The Impact of the Postpartum “Doing-the-Month” Practice on Human Milk Microbiota: A Pilot Study in Taiwan
Abstract
1. Introduction
2. Materials and Methods
2.1. Milk Sample and Practice of “Doing-the-Month” in Postpartum Care Centers
2.2. DNA Extraction
2.3. Milk Microbiota Analysis
2.4. Statistical Analysis
3. Results
3.1. Information for Milk Groups
3.2. Prevalence of Lactobacillus or Bifidobacteria in the Three Milk Groups
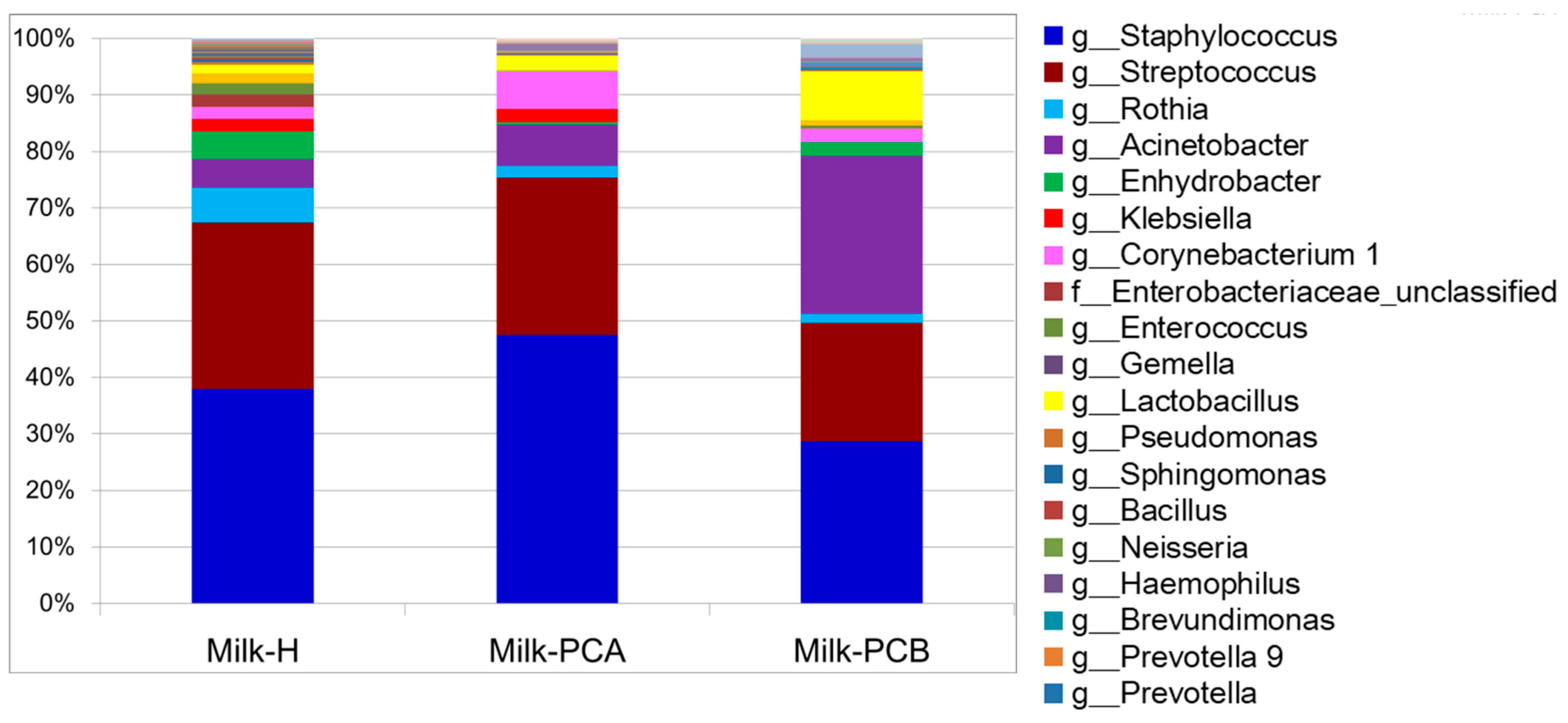
3.3. Proportion of Major and Minor Bacterial Genera in the Milk Groups
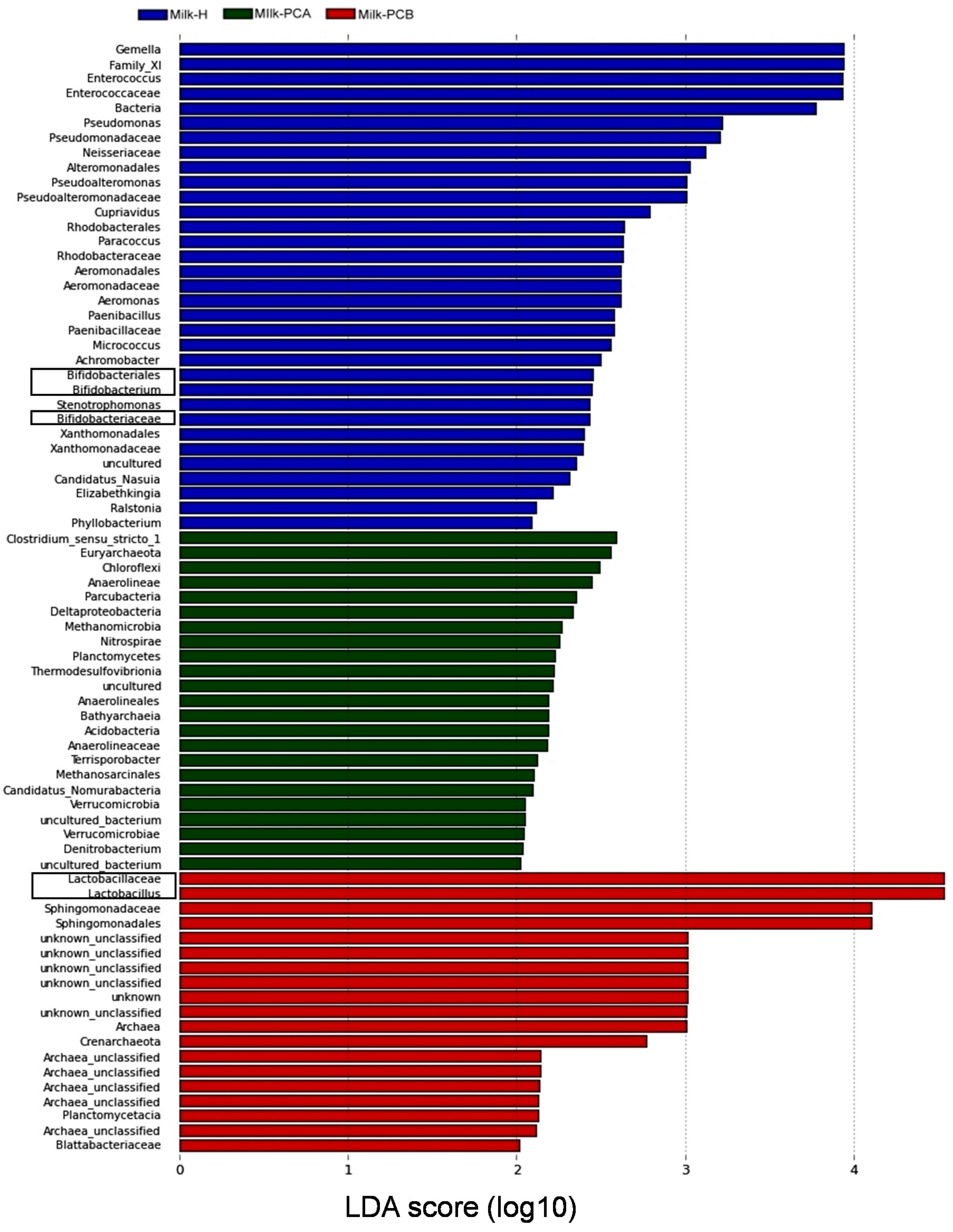
3.4. Linear Discriminant Analysis Effect Size (LEfSe) Analysis to Identify Differences in Abundant Microbial Taxa between the Three Milk Groups
3.5. Cladogram Analysis to Identify the Greatest Differences in Microbial Taxa between the Milk Groups
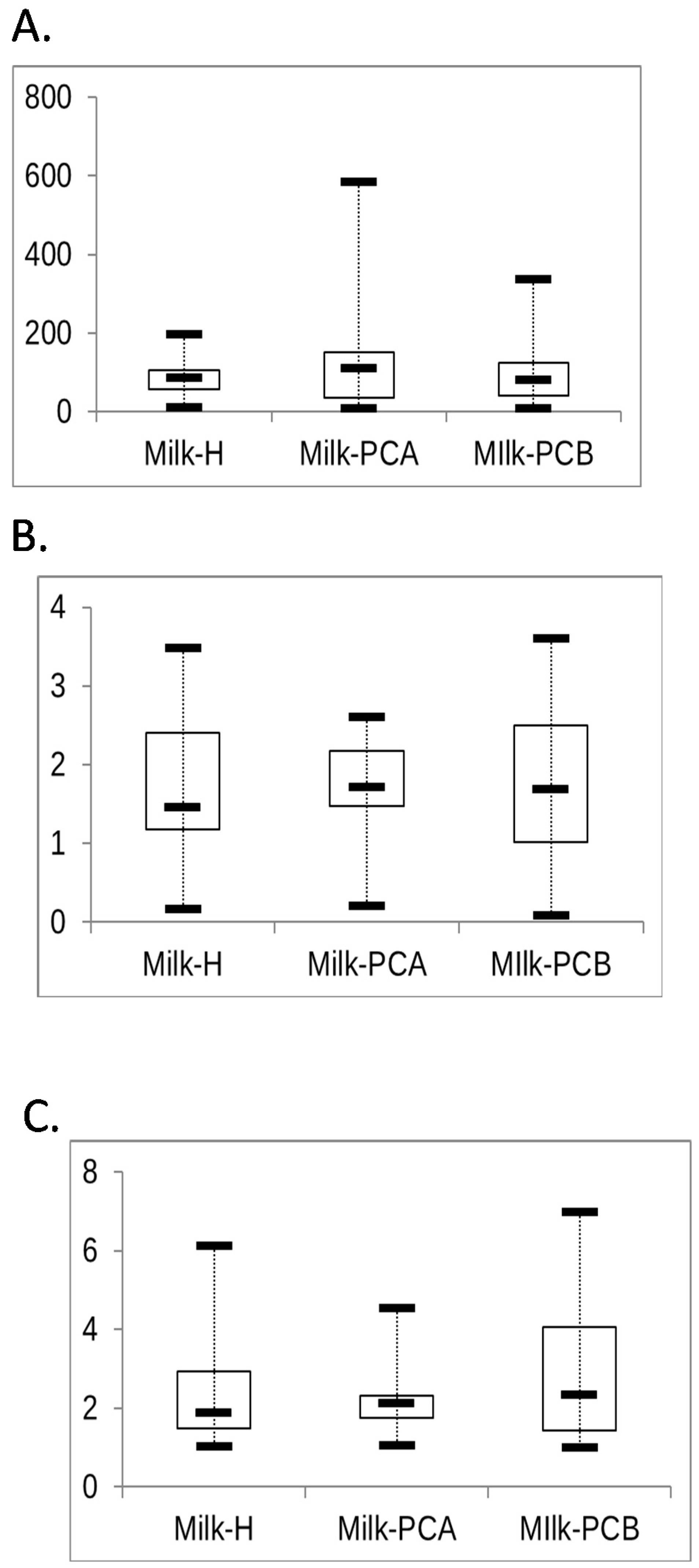
3.6. Analysis of the Similarity and Diversity of Milk Microbial Communities between “Doing-the-Month” and “Non-Doing-the-Month” Groups
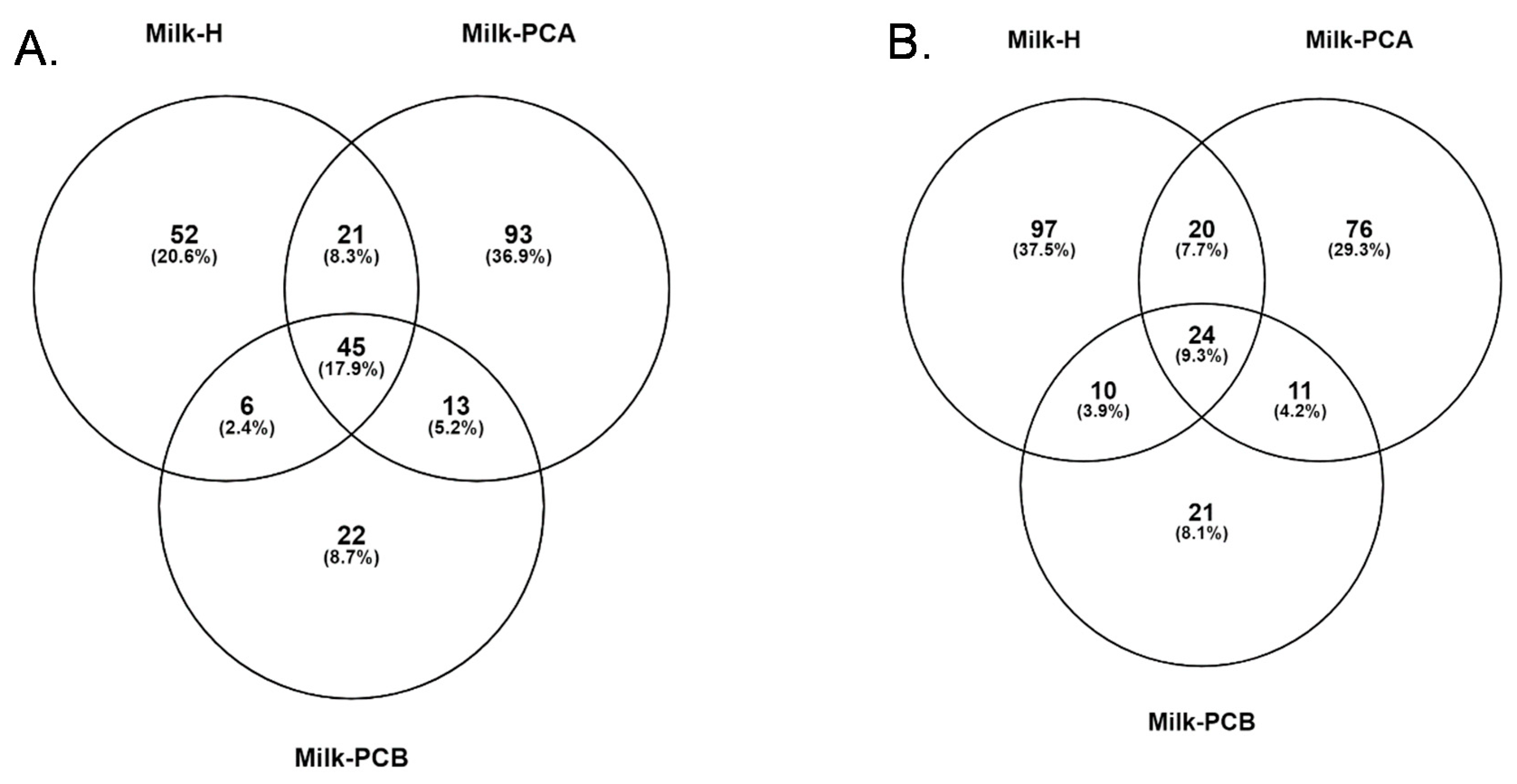
3.7. Common and Different Bacterial Genera and Species between the Three Milk Groups
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef]
- Bardanzellu, F.; Fanos, V.; Reali, A. “Omics” in Human Colostrum and Mature Milk: Looking to Old Data with New Eyes. Nutrients 2017, 9, 843. [Google Scholar] [CrossRef]
- Duijts, L.; Jaddoe, V.W.; Hofman, A.; Moll, H.A. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics 2010, 126, e18–e25. [Google Scholar] [CrossRef]
- Meinzen-Derr, J.; Poindexter, B.; Wrage, L.; Morrow, A.L.; Stoll, B.; Donovan, E.F. Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J. Perinatol. 2009, 29, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, K.; Carroll, K. An exclusively human milk diet reduces necrotizing enterocolitis. Breastfeed Med. 2014, 9, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Alarcon, M.; Villalpando, S.; Fajardo, A. Breast-feeding lowers the frequency and duration