Recent Advances in Novel Antiviral Therapies against Human Adenovirus
Abstract
:1. Introduction
2. HAdV Biology: The Viral Genome
2.1. The Early Genes
2.2. The Late Genes
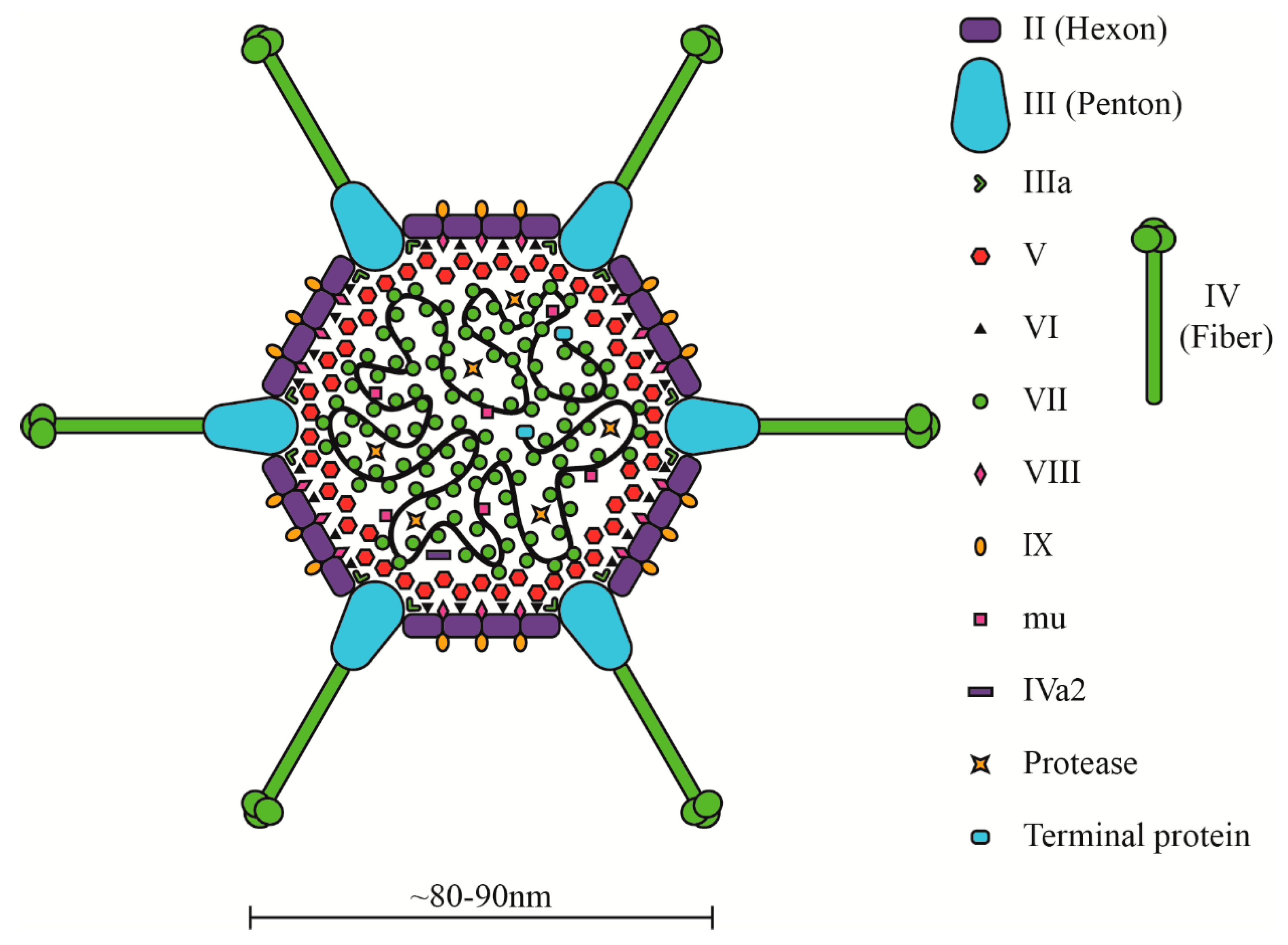
3. HAdV Biology: The Virion
4. Infection and Virus Lifecycle
5. HAdV-Induced Disease and Current Treatment Options
5.1. HAdV Infections
5.2. Current Antiviral Therapies
5.3. Anti-HAdV Vaccines
6. Discovery of New Antiviral Therapies
6.1. Nucleoside/Nucleotide Analogues
6.2. HDAC Inhibitors and Inhibitors of Other Epigenetic Regulatory Proteins
6.3. Steroid-Based Compounds
6.4. Other Compounds with Anti-HAdV Activity
6.5. Vaccines and Biological Antivirals against HAdV
6.6. Small Molecule Library Screens
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Hoke, C.H., Jr.; Snyder, C.E., Jr. History of the restoration of adenovirus type 4 and type 7 vaccine, live oral (Adenovirus Vaccine) in the context of the department of defense acquisition system. Vaccine 2013, 31, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Hilleman, M.R.; Werner, J.H. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 1954, 85, 183–188. [Google Scholar] [CrossRef]
- Dhingra, A.; Hage, E.; Ganzenmueller, T.; Bottcher, S.; Hofmann, J.; Hamprecht, K.; Obermeier, P.; Rath, B.; Hausmann, F.; Dobner, T.; et al. Molecular evolution of Human Adenovirus (HAdV) species C. Sci. Rep. 2019, 9, 1039. [Google Scholar] [CrossRef]
- Whyte, P.; Williamson, N.M.; Harlow, E. Cellular targets for transformation by the adenovirus E1A proteins. Cell 1989, 56, 67–75. [Google Scholar] [CrossRef]
- Eckner, R.; Ewen, M.E.; Newsome, D.; Gerdes, M.; DeCaprio, J.A.; Lawrence, J.B.; Livingston, D.M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994, 8, 869–884. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, M.; Gerber, J.; Drapkin, R.; Sif, S.; Ikura, T.; Ogryzko, V.; Lane, W.S.; Nakatani, Y.; Livingston, D.M. The p400 complex is an essential E1A transformation target. Cell 2001, 106, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Chow, L.T.; Gelinas, R.E.; Broker, T.R.; Roberts, R.J. An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell 1977, 12, 1–8. [Google Scholar] [CrossRef]
- Flint, J. Chapter 3: Organization of the Adenoviral Genome. In Adenoviruses: Basic Biology to Gene Therapy; Seth, P., Ed.; R.G. Landes Company: Austin, TX, USA, 1999; pp. 17–30. [Google Scholar]
- Berk, A.J. Adenoviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 2355–2394. [Google Scholar]
- Ben-Israel, H.; Kleinberger, T. Adenovirus and cell cycle control. Front. Biosci. J. Virtual Libr. 2002, 7, d1369–d1395. [Google Scholar] [CrossRef]
- Musher, D.M. How contagious are common respiratory tract infections? N. Engl. J. Med. 2003, 348, 1256–1266. [Google Scholar] [CrossRef]
- Joffe, M.; Wagner, S.D.; Tang, J.W. Case report: A fatal case of disseminated adenovirus infection in a non-transplant adult haematology patient. BMC Infect. Dis. 2018, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Kojaoghlanian, T.; Flomenberg, P.; Horwitz, M.S. The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 2003, 13, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Ying, B.; Tollefson, A.E.; Spencer, J.F.; Balakrishnan, L.; Dewhurst, S.; Capella, C.; Buller, R.M.; Toth, K.; Wold, W.S. Ganciclovir inhibits human adenovirus replication and pathogenicity in permissive immunosuppressed Syrian hamsters. Antimicrob. Agents Chemother. 2014, 58, 7171–7181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keyes, A.; Mathias, M.; Boulad, F.; Lee, Y.J.; Marchetti, M.A.; Scaradavou, A.; Spitzer, B.; Papanicolaou, G.A.; Wieczorek, I.; Busam, K.J. Cutaneous involvement of disseminated adenovirus infection in an allogeneic stem cell transplant recipient. Br. J. Dermatol. 2016, 174, 885–888. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, L.; De Clercq, E.; Naesens, L. Clinical features and treatment of adenovirus infections. Rev. Med. Virol. 2008, 18, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Ronchi, A.; Doern, C.; Brock, E.; Pugni, L.; Sanchez, P.J. Neonatal adenoviral infection: A seventeen year experience and review of the literature. J. Pediatrics 2014, 164, 529–535.e4. [Google Scholar] [CrossRef]
- Binder, A.M.; Biggs, H.M.; Haynes, A.K.; Chommanard, C.; Lu, X.; Erdman, D.D.; Watson, J.T.; Gerber, S.I. Human adenovirus surveillance—United States, 2003–2016. Morbidity Mortal. Wkly. Rep. 2017, 66, 1039–1042. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.; Pereda, M.A.; Bala, N.; Nagarajan, S. In-hospital mortality of hematopoietic stem cell transplantation among children with nonmalignancies: A nationwide study in the United States from 2000 to 2012. Pediatric Blood Cancer 2019, 66, e27626. [Google Scholar] [CrossRef]
- Lee, B.; Park, E.; Ha, J.; Ha, I.S.; Il Cheong, H.; Kang, H.G. Disseminated adenovirus infection in a 10-year-old renal allograft recipient. Kidney Res. Clin. Pract. 2018, 37, 414–417. [Google Scholar] [CrossRef] [Green Version]
