some mutated variants of type A. Biochim. Biophys. Acta 2014, 1837, 999–1003. [Google Scholar] [CrossRef]
- Ducluzeau, A.L.; Schoepp-Cothenet, B.; van Lis, R.; Baymann, F.; Russell, M.J.; Nitschke, W. The evolution of respiratory O2/NO reductases: An out-of-the-phylogenetic-box perspective. J. R. Soc. Interface 2014, 11, 20140196. [Google Scholar] [CrossRef]
- Xu, K.D.; Stewart, P.S.; Xia, F.; Huang, C.T.; McFeters, G.A. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 1998, 64, 4035–4039. [Google Scholar] [CrossRef]
- Alvarez-Ortega, C.; Harwood, C.S. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 2007, 65, 153–165. [Google Scholar] [CrossRef]
- Kucera, I.; Kozak, L.; Dadak, V. Aerobic dissimilatory reduction of nitrite by cells of Paracoccus denitrificans: The role of nitric oxide. Biochim. Biophys. Acta 1987, 894, 120–126. [Google Scholar] [CrossRef]
- Conthe, M.; Parchen, C.; Stouten, G.; Kleerebezem, R.; van Loosdrecht, M.C.M. O2 versus N2O respiration in a continuous microbial enrichment. Appl. Microbiol. Biotechnol. 2018, 102, 8943–8950. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Peng, Y.; Li, B.; Guo, J.; Yang, Q.; Wang, S. Biological removal of nitrogen from wastewater. Rev. Environ. Contam. Toxicol. 2008, 192, 159–195. [Google Scholar] [PubMed]
- Mpongwana, N.; Ntwampe, S.K.O.; Omodanisi, E.I.; Chidi, B.S.; Razanamahandry, L.C. Sustainable approach to eradicate the inhibitory effect of free-cyanide on simultaneous nitrification and aerobic denitrification during wastewater treatment. Sustainability 2019, 11, 6180. [Google Scholar] [CrossRef]
- Demone, J.J.; Wan, S.; Nourimand, M.; Hansen, A.E.; Shu, Q.Y.; Altosaar, I. New breeding techniques for greenhouse gas (GHG) mitigation: Plants may express nitrous oxide reductase. Climate 2018, 6, 80. [Google Scholar] [CrossRef]
- Bouchal, P.; Struharova, I.; Budinska, E.; Sedo, O.; Vyhlidalova, T.; Zdrahal, Z.; van Spanning, R.; Kucera, I. Unraveling an FNR based regulatory circuit in Paracoccus denitrificans using a proteomics-based approach. Biochim. Biophys. Acta 2010, 1804, 1350–1358. [Google Scholar] [CrossRef]
- Qu, Z.; Bakken, L.R.; Molstad, L.; Frostegard, A.; Bergaust, L.L. Transcriptional and metabolic regulation of denitrification in Paracoccus denitrificans allows low but significant activity of nitrous oxide reductase under oxic conditions. Environ. Microbiol. 2016, 18, 2951–2963. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kučera, I.; Sedláček, V. Involvement of the cbb3-Type Terminal Oxidase in Growth Competition of Bacteria, Biofilm Formation, and in Switching between Denitrification and Aerobic Respiration. Microorganisms 2020, 8, 1230. https://doi.org/10.3390/microorganisms8081230
Kučera I, Sedláček V. Involvement of the cbb3-Type Terminal Oxidase in Growth Competition of Bacteria, Biofilm Formation, and in Switching between Denitrification and Aerobic Respiration. Microorganisms. 2020; 8(8):1230. https://doi.org/10.3390/microorganisms8081230
Chicago/Turabian StyleKučera, Igor, and Vojtěch Sedláček. 2020. "Involvement of the cbb3-Type Terminal Oxidase in Growth Competition of Bacteria, Biofilm Formation, and in Switching between Denitrification and Aerobic Respiration" Microorganisms 8, no. 8: 1230. https://doi.org/10.3390/microorganisms8081230
APA StyleKučera, I., & Sedláček, V. (2020). Involvement of the cbb3-Type Terminal Oxidase in Growth Competition of Bacteria, Biofilm Formation, and in Switching between Denitrification and Aerobic Respiration. Microorganisms, 8(8), 1230. https://doi.org/10.3390/microorganisms8081230