of acute respiratory infection and diarrhea in infants under six months of age. J. Nutr. 1997, 127, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Sadeharju, K.; Knip, M.; Virtanen, S.M.; Savilahti, E.; Tauriainen, S.; Koskela, P.; Akerblom, H.K.; Hyoty, H.; Finnish, T.S.G. Maternal antibodies in breast milk protect the child from enterovirus infections. Pediatrics 2007, 119, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Munblit, D.; Verhasselt, V. Allergy prevention by breastfeeding: Possible mechanisms and evidence from human cohorts. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 427–433. [Google Scholar] [CrossRef]
- Fernandez, L.; Arroyo, R.; Espinosa, I.; Marin, M.; Jimenez, E.; Rodriguez, J.M. Probiotics for human lactational mastitis. Benef. Microbes 2014, 5, 169–183. [Google Scholar] [CrossRef]
- Jiménez, E.; Fernández, L.; Maldonado, A.; Martín, R.; Olivares, M.; Xaus, J.; Rodríguez, J.J.a.E.M. Oral administration of Lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl. Environ. Microbiol. 2008, 74, 4650–4655. [Google Scholar] [CrossRef]
- Arroyo, R.; Martín, V.; Maldonado, A.; Jiménez, E.; Fernández, L.; Rodríguez, J.M.J.C.I.D. Treatment of infectious mastitis during lactation: Antibiotics versus oral administration of Lactobacilli isolated from breast milk. Clin. Infect. Dis. 2010, 50, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Factors Influencing the Intestinal Microbiome during the First Year of Life. Pediatr. Infect. Dis. J. 2018, 37, e315–e335. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Curley, D.; O’callaghan, T.F.; O’shea, C.A.; Dempsey, E.M.; O’toole, P.W.; Ross, R.P.; Ryan, C.A.; Stanton, C. The Composition of Human Milk and Infant Faecal Microbiota over the First Three Months of Life: A Pilot Study. Sci. Rep. 2017, 7, 40597. [Google Scholar] [CrossRef]