- Khanal, S.; Ghimire, P.; Dhamoon, A.S. The repertoire of adenovirus in human disease: The innocuous to the deadly. Biomedicines 2018, 6, 30. [Google Scholar] [CrossRef] [Green Version]
- Scott, M.K.; Chommanard, C.; Lu, X.; Appelgate, D.; Grenz, L.; Schneider, E.; Gerber, S.I.; Erdman, D.D.; Thomas, A. Human adenovirus associated with severe respiratory infection, Oregon, USA, 2013–2014. Emerg. Infect. Dis. 2016, 22, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Zalisnak, D.; Rapuano, C.; Sheppard, J.D.; Davis, A.R. Adenovirus ocular infections: Prevalence, pathology, pitfalls, and practical pointers. Eye Contact Lens 2018, 44 (Suppl. 1), S1–S7. [Google Scholar] [CrossRef]
- Lopez, S.M.C.; Michaels, M.G.; Green, M. Adenovirus infection in pediatric transplant recipients: Are effective antiviral agents coming our way? Curr. Opin. Organ Transplant. 2018, 23, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, S.; Berger, M.; Tridello, G.; Mikulska, M.; Ward, K.N.; Ljungman, P.; Van Der Werf, S.; Averbuch, D.; Styczynski, J. A survey on incidence and management of adenovirus infection after allogeneic HSCT. Bone Marrow Transplant. 2018. [Google Scholar] [CrossRef] [PubMed]
- Caruso Brown, A.E.; Cohen, M.N.; Tong, S.; Braverman, R.S.; Rooney, J.F.; Giller, R.; Levin, M.J. Pharmacokinetics and safety of intravenous cidofovir for life-threatening viral infections in pediatric hematopoietic stem cell transplant recipients. Antimicrob. Agents Chemother. 2015, 59, 3718–3725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zecca, M.; Wynn, R.; Dalle, J.H.; Feuchtinger, T.; Vainorius, E.; Brundage, T.M.; Chandak, A.; Mozaffari, E.; Nichols, G.; Locatelli, F. Association between adenovirus viral load and mortality in pediatric allo-HCT recipients: The multinational AdVance study. Bone Marrow Transplant. 2019. [Google Scholar] [CrossRef] [Green Version]
- Dehghan, S.; Seto, J.; Liu, E.B.; Ismail, A.M.; Madupu, R.; Heim, A.; Jones, M.S.; Dyer, D.W.; Chodosh, J.; Seto, D. A zoonotic adenoviral human pathogen emerged through genomic recombination among human and nonhuman simian hosts. J. Virol. 2019, 93, e00564-19. [Google Scholar] [CrossRef] [Green Version]
- Yoshitomi, H.; Sera, N.; Gonzalez, G.; Hanaoka, N.; Fujimoto, T. First isolation of a new type of human adenovirus (genotype 79), species Human mastadenovirus B (B2) from sewage water in Japan. J. Med Virol. 2017, 89, 1192–1200. [Google Scholar] [CrossRef]
- Kajan, G.L.; Kajon, A.E.; Pinto, A.C.; Bartha, D.; Arnberg, N. The complete genome sequence of human adenovirus 84, a highly recombinant new Human mastadenovirus D type with a unique fiber gene. Virus Res. 2017, 242, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Mao, N.; Zhu, Z.; Rivailler, P.; Chen, M.; Fan, Q.; Huang, F.; Xu, W. Whole genomic analysis of two potential recombinant strains within Human mastadenovirus species C previously found in Beijing, China. Sci. Rep. 2017, 7, 15380. [Google Scholar] [CrossRef] [Green Version]
- Kaján, G.L.; Lipiec, A.; Bartha, D.; Allard, A.; Arnberg, N. A multigene typing system for human adenoviruses reveals a new genotype in a collection of Swedish clinical isolates. PLoS ONE 2018, 13, e0209038. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.P.; Mathews, M.B. DNA replication and the early to late transition in adenovirus infection. Cell 1980, 22, 523–533. [Google Scholar] [CrossRef]
- Chroboczek, J.; Bieber, F.; Jacrot, B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology 1992, 186, 280–285. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, M.; Pettersson, U. A new look at adenovirus splicing. Virology 2014, 456–457, 329–341. [Google Scholar] [CrossRef] [Green Version]
- King, C.R.; Zhang, A.; Tessier, T.M.; Gameiro, S.F.; Mymryk, J.S. Hacking the cell: Network intrusion and exploitation by adenovirus E1A. mBio 2018, 9, e00390-18. [Google Scholar] [CrossRef] [Green Version]
- Horwitz, M.S. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 2004, 6 (Suppl. 1), S172–S183. [Google Scholar] [CrossRef]
- Weitzman, M.D. Functions of the adenovirus E4 proteins and their impact on viral vectors. Front. Biosci. J. Virtual Libr. 2005, 10, 1106–1117. [Google Scholar] [CrossRef] [Green Version]
- Berk, A.J. Adenovirus promoters and E1A transactivation. Annu. Rev. Genet. 1986, 20, 45–79. [Google Scholar] [CrossRef]
- Stephens, C.; Harlow, E. Differential splicing yields novel adenovirus 5 E1A mRNAs that encode 30 kd and 35 kd proteins. EMBO J. 1987, 6, 2027–2035. [Google Scholar] [CrossRef]
- Pelka, P.; Ablack, J.N.; Torchia, J.; Turnell, A.S.; Grand, R.J.; Mymryk, J.S. Transcriptional control by adenovirus E1A conserved region 3 via p300/CBP. Nucleic Acids Res. 2009, 37, 1095–1106. [Google Scholar] [CrossRef]
