Abstract
Marine anaerobic methane oxidation (AOM) is generally assumed to be coupled to sulfate reduction, via a consortium of anaerobic methane-oxidizing archaea (ANME) and sulfate-reducing bacteria (SRB). ANME-1 are, however, often found as single cells, or only loosely aggregated with SRB, suggesting they perform a form of AOM independent of sulfate reduction. Oxidized metals and humic substances have been suggested as potential electron acceptors for ANME, but up to now, AOM linked to reduction of these compounds has only been shown for the ANME-2 and ANME-3 clades. Here, the effect of the electron acceptors anthraquinone-disulfonate (AQDS), a humic acids analog, and Fe3+ on anaerobic methane oxidation were assessed by incubation experiments with anoxic Black Sea water containing ANME-1b. Incubation experiments with 13C-methane and AQDS showed a stimulating effect of AQDS on methane oxidation. Fe3+ enhanced the ANME-1b abundance but did not substantially increase methane oxidation. Sodium molybdate, which was added as an inhibitor of sulfate reduction, surprisingly enhanced methane oxidation, possibly related to the dominant abundance of Sulfurospirillum in those incubations. The presented data suggest the potential involvement of ANME-1b in AQDS-enhanced anaerobic methane oxidation, possibly via electron shuttling to AQDS or via interaction with other members of the microbial community.
1. Introduction
Methane is a potent greenhouse gas (warming potential 34 times greater than CO2 (Forster et al. 2007), and its atmospheric concentrations is rapidly increasing [1]). There is a large and continuous production of methane in anaerobic marine sediments by methanogenic archaea. However, most of this methane is converted into carbon dioxide by oxidation, when methane is still in the sediment. It is estimated that marine anaerobic methane oxidizers consume 70–300 Tg CH4 year−1, reducing the atmospheric methane budget by 10–60% [2,3]. The methane that is not oxidized in the sediments, gets released into the water column, via diffusion or bubbling. From there, it can be emitted into the atmosphere. Methane oxidation in the water column can also (partially) consume this methane and thus forms an additional filter to prevent methane emission from marine systems.
To be thermodynamically favorable, anaerobic methane oxidation (AOM) needs to be coupled to the reduction of another compound. In marine settings, this compound is generally sulfate, and anaerobic methane oxidation is typically performed by a consortium of anaerobic methane-oxidizing archaea (ANME) and sulfate-reducing bacteria (SRB) [4]. Methane oxidation coupled to sulfate reduction yields a Gibbs free energy yield of only −17 kJ mol−1, which is near the minimum requirement for life, possibly being one of the factors explaining the slow growth rates of anaerobic marine methane oxidizers (doubling times 2–7 months [5,6]. Theoretically, methane oxidation coupled to the reduction of other compounds, such as Fe3+, nitrate, or nitrite, has a substantially higher energy yield [7]. Metal-oxide dependent AOM by ANME has been detected in enrichment cultures [8], but despite the thermodynamic advantages, observations of AOM not coupled to sulfate reduction in natural situations have been scarce. Egger et al. (2014) demonstrated iron oxide-mediated AOM was likely to occur in sediments of the Bothnian Sea (Northeast Baltic), but the microorganisms involved were not identified [9]. Scheller et al. (2016) showed that ANME were capable of performing methane oxidation with Fe3+ and 9,10-anthraquinone-2,6-disulfonate (AQDS), which was used as a humic acid analog [10]. Iron-mediated AOM, catalyzed by humic substances, was also detected by Valenzuela et al. (2019) [11]. Bai et al. (2019) also showed nitrate-reducing ANME were capable of using AQDS as an electron acceptor [12].
ANME oxidize methane via a reversed methanogenesis pathway (e.g. [13,14]). Three clades of ANME including several subclasses are recognized, all related to different groups of methanogens, namely ANME-1, ANME-2, and ANME-3 [13,15,16,17]. All clades are regularly found in syntrophy with sulfate-reducing bacteria (SRB), but whereas ANME-2 and ANME-3 are generally found in aggregates together with SRB cells, ANME-1 are often observed as single cells or loose aggregates [18]. The mechanism behind the ANME–SRB syntrophy is still under debate. A relationship based on the exchange of reaction products has been proposed [18,19,20,21], but other studies also suggested direct interspecies electron transfer [22,23]. ANME has been shown to be capable of forming intracellular wiring, creating a cell-to-cell connection that could allow a direct shuttling of electrons [22,23,24,25]. In this regard, the electron shuttling to abiotic particles such as oxidized metals or AQDS has only been observed in ANME of the clade ANME-2 [10,12].
The Black Sea is rich in methane due to the release from numerous cold seeps, and despite active methane oxidation in the sediments, water column methane concentrations below the chemocline are ca. 10–15 µM [26,27]. ANME-1 have been detected in the Black Sea water column before [28,29,30,31]. The 13C-depleted stable carbon isotopic composition of a- and monocyclic biphytanes derived from the characteristic membrane lipids of ANME-1 revealed that these archaea actively consume methane in the water column [29]. ANME have been suggested to play a major role in decreasing water column methane concentrations [31]. Although ANME-1 have often been observed in environments low in sulfate, and without a syntrophic SRB partner, their methane oxidation pathway independent of sulfate reduction remains unknown [32]. Previous studies have suggested a decoupling between AOM and sulfate reduction in the Black Sea water column, as no substantial stable carbon isotope depletion of SRB phospholipid fatty acid could be detected [29]. As AOM coupled to the reduction of alternative electron acceptors, such as humic substances or Fe3+, has a much higher (theoretical) energy gain than sulfate-mediated AOM, the availability of these electron acceptors could theoretically make AOM more thermodynamically favorable for ANME-1b [7].