- Martin, R.; Jimenez, E.; Heilig, H.; Fernandez, L.; Marin, M.L.; Zoetendal, E.G.; Rodriguez, J.M. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl. Environ. Microbiol. 2009, 75, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Gronlund, M.M.; Gueimonde, M.; Laitinen, K.; Kociubinski, G.; Gronroos, T.; Salminen, S.; Isolauri, E. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin. Exp. Allergy 2007, 37, 1764–1772. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Maldonado-Barragan, A.; Moles, L.; Rodriguez-Banos, M.; Campo, R.D.; Fernandez, L.; Rodriguez, J.M.; Jimenez, E. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 2012, 28, 36–44. [Google Scholar] [CrossRef]
- Jimenez, E.; Martin, R.; Maldonado, A.; Martin, V.; Gomez De Segura, A.; Fernandez, L.; Rodriguez, J.M. Complete genome sequence of Lactobacillus salivarius CECT 5713, a probiotic strain isolated from human milk and infant feces. J. Bacteriol. 2010, 192, 5266–5267. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C.J.B.J.O.N. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 2013, 110, 1253–1262. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. Breast milk microbiota: A complex microbiome with multiple impacts and conditioning factors. J. Infect. 2020, 81, 17–47. [Google Scholar] [CrossRef]
- Abrahamsson, T.R.; Sinkiewicz, G.; Jakobsson, T.; Fredrikson, M.; Bjorksten, B. Probiotic lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J. Pediatr. Gastroenterol. Nutr. 2009, 49, 349–354. [Google Scholar] [CrossRef]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; De Souza, R.J.; Becker, A.B.; Mandhane, P.J. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 2019, 25, 324–335.e4. [Google Scholar] [CrossRef] [PubMed]
- Holroyd, E.; Twinn, S.; Yim, I.W. Exploring Chinese women’s cultural beliefs and behaviours regarding the practice of “doing the month”. Women Health 2005, 40, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C. Chinese values, health and nursing. J. Adv. Nurs. 2001, 36, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Callister, L.C. Doing the month: Chinese postpartum practices. MCN Am. J. Matern Child. Nurs. 2006, 31, 390. [Google Scholar] [CrossRef]