- Swaminathan, S.; Thimmapaya, B. Transactivation of adenovirus E2-early promoter by E1A and E4 6/7 in the context of viral chromosome. J. Mol. Biol. 1996, 258, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, D.; Ghosh, M.K.; Mal, A.; Harter, M.L. Inactivation of p21 by E1A leads to the induction of apoptosis in DNA-damaged cells. J. Virol. 2001, 75, 9844–9856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Green, M.R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature 1994, 368, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Parks, C.L.; Shenk, T. Activation of the adenovirus major late promoter by transcription factors MAZ and Sp1. J. Virol. 1997, 71, 9600–9607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisch, S.M.; Mymryk, J.S. Adenovirus-5 E1A: Paradox and paradigm. Nat. Rev. Mol. Cell Biol. 2002, 3, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Gallimore, P.H.; Turnell, A.S. Adenovirus E1A: Remodelling the host cell, a life or death experience. Oncogene 2001, 20, 7824–7835. [Google Scholar] [CrossRef] [Green Version]
- Badr, K.R.; Parente-Rocha, J.A.; Baeza, L.C.; Ficcadori, F.S.; Souza, M.; Soares, C.M.; Guissoni, A.C.P.; Almeida, T.N.; Cardoso, D.D. Quantitative proteomic analysis of A549 cells infected with human adenovirus type 2. J. Med. Virol. 2019. [Google Scholar] [CrossRef]
- Blackford, A.N.; Grand, R.J.A. Adenovirus E1B 55-kilodalton protein: Multiple roles in viral infection and cell transformation. J. Virol. 2009, 83, 4000–4012. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, R.; Huang, W.; Finnen, R.; Bragg, C.; Flint, S.J. Adenovirus E1B 55-kilodalton protein is required for both regulation of mRNA export and efficient entry into the late phase of infection in normal human fibroblasts. J. Virol. 2006, 80, 964–974. [Google Scholar] [CrossRef] [Green Version]
- Caravokyri, C.; Leppard, K.N. Human adenovirus type 5 variants with sequence alterations flanking the E2A gene: Effects on E2 expression and DNA replication. Virus Genes 1996, 12, 65–75. [Google Scholar] [CrossRef]
- Saha, B.; Parks, R.J. Human adenovirus type 5 vectors deleted of early region 1 (E1) undergo limited expression of early replicative E2 proteins and DNA replication in non-permissive cells. PLoS ONE 2017, 12, e0181012. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Kanellopoulos, P.N.; Loonstra, A.K.; van Oosterhout, J.A.; Leonard, K.; Tucker, P.A.; van der Vliet, P.C. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 1997, 16, 1455–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jong, R.N.; Meijer, L.A.T.; van der Vliet, P.C. DNA binding properties of the adenovirus DNA replication priming protein pTP. Nucleic Acids Res. 2003, 31, 3274–3286. [Google Scholar] [CrossRef] [PubMed]
- Hoeben, R.C.; Uil, T.G. Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a013003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, K.; Guggenheimer, R.A.; Hurwitz, J. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. Natl. Acad. Sci. USA 1983, 80, 6177–6181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Mermod, N.; Horwitz, M.S. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J. Biol. Chem. 1990, 265, 18634–18642. [Google Scholar] [PubMed]
- Mangel, W.F.; Toledo, D.L.; Ding, J.; Sweet, R.M.; McGrath, W.J. Temporal and spatial control of the adenovirus proteinase by both a peptide and the viral DNA. Trends Biochem. Sci. 1997, 22, 393–398. [Google Scholar] [CrossRef]
- Arnberg, N. Adenovirus E3 protein modulates leukocyte functions. Proc. Natl. Acad. Sci. USA 2013, 110, 19976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Martin, N.; Ramani, S.R.; Hackney, J.A.; Tom, I.; Wranik, B.J.; Chan, M.; Wu, J.; Paluch, M.T.; Takeda, K.; Hass, P.E.; et al. The extracellular interactome of the human adenovirus family reveals diverse strategies for immunomodulation. Nat. Commun. 2016, 7, 11473. [Google Scholar] [CrossRef] [Green Version]
- Tollefson, A.E.; Scaria, A.; Hermiston, T.W.; Ryerse, J.S.; Wold, L.J.; Wold, W.S. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 1996, 70, 2296–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vink, E.I.; Zheng, Y.; Yeasmin, R.; Stamminger, T.; Krug, L.T.; Hearing, P. Impact of adenovirus E4-ORF3 oligomerization and protein localization on cellular gene expression. Viruses 2015, 7, 2428–2449. [Google Scholar] [CrossRef] [PubMed]
- Stracker, T.H.; Carson, C.T.; Weitzman, M.D. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 2002, 418, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Ahi, Y.S.; Mittal, S.K. Components of adenovirus genome packaging. Front. Microbiol. 2016, 7, 1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, C.S. The structure and function of the adenovirus major late promoter. Curr. Top. Microbiol. Immunol. 2003, 272, 213–249. [Google Scholar]
- Prescott, J.C.; Falck-Pedersen, E. Varied poly(A) site efficiency in the adenovirus major late transcription unit. J. Biol. Chem. 1992, 267, 8175–8181. [Google Scholar]
- Morris, S.J.; Scott, G.E.; Leppard, K.N. Adenovirus late-phase infection is controlled by a novel L4 promoter. J. Virol. 2010, 84, 7096–7104. [Google Scholar] [CrossRef] [Green Version]
- Saha, B.; Wong, C.M.; Parks, R.J. The adenovirus genome contributes to the structural stability of the virion. Viruses 2014, 6, 3563–3583. [Google Scholar] [CrossRef] [Green Version]
- Parks, R.J. Adenovirus protein IX: A new look at an old protein. Mol. Ther. J. Am. Soc. Gene Ther. 2005, 11, 19–25. [Google Scholar] [CrossRef]
- O’Malley, R.P.; Mariano, T.M.; Siekierka, J.; Mathews, M.B. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell 1986, 44, 391–400. [Google Scholar] [CrossRef]
- Aparicio, O.; Razquin, N.; Zaratiegui, M.; Narvaiza, I.; Fortes, P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 2006, 80, 1376–1384. [Google Scholar] [CrossRef] [Green Version]
- Vachon, V.K.; Conn, G.L. Adenovirus VA RNA: An essential pro-viral non-coding RNA. Virus Res. 2016, 212, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Tollefson, A.E.; Ying, B.; Doronin, K.; Sidor, P.D.; Wold, W.S. Identification of a new human adenovirus protein encoded by a novel late l-strand transcription unit. J. Virol. 2007, 81, 12918–12926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, B.; Tollefson, A.E.; Wold, W.S.M. Identification of a previously unrecognized promoter that drives expression of the UXP transcription unit in the human adenovirus type 5 genome. J. Virol. 2010, 84, 11470–11478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Jin, L.; Koh, S.B.; Atanasov, I.; Schein, S.; Wu, L.; Zhou, Z.H. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 2010, 329, 1038–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, V.S.; Natchiar, S.K.; Stewart, P.L.; Nemerow, G.R. Crystal structure of human adenovirus at 3.5 A resolution. Science 2010, 329, 1071–1075. [Google Scholar] [CrossRef] [Green Version]
- Nemerow, G.R.; Stewart, P.L.; Reddy, V.S. Structure of human adenovirus. Curr. Opin. Virol. 2012, 2, 115–121. [Google Scholar] [CrossRef] [Green Version]
- San Martín, C. Latest insights on adenovirus structure and assembly. Viruses 2012, 4, 847–877. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, P.K.; Vayda, M.E.; Flint, S.J. Interactions among the three adenovirus core proteins. J. Virol. 1985, 55, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Korn, R.; Horwitz, M.S. Adenovirus DNA synthesis in vitro is inhibited by the virus-coded major core protein. Virology 1986, 150, 342–351. [Google Scholar] [CrossRef]
- Anderson, C.W.; Young, M.E.; Flint, S.J. Characterization of the adenovirus 2 virion protein, mu. Virology 1989, 172, 506–512. [Google Scholar] [CrossRef]
- Pérez-Vargas, J.; Vaughan, R.C.; Houser, C.; Hastie, K.M.; Kao, C.C.; Nemerow, G.R. Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. J. Virol. 2014, 88, 9287–9296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, D.A.; Russell, W.C. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 1998, 79, 1671–1675. [Google Scholar] [CrossRef] [PubMed]
- Everitt, E.; Lutter, L.; Philipson, L. Structural proteins of adenoviruses: XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic lodination, immunoprecipitation and chemical cross-linking. Virology 1975, 67, 197–208. [Google Scholar] [CrossRef]
- Christensen, J.B.; Byrd, S.A.; Walker, A.K.; Strahler, J.R.; Andrews, P.C.; Imperiale, M.J. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol. 2008, 82, 9086–9093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vellinga, J.; Van der Heijdt, S.; Hoeben, R.C. The adenovirus capsid: Major progress in minor proteins. J. Gen. Virol. 2005, 86, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bergelson, J.M. Adenovirus receptors. J. Virol. 2005, 79, 12125–12131. [Google Scholar] [CrossRef] [Green Version]
- Bergelson, J.M.; Cunningham, J.A.; Droguett, G.; Kurt-Jones, E.A.; Krithivas, A.; Hong, J.S.; Horwitz, M.S.; Crowell, R.L.; Finberg, R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 1997, 275, 1320–1323. [Google Scholar] [CrossRef]
- Lyle, C.; McCormick, F. Integrin αvβ5 is a primary receptor for adenovirus in CAR-negative cells. Virol. J. 2010, 7, 148. [Google Scholar] [CrossRef] [Green Version]
- Wiethoff, C.M.; Wodrich, H.; Gerace, L.; Nemerow, G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 2005, 79, 1992–2000. [Google Scholar] [CrossRef] [Green Version]
- Wickham, T.J.; Mathias, P.; Cheresh, D.A.; Nemerow, G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 1993, 73, 309–319. [Google Scholar] [CrossRef]
- Wickham, T.J.; Filardo, E.J.; Cheresh, D.A.; Nemerow, G.R. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 1994, 127, 257–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leopold, P.L.; Ferris, B.; Grinberg, I.; Worgall, S.; Hackett, N.R.; Crystal, R.G. Fluorescent virions: Dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 1998, 9, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Flatt, J.W.; Butcher, S.J. Adenovirus flow in host cell networks. Open Biol. 2019, 9, 190012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strunze, S.; Engelke, M.F.; Wang, I.H.; Puntener, D.; Boucke, K.; Schleich, S.; Way, M.; Schoenenberger, P.; Burckhardt, C.J.; Greber, U.F. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 2011, 10, 210–223. [Google Scholar] [CrossRef] [Green Version]