- Chuang, C.-H.; Chang, P.-J.; Hsieh, W.-S.; Tsai, Y.-J.; Lin, S.-J.; Chen, P.-C. Chinese herbal medicine use in Taiwan during pregnancy and the postpartum period: A population-based cohort study. Int. J. Nurs. Stud. 2009, 46, 787–795. [Google Scholar] [CrossRef]
- Ho, M.; Li, T.C.; Su, S.Y. The Association between Traditional Chinese Dietary and Herbal Therapies and Uterine Involution in Postpartum Women. Evid. Based Complement. Altern. Med. 2011, 2011, 918291. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Maloni, J.A.; Petrini, M.A. Effect of postpartum practices of doing the month on Chinese women’s physical and psychological health. Biol. Res. Nurs. 2014, 16, 55–63. [Google Scholar] [CrossRef]
- Leung, S.S.; Arthur, D.; Martinson, I.M. Perceived stress and support of the Chinese postpartum ritual “doing the month”. Health Care Women Int. 2005, 26, 212–224. [Google Scholar] [CrossRef]
- Tang, L.; Lee, A.H.; Binns, C.W.; VanHui, Y.; Yau, K.K.W. Consumption of Chinese herbal medicines during pregnancy and postpartum: A prospective cohort study in China. Midwifery 2016, 34, 205–210. [Google Scholar] [CrossRef]
- Holroyd, E.; Katie, F.K.; Chun, L.S.; Ha, S.W. “Doing the month”: An exploration of postpartum practices in Chinese women. Health Care Women Int. 1997, 18, 301–313. [Google Scholar] [CrossRef]
- Albesharat, R.; Ehrmann, M.A.; Korakli, M.; Yazaji, S.; Vogel, R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011, 34, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.W.; Lin, Y.L.; Huang, M.S. Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J. Food Drug Anal. 2018, 26, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Lozupone, C.; Knight, R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Boix-Amoros, A.; Collado, M.C.; Mira, A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front. Microbiol. 2016, 7, 492. [Google Scholar] [CrossRef]
- Jeurink, P.V.; van Bergenhenegouwen, J.; Jiménez, E.; Knippels, L.M.J.; Fernández, L.; Garssen, J.; Knol, J.; Rodríguez, J.M.; Martín, R. Human milk: A source of more life than we imagine. Benef. Microbes 2013, 4, 17–30. [Google Scholar] [CrossRef]
- Martín, R.; Heilig, G.H.J.; Zoetendal, E.G.; Smidt, H.; Rodríguez, J.M. Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J. Appl. Microbiol. 2007, 103, 2638–2644. [Google Scholar]
- Gueimonde, M.; Laitinen, K.; Salminen, S.; Isolauri, E. Breast milk: A source of bifidobacteria for infant gut development and maturation? Neonatology 2007, 92, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-W.; Watanabe, K.; Hsu, C.-C.; Chao, S.-H.; Yang, Z.-H.; Lin, Y.-J.; Chen, C.-C.; Cao, Y.-M.; Huang, H.-C.; Chang, C.-H.; et al. Bacterial Composition and Diversity in Breast Milk Samples from Mothers Living in Taiwan and Mainland China. Front. Microbiol. 2017, 8, 965. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 1998, 84, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73, 386S–392S. [Google Scholar] [CrossRef]
- Martin, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef]
- Lawrence, R.A.; Lawrence, R.M. Breastfeeding E-Book: A Guide for the Medical Professional (Expert Consult-Online and Print); Elsevier Health Sciences: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Kent, J.C.; Mitoulas, L.R.; Cregan, M.D.; Ramsay, D.T.; Doherty, D.A.; Hartmann, P.E. Volume and frequency of breastfeedings and fat content of breast milk throughout the day. Pediatrics 2006, 117, e387–e395. [Google Scholar] [CrossRef]
- Huang, M.S.; Cheng, C.-C.; Tseng, S.-Y.; Lin, Y.-L.; Lo, H.-M.; Chen, P.-W. Most commensally bacterial strains in human milk of healthy mothers display multiple antibiotic resistance. Microbiologyopen 2019, 8, e00618. [Google Scholar] [CrossRef]
- Heikkilä, M.P.; Saris, P. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 2003, 95, 471–478. [Google Scholar] [CrossRef]