- Greber, U.F.; Suomalainen, M.; Stidwill, R.P.; Boucke, K.; Ebersold, M.W.; Helenius, A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997, 16, 5998–6007. [Google Scholar] [CrossRef] [Green Version]
- Greber, U.F.; Willetts, M.; Webster, P.; Helenius, A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 1993, 75, 477–486. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, T.; Haruki, H.; Nagata, K. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res. 2011, 39, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, T.; Nagata, K. Replication-uncoupled histone deposition during adenovirus DNA replication. J. Virol. 2012, 86, 6701–6711. [Google Scholar] [CrossRef] [Green Version]
- Giberson, A.; Saha, B.; Campbell, K.; Christou, C.; Poulin, K.L.; Parks, R.J. Human adenoviral DNA association with nucleosomes containing histone variant H3.3 during the early phase of infection is not dependent on viral transcription or replication. Biochem. Cell Biol. 2018. [Google Scholar] [CrossRef]
- Tate, V.E.; Philipson, L. Parental adenovirus DNA accumulates in nucleosome-like structures in infected cells. Nucleic Acids Res. 1979, 6, 2769–2785. [Google Scholar] [CrossRef] [Green Version]
- Daniell, E.; Groff, D.E.; Fedor, M.J. Adenovirus chromatin structure at different stages of infection. Mol. Cell. Biol. 1981, 1, 1094–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, A.L.; Bouton, A.H.; Hodge, L.D.; Miller, O.L., Jr. Visualization of the major late R strand transcription unit of adenovirus serotype 2. J. Mol. Biol. 1981, 147, 269–295. [Google Scholar] [CrossRef]
- Stuiver, M.H.; Bergsma, W.G.; Arnberg, A.C.; van Amerongen, H.; van Grondelle, R.; van der Vliet, P.C. Structural alterations of double-stranded DNA in complex with the adenovirus DNA-binding protein. Implications for its function in DNA replication. J. Mol. Biol. 1992, 225, 999–1011. [Google Scholar] [CrossRef]
- Wodrich, H.; Guan, T.; Cingolani, G.; von Seggern, D.; Nemerow, G.; Gerace, L. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003, 22, 6245–6255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangel, W.F.; San Martin, C. Structure, function and dynamics in adenovirus maturation. Viruses 2014, 6, 4536–4570. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.T.; Schneider, R.J. Adenovirus inhibition of cellular protein synthesis is prevented by the drug 2-aminopurine. Proc. Natl. Acad. Sci. USA 1990, 87, 7115–7119. [Google Scholar] [CrossRef] [Green Version]
- Kajon, A.E.; Lamson, D.M.; Bair, C.R.; Lu, X.; Landry, M.L.; Menegus, M.; Erdman, D.D.; St George, K. Adenovirus type 4 respiratory infections among civilian adults, Northeastern United States, 2011–2015(1). Emerg. Infect. Dis. 2018, 24, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, K.L.; Richardson, S.E.; MacGregor, D.; Mahant, S.; Raghuram, K.; Bitnun, A. Adenovirus-Associated Central Nervous System Disease in Children. J. Pediatrics 2019, 205, 130–137. [Google Scholar] [CrossRef]
- Crenshaw, B.J.; Jones, L.B.; Bell, C.R.; Kumar, S.; Matthews, Q.L. Perspective on adenoviruses: Epidemiology, pathogenicity, and gene therapy. Biomedicines 2019, 7, 61. [Google Scholar] [CrossRef] [Green Version]
- Bhatti, Z.; Dhamoon, A. Fatal adenovirus infection in an immunocompetent host. Am. J. Emerg. Med. 2017, 35, 1034.e1031–1034.e1032. [Google Scholar] [CrossRef]
- Kajon, A.E.; Ison, M.G. Severe infections with human adenovirus 7d in 2 adults in family, Illinois, USA, 2014. Emerg. Infect. Dis. J. 2016, 22, 730. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tian, X. Vaccine development for human mastadenovirus. J. Thorac. Dis. 2018, 10, S2280–S2294. [Google Scholar] [CrossRef] [PubMed]
- Jobran, S.; Kattan, R.; Shamaa, J.; Marzouqa, H.; Hindiyeh, M. Adenovirus respiratory tract infections in infants: A retrospective chart-review study. Lancet 2018, 391 (Suppl. 2), S43. [Google Scholar] [CrossRef]
- Lion, T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 2014, 27, 441–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, F.-Z.; Shen, X.-X.; Li, G.-X.; Zhao, L.; Chen, C.; Duan, S.-X.; Guo, J.-Y.; Zhao, M.-C.; Yan, T.-F.; Qi, J.-J.; et al. Adenovirus associated with acute diarrhea: A case-control study. BMC Infect. Dis. 2018, 18, 450. [Google Scholar] [CrossRef]
- Naesens, L.; Lenaerts, L.; Andrei, G.; Snoeck, R.; van Beers, D.; Holy, A.; Balzarini, J.; de Clercq, E. Antiadenovirus activities of several classes of nucleoside and nucleotide analogues. Antimicrob. Agents Chemother. 2005, 49, 1010–1016. [Google Scholar] [CrossRef] [Green Version]