- Derrien, M.; Vlieg, J.E.V. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef]
- Derrien, M.; Veiga, P. Rethinking Diet to Aid Human-Microbe Symbiosis. Trends Microbiol. 2017, 25, 100–112. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.T.; Chen, C.; Newburg, D.S. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 2013, 23, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Asakuma, S.; Hatakeyama, E.; Urashima, T.; Yoshida, E.; TKatayama, T.; Yamamoto, K.; Kumagai, H.; Ashida, H.; Hirose, J.; Kitaoka, M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 2011, 286, 34583–34592. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, S.S. Challenges and Pitfalls in Human Milk Oligosaccharide Analysis. Nutrients 2019, 11, 2684. [Google Scholar] [CrossRef]
- Thurl, S.; Munzert, M.; Henker, J.; Boehm, G.; Müller-Werner, B.; Jelinek, J.; Stahl, B. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 2010, 104, 1261–1271. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Kunz, C.; Rudloff, S.; García-Mantrana, I.; Crehuá-Gaudiza, E.; Martínez-Costa, C.; Collado, M.C. Association of maternal secretor status and human milk oligosaccharides with milk microbiota: An observational pilot study. J. Pediatr. Gastroenterol. Nutr. 2019, 68, 256–263. [Google Scholar] [CrossRef]
- Khodayar-Pardo, P.; Mira-Pascual, L.; Collado, M.C.; Martínez-Cost, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 2014, 34, 599–605. [Google Scholar] [CrossRef]
- Chien, L.Y.; Tai, C.-J.; Ko, Y.-L.; Huang, C.-H.; Sheu, S.-J. Adherence to “Doing-the-month” practices is associated with fewer physical and depressive symptoms among postpartum women in Taiwan. Res. Nurs. Health 2006, 29, 374–383. [Google Scholar] [CrossRef]
- Browne, P.D.; Aparicio, M.; Alba, C.; Hechler, C.; Beijers, R.; Rodrígue, J.M.; Fernández, L.; de Weerth, C. Human milk microbiome and maternal postnatal psychosocial distress. Front. Microbiol. 2019, 10, 2333. [Google Scholar] [CrossRef]
- Alexandraki, V.; Kazou, M.; Angelopoulou, A.; Arena, M.P.; Capozzi, V.; Russo, P.; Fiocco, D.; Spano, G.; Papadimitriou, K.; Tsakalidou, E. The microbiota of non-cow milk and products. In Non-Bovine Milk and Milk Products; Elsevier: Amsterdam, The Netherlands, 2016; pp. 117–159. [Google Scholar]
- Costello, E.K.; Carlisle, E.M.; Bik, E.M.; Morowitz, M.J.; Relman, D.A. Microbiome assembly across multiple body sites in low-birthweight infants. mBio 2013, 4, e00782-13. [Google Scholar] [CrossRef]
- Zaura, E.; Nicu, E.A.; Krom, B.P.; Keijser, B.J.F. Acquiring and maintaining a normal oral microbiome: Current perspective. Front Cell Infect. Microbiol. 2014, 4, 85. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 2016, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Moossavi, S.; Atakora, F.; Miliku, K.; Sepehri, S.; Robertson, B.; Duan, Q.L.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; Moraes, T.J.; et al. Integrated Analysis of Human Milk Microbiota With Oligosaccharides and Fatty Acids in the CHILD Cohort. Front. Nutr. 2019, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.L.; Hosid, S.; Ioshikhes, I.; Altosaar, I. Human milk metagenome: A functional capacity analysis. BMC Microbiol. 2013, 13, 116. [Google Scholar] [CrossRef]
- Jimenez, E.; de Andrés, J.; Manrique, M.; Pareja-Tobes, P.; Tobes, R.; Martínez-Blanch, J.F.; Codoñer, F.M.; Ramón, D.; Fernández, L.; Rodríguez, J.M. Metagenomic Analysis of Milk of Healthy and Mastitis-Suffering Women. J. Hum. Lact. 2015, 31, 406–415. [Google Scholar] [CrossRef]
- Togo, A.H.; Grine, G.; Khelaifia, S.; des Robert, C.; Brevaut, V.; Caputo, A.; Baptiste, E.; Bonnet, M.; Levasseur, A.; Drancourt, M.; et al. Culture of Methanogenic Archaea from Human Colostrum and Milk. Sci. Rep. 2019, 9, 18653. [Google Scholar] [CrossRef]
- Toscano, M.; De Grandi, R.; Peroni, D.G.; Grossi, E.; Facchin, V.; Comberiati, P.; Drago, L. Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol. 2017, 17, 205. [Google Scholar] [CrossRef]