- Detweiler, C.J.; Mueller, S.B.; Sung, A.D.; Saullo, J.L.; Prasad, V.K.; Cardona, D.M. Brincidofovir (CMX001) toxicity associated with epithelial apoptosis and crypt drop out in a hematopoietic cell transplant patient: Challenges in distinguishing drug toxicity From GVHD. J. Pediatric Hematol. Oncol. 2018, 40, e364–e368. [Google Scholar] [CrossRef]
- Ramsay, I.D.; Attwood, C.; Irish, D.; Griffiths, P.D.; Kyriakou, C.; Lowe, D.M. Disseminated adenovirus infection after allogeneic stem cell transplant and the potential role of brincidofovir—Case series and 10 year experience of management in an adult transplant cohort. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2017, 96, 73–79. [Google Scholar] [CrossRef]
- Florescu, D.F.; Schaenman, J.M. Adenovirus in solid organ transplant recipients: Guidelines from the American society of transplantation infectious diseases community of practice. Clin. Transplant. 2019, e13527. [Google Scholar] [CrossRef]
- Mentel, R.; Wegner, U. Evaluation of the efficacy of 2′,3′-dideoxycytidine against adenovirus infection in a mouse pneumonia model. Antivir. Res. 2000, 47, 79–87. [Google Scholar] [CrossRef]
- Ison, M.G. Adenovirus infections in transplant recipients. Clin. Infect. Dis. 2006, 43, 331–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waye, M.M.Y.; Sing, C.W. Anti-viral drugs for human adenoviruses. Pharmaceuticals 2010, 3, 3343–3354. [Google Scholar] [CrossRef]
- Hartline, C.B.; Gustin, K.M.; Wan, W.B.; Ciesla, S.L.; Beadle, J.R.; Hostetler, K.Y.; Kern, E.R. Ether lipid-ester prodrugs of acyclic nucleoside phosphonates: Activity against adenovirus replication in vitro. J. Infect. Dis. 2005, 191, 396–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, B.; Parks, R.J. Identification of human adenovirus replication inhibitors from a library of small molecules targeting cellular epigenetic regulators. Virology 2020. [Google Scholar] [CrossRef]
- Saha, B.; Varette, O.; Stanford, W.L.; Diallo, J.-S.; Parks, R.J. Development of a novel screening platform for the identification of small molecule inhibitors of human adenovirus. Virology 2019, 538, 24–34. [Google Scholar] [CrossRef]
- Mentel, R.; Kinder, M.; Wegner, U.; von Janta-Lipinski, M.; Matthes, E. Inhibitory activity of 3′-fluoro-2′ deoxythymidine and related nucleoside analogues against adenoviruses in vitro. Antivir. Res. 1997, 34, 113–119. [Google Scholar] [CrossRef]
- Hoti, N.; Chowdhury, W.; Hsieh, J.T.; Sachs, M.D.; Lupold, S.E.; Rodriguez, R. Valproic acid, a histone deacetylase inhibitor, is an antagonist for oncolytic adenoviral gene therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2006, 14, 768–778. [Google Scholar] [CrossRef]
- Saha, B.; Parks, R.J. Histone deacetylase inhibitor suberoylanilide hydroxamic acid suppresses human adenovirus gene expression and replication. J. Virol. 2019, 93, e00088-19. [Google Scholar] [CrossRef] [Green Version]
- Arbuckle, J.H.; Gardina, P.J.; Gordon, D.N.; Hickman, H.D.; Yewdell, J.W.; Pierson, T.C.; Myers, T.G.; Kristie, T.M. Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. mBio 2017, 8, e01141-17. [Google Scholar] [CrossRef] [Green Version]
- Grosso, F.; Stoilov, P.; Lingwood, C.; Brown, M.; Cochrane, A. Suppression of adenovirus replication by cardiotonic steroids. J. Virol. 2017, 91, e01623-16. [Google Scholar] [CrossRef] [Green Version]
- Marrugal-Lorenzo, J.A.; Serna-Gallego, A.; Gonzalez-Gonzalez, L.; Bunuales, M.; Poutou, J.; Pachon, J.; Gonzalez-Aparicio, M.; Hernandez-Alcoceba, R.; Sanchez-Cespedes, J. Inhibition of adenovirus infection by mifepristone. Antivir. Res. 2018, 159, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Romanutti, C.; Bruttomesso, A.C.; Castilla, V.; Galagovsky, L.R.; Wachsman, M.B. Anti-adenovirus activity of epiandrosterone and dehydroepiandrosterone derivatives. Chemotherapy 2010, 56, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Cespedes, J.; Moyer, C.L.; Whitby, L.R.; Boger, D.L.; Nemerow, G.R. Inhibition of adenovirus replication by a trisubstituted piperazin-2-one derivative. Antivir. Res. 2014, 108, 65–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Cespedes, J.; Martinez-Aguado, P.; Vega-Holm, M.; Serna-Gallego, A.; Candela, J.I.; Marrugal-Lorenzo, J.A.; Pachon, J.; Iglesias-Guerra, F.; Vega-Perez, J.M. New 4-Acyl-1-phenylaminocarbonyl-2-phenylpiperazine derivatives as potential inhibitors of adenovirus infection. synthesis, biological evaluation, and structure-activity relationships. J. Med. Chem. 2016, 59, 5432–5448. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Q.; Zhou, Z.; Liu, M.; Chen, Y.; Li, J.; Xu, L.; Guo, J.; Li, Q.; Yang, J.; et al. Retinoic acid receptor beta, a potential therapeutic target in the inhibition of adenovirus replication. Antivir. Res. 2018, 152, 84–93. [Google Scholar] [CrossRef]
- King, C.R.; Tessier, T.M.; Dodge, M.J.; Weinberg, J.B.; Mymryk, J.S. Inhibition of human adenovirus replication by the importin α/β1 nuclear import inhibitor ivermectin. J. Virol. 2020. [Google Scholar] [CrossRef]
- Georgi, F.; Andriasyan, V.; Witte, R.; Murer, L.; Hemmi, S.; Yu, L.; Grove, M.; Meili, N.; Kuttler, F.; Yakimovich, A.; et al. The FDA-approved drug Nelfinavir inhibits lytic cell-free, but not cell-associated non-lytic transmission of human adenovirus. Antimicrob. Agents Chemother. 2020, 1002–1020. [Google Scholar] [CrossRef]
- McGrath, W.J.; Graziano, V.; Zabrocka, K.; Mangel, W.F. First generation inhibitors of the adenovirus proteinase. FEBS Lett. 2013, 587, 2332–2339. [Google Scholar] [CrossRef] [Green Version]
- Giberson, A.N.; Davidson, A.R.; Parks, R.J. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res. 2012, 40, 2369–2376. [Google Scholar] [CrossRef] [Green Version]
- Hsu, E.; Pennella, M.A.; Zemke, N.R.; Eng, C.; Berk, A.J. Adenovirus E1A activation domain regulates H3 acetylation affecting varied steps in transcription at different viral promoters. J. Virol. 2018. [Google Scholar] [CrossRef]
- Gupta, A.; Jha, S.; Engel, D.A.; Ornelles, D.A.; Dutta, A. Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator EIA. Oncogene 2012, 32, 5017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soriano, A.M.; Crisostomo, L.; Mendez, M.; Graves, D.; Frost, J.R.; Olanubi, O.; Whyte, P.F.; Hearing, P.; Pelka, P. Adenovirus 5 E1A interacts with E4orf3 to regulate viral chromatin organization. J. Virol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Milavetz, B.I.; Balakrishnan, L. Viral epigenetics. Methods Mol. Biol. 2015, 1238, 569–596. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Vogel, J.L.; Narayanan, A.; Peng, H.; Kristie, T.M. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 2009, 15, 1312–1317. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Vogel, J.L.; Arbuckle, J.H.; Rai, G.; Jadhav, A.; Simeonov, A.; Maloney, D.J.; Kristie, T.M. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci. Transl. Med. 2013, 5, 165–167. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Quenelle, D.; Vogel, J.L.; Mascaro, C.; Ortega, A.; Kristie, T.M. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 2013, 4, e00558-12. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, M.; Li, J.; Yang, G.; Huang, Q.; Li, J.; Wang, H.; He, S.; Li, E. Histone deacetylase inhibitors promote latent adenovirus reactivation from tonsillectomy specimens. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Shi, Y.J.; Matson, C.; Lan, F.; Iwase, S.; Baba, T.; Shi, Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 2005, 19, 857–864. [Google Scholar] [CrossRef]
- Ferrari, R.; Pellegrini, M.; Horwitz, G.A.; Xie, W.; Berk, A.J.; Kurdistani, S.K. Epigenetic reprogramming by adenovirus e1a. Science 2008, 321, 1086–1088. [Google Scholar] [CrossRef] [Green Version]
- Horwitz, G.A.; Zhang, K.; McBrian, M.A.; Grunstein, M.; Kurdistani, S.K.; Berk, A.J. Adenovirus small e1a alters global patterns of histone modification. Science 2008, 321, 1084–1085. [Google Scholar] [CrossRef] [Green Version]
- Amarelle, L.; Lecuona, E. The antiviral effects of Na,K-ATPase inhibition: A minireview. Int. J. Mol. Sci. 2018, 19, 2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodson, A.W.; Taylor, T.J.; Knipe, D.M.; Coen, D.M. Inhibitors of the sodium potassium ATPase that impair herpes simplex virus replication identified via a chemical screening approach. Virology 2007, 366, 340–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, C.T.; Hsu, J.T.; Hsieh, H.P.; Lin, P.H.; Chen, T.C.; Kao, C.L.; Lee, C.N.; Chang, S.Y. Anti-HSV activity of digitoxin and its possible mechanisms. Antivir. Res. 2008, 79, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Tollefson, A.E.; Ying, B.; Spencer, J.F.; Sagartz, J.E.; Wold, W.S.M.; Toth, K. Pathology in permissive syrian hamsters after infection with species c Human Adenovirus (HAdV-C) is the result of virus replication: HAdV-C6 replicates more and causes more pathology than HAdV-C5. J. Virol. 2017, 91, e00284-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenaerts, L.; Naesens, L. Antiviral therapy for adenovirus infections. Antivir. Res. 2006, 71, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Kuschner, R.A.; Russell, K.L.; Abuja, M.; Bauer, K.M.; Faix, D.J.; Hait, H.; Henrick, J.; Jacobs, M.; Liss, A.; Lynch, J.A.; et al. A phase 3, randomized, double-blind, placebo-controlled study of the safety and efficacy of the live, oral adenovirus type 4 and type 7 vaccine, in U.S. military recruits. Vaccine 2013, 31, 2963–2971. [Google Scholar] [CrossRef]
- Bradley, R.R.; Lynch, D.M.; Iampietro, M.J.; Borducchi, E.N.; Barouch, D.H. Adenovirus serotype 5 neutralizing antibodies target both hexon and fiber following vaccination and natural infection. J. Virol. 2012, 86, 625–629. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhou, Z.; Tian, X.; Liu, W.; Xu, D.; Fan, Y.; Liao, J.; Gu, S.; Li, X.; Zhou, R. A recombinant trivalent vaccine candidate against human adenovirus types 3, 7, and 55. Vaccine 2018, 36, 2199–2206. [Google Scholar] [CrossRef]