- Hung, C.H.; Yu, C.-Y.; Liu, C.-F.; Stocker, J. Maternal satisfaction with postpartum nursing centers. Res. Nurs. Health 2010, 33, 345–354. [Google Scholar] [CrossRef]


| Milk Origin/Group | Postpartum Care Center A (Milk-PCA) | Postpartum Care Center B (Milk-PCB) | Obstetrics Department (Milk-H) |
|---|---|---|---|
| Sample size | 14 | 27 | 46 |
| Age for mothers who enrolled in study, year | 22 to 43 | 22 to 38 | 17 to 43 |
| Duration for Mother stayed in care centers, day | ≥20 (20 to 30) | ≥20 (20 to 30) | 0 |
| Services in care centers | |||
| Meal plan | Dietary therapy and herbal therapy a | Dietary therapy and herbal therapy a | None |
| Meal frequency | 3 to 5 meals/day | 5 meals/day | None |
| Content for dietary and herbal therapy |
|
|
| Type of Milk Group | Occurrence, % a | |
|---|---|---|
| Lactobacillus Bacterium | Bifidobacterial Bacterium | |
| Milk-H b (n = 46) | 86.9 | 32.6 |
| Milk-PCA b (n = 14) | 100 | 62.9 |
| Milk-PCB b (n = 27) | 92.8 | 57.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.-W.; Kuo, Y.-H.; Lin, Y.-L. The Impact of the Postpartum “Doing-the-Month” Practice on Human Milk Microbiota: A Pilot Study in Taiwan. Microorganisms 2020, 8, 1283. https://doi.org/10.3390/microorganisms8091283
Chen P-W, Kuo Y-H, Lin Y-L. The Impact of the Postpartum “Doing-the-Month” Practice on Human Milk Microbiota: A Pilot Study in Taiwan. Microorganisms. 2020; 8(9):1283. https://doi.org/10.3390/microorganisms8091283
Chicago/Turabian StyleChen, Po-Wen, Yu-Hsien Kuo, and Yi-Ling Lin. 2020. "The Impact of the Postpartum “Doing-the-Month” Practice on Human Milk Microbiota: A Pilot Study in Taiwan" Microorganisms 8, no. 9: 1283. https://doi.org/10.3390/microorganisms8091283
APA StyleChen, P.-W., Kuo, Y.-H., & Lin, Y.-L. (2020). The Impact of the Postpartum “Doing-the-Month” Practice on Human Milk Microbiota: A Pilot Study in Taiwan. Microorganisms, 8(9), 1283. https://doi.org/10.3390/microorganisms8091283