- Eckstein, A.; Grossl, T.; Geisler, A.; Wang, X.; Pinkert, S.; Pozzuto, T.; Schwer, C.; Kurreck, J.; Weger, S.; Vetter, R.; et al. Inhibition of adenovirus infections by siRNA-mediated silencing of early and late adenoviral gene functions. Antivir. Res. 2010, 88, 86–94. [Google Scholar] [CrossRef]
- Zhao, H.; Granberg, F.; Elfineh, L.; Pettersson, U.; Svensson, C. Strategic attack on host cell gene expression during adenovirus infection. J. Virol. 2003, 77, 11006–11015. [Google Scholar] [CrossRef] [Green Version]
- Prusinkiewicz, M.A.; Mymryk, J.S. Metabolic reprogramming of the host cell by human adenovirus infection. Viruses 2019, 11, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Chen, M.; Valdes, A.; Lind, S.B.; Pettersson, U. Transcriptomic and proteomic analyses reveal new insights into the regulation of immune pathways during adenovirus type 2 infection. BMC Microbiol. 2019, 19, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stray, S.J.; Johnson, J.M.; Kopek, B.G.; Zlotnick, A. An in vitro fluorescence screen to identify antivirals that disrupt hepatitis B virus capsid assembly. Nat. Biotechnol. 2006, 24, 358–362. [Google Scholar] [CrossRef]
- Nun, T.K.; Kroll, D.J.; Oberlies, N.H.; Soejarto, D.D.; Case, R.J.; Piskaut, P.; Matainaho, T.; Hilscher, C.; Wang, L.; Dittmer, D.P.; et al. Development of a fluorescence-based assay to screen antiviral drugs against Kaposi’s sarcoma associated herpesvirus. Mol. Cancer Ther. 2007, 6, 2360–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luthra, P.; Liang, J.; Pietzsch, C.A.; Khadka, S.; Edwards, M.R.; Wei, S.; De, S.; Posner, B.; Bukreyev, A.; Ready, J.M.; et al. A high throughput screen identifies benzoquinoline compounds as inhibitors of Ebola virus replication. Antivir. Res. 2018, 150, 193–201. [Google Scholar] [CrossRef]
- Johansen, L.M.; Brannan, J.M.; Delos, S.E.; Shoemaker, C.J.; Stossel, A.; Lear, C.; Hoffstrom, B.G.; Dewald, L.E.; Schornberg, K.L.; Scully, C.; et al. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 2013, 5, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Hartline, C.B.; Keith, K.A.; Eagar, J.; Harden, E.A.; Bowlin, T.L.; Prichard, M.N. A standardized approach to the evaluation of antivirals against DNA viruses: Orthopox-, adeno-, and herpesviruses. Antivir. Res. 2018, 159, 104–112. [Google Scholar] [CrossRef]
- Duffy, M.R.; Parker, A.L.; Kalkman, E.R.; White, K.; Kovalskyy, D.; Kelly, S.M.; Baker, A.H. Identification of novel small molecule inhibitors of adenovirus gene transfer using a high throughput screening approach. J. Control. Release Off. J. Control. Release Soc. 2013, 170, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Andersson, E.K.; Strand, M.; Edlund, K.; Lindman, K.; Enquist, P.-A.; Spjut, S.; Allard, A.; Elofsson, M.; Mei, Y.-F.; Wadell, G. Small-molecule screening using a whole-cell viral replication reporter gene assay identifies 2-{[2-(benzoylamino)benzoyl]amino}-benzoic acid as a novel antiadenoviral compound. Antimicrob. Agents Chemother. 2010, 54, 3871–3877. [Google Scholar] [CrossRef] [Green Version]
- Russell, K.L.; Hawksworth, A.W.; Ryan, M.A.K.; Strickler, J.; Irvine, M.; Hansen, C.J.; Gray, G.C.; Gaydos, J.C. Vaccine-preventable adenoviral respiratory illness in US military recruits, 1999–2004. Vaccine 2006, 24, 2835–2842. [Google Scholar] [CrossRef] [Green Version]

| Category | Compound | Reference(s) |
|---|---|---|
| Nucleoside/nucleotide analogues | Zalcitabine | [118,122,123] |
| Alvoudine | [118] | |
| Stavudine | [124] | |
| Acyclic nucleoside phosphonate analogues of cidofovir | [118] | |
| Various ether lipid-ester prodrugs of cidofovir | [125] | |
| Gemcitabine | [126] | |
| Cytarabine | [127] | |
| Other nucleoside/nucleotide analogues | [128] | |
| Inhibitors of epigenetic regulators | Valproic acid | [129] |
| Vorinostat | [130] | |
| Trichostatin A | [130] | |
| Other HDAC inhibitors | [130] | |
| Chaetocin | [126] | |
| GSK126 | [131] | |
| GSK343 | [131] | |
| Lestaurtinib | [126] | |
| Steroid-based compounds | Digoxin | [127,132] |
| Digitoxin | [132] | |
| Digitoxigenin | [127] | |
| Lanatoside C | [127] | |
| Dexamethasone | [127] | |
| Flunisolide | [127] | |
| Mifepristone | [133] | |
| Epiandrosterone and derivatives | [134] | |
| Other compounds | Piperazine derivatives | [135,136] |
| Tazarotene | [137] | |
| Ivermectin | [138] | |
| Protease inhibitors | [139,140] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, B.; Parks, R.J. Recent Advances in Novel Antiviral Therapies against Human Adenovirus. Microorganisms 2020, 8, 1284. https://doi.org/10.3390/microorganisms8091284
Saha B, Parks RJ. Recent Advances in Novel Antiviral Therapies against Human Adenovirus. Microorganisms. 2020; 8(9):1284. https://doi.org/10.3390/microorganisms8091284
Chicago/Turabian StyleSaha, Bratati, and Robin J. Parks. 2020. "Recent Advances in Novel Antiviral Therapies against Human Adenovirus" Microorganisms 8, no. 9: 1284. https://doi.org/10.3390/microorganisms8091284