To explore the metabolic versatility of ANME, the ANME-1b subgroup present in the anoxic water column of the Black Sea was studied. Suspended particulate matter (SPM) from the water column was collected and used for incubation studies with 13CH4, exploring the response of the microbial community to AQDS and Fe3+ in the presence of sodium molybdate (an inhibitor of sulfate reduction). The 13CO2 concentration was followed over time as a measure for methane oxidation. The microbial diversity at the end of the incubation experiments was analyzed by 16S rRNA gene sequencing to assess changes in the community composition under different conditions.
2. Materials and Methods
2.1. Sample Collection
Sampling was performed during cruise 64PE444 on R/V Pelagia in August 2018 at station 42° 53.8’ N 30° 40.7’ E. The conditions in the water column at the moment of sampling are shown in Fig. S1. Water samples were taken using a conductivity-temperature-density (CTD) system equipped with Niskin sampling bottles. Samples for nutrient analysis were collected directly after CTD recovery. A constant N2 flow during CTD sampling was used to retain anoxic conditions. N2 flushed pressure bottles (1 L) were filled with water collected at 1000 m depth by piercing the butyl stoppers with a needle.
Water column SPM from a depth of 1000 m was collected onto GF75 pore size 0.3 µm glass fiber filters (Advantec, Dublin, CA, USA) using a McLane WTS-LV in situ pump (McLane, East Falmouth, MA, USA), completed with a special filter head for anoxic sampling. Pumps were left in the water column to filter for 6 h. After pump recovery, the filter heads were transported to an anoxic glove bag, which was flushed with N2 three times before the overlying anoxic water was removed from the filters. Filters were then transferred to 1 L glass bottles filled with anoxic water collected from 1000 m, closed, and stored in the dark at 4–10 °C for 60 days until incubations were set up in the laboratory. Another filter was directly stored at −80 °C for analysis of the in situ microbial community. Water column samples for nutrient and DNA analysis were collected as described in Sollai et al. (2019) and Suominen et al. (2020) [30,33].
2.2. Incubations with Suspended Particulate Matter
To set up the incubation experiments, water column SPM was retrieved by scraping off the top layer of the glass fiber filters under anoxic conditions inside an anaerobic glove bag (Sigma Aldrich, St. Louis, MI, USA) under N2 atmosphere, and subsequently resuspended in 1 L of anoxic artificial seawater (commercially available mixture of sea salts, Sigma Aldrich, containing 28 mM SO42− but no sulfide) in incubation bottles of 1.2 L. All media in the anoxic bottles was boiled and bubbled with nitrogen for 20 min to remove residual oxygen, after which the bottles were closed, crimp sealed, and the headspace was flushed and exchanged with N2 gas using a GRInstruments (Wijk bij Duurstede, the Netherlands) automatic gas exchanger. Due to the used method, fibers of the filter were present in the incubation bottles. 15N-ammonium chloride (0.016 g) was also added for stable isotope activity measurements, but in the end, 15N incorporation was not measured. 10 mL 13CH4 (99% labeled; Sigma-Aldrich, St. Louis, MI, USA, resulting in a methane concentration of 500 µM) was added in order to follow methane-derived 13C over the course of the experiments. Depending on the type of incubation, 4.1 g sodium molybdate (Sigma-Aldrich), 0.03 g iron(III) citrate (Sigma-Aldrich), or 1.65 g anthraquinone-2,7-disulfonic acid disodium salt (AQDS, TCI Chemicals, Tokyo, Japan) or a combination of these (Table S1) was added to the medium. Autoclaved artificial seawater was used as an abiotic control to assess abiotic variations and instrument variability of the measured parameters. All experiments were performed in duplicate. The bottles were incubated in the dark at 10 °C for 58 days. Every 14 days, the bottles were shaken, and headspace gas was withdrawn for analysis. At the termination of the incubations, 10 mL of the medium was collected for nutrient analysis, stored at −20 °C until analysis, and processed as previously described [30]. The remaining medium was filtered over 0.3 µm GF75 filters (Advantec, Dublin, CA, US) for DNA analysis and was stored at −80 °C.
2.3. 13CO2 Analysis
13C-labeled carbon dioxide concentrations in the headspace of the incubation bottles were measured using a gas chromatograph (GC) equipped with a mass spectrometer (MS) (Agilent, Santa Clara, CA, USA, 7890B GC with 5975C MSD) in analytical triplicates. To study and compare the relatively small production or consumption of these compounds in the different incubation experiments with slightly different starting concentrations, the data of each individual incubation bottle was normalized on the starting value (t0) as 100%.
2.4. DNA Extraction and Analysis
DNA was extracted from the filters using the PowerSoil DNA extraction kit (MoBio Laboratories, Carlsbad, CA, USA) and stored at −80 °C until further analysis. The general 16S rRNA archaeal and bacteria primer pair 515F and 806RB targeting the V4 region [34] were used for the 16S rRNA gene amplicon sequencing and analysis, as described in Besseling et al. (2018) [35]. PCR products were gel purified using the QIAquick Gel-Purification kit (Qiagen, Hilden, Germany), pooled, and diluted. Sequencing was performed by the Utrecht Sequencing Facility (Utrecht, the Netherlands), using an Illumina MiSeq sequencing platform. Analysis of the 16S rRNA gene amplicon sequences was performed with the Cascabel pipeline [36], including quality assessment by FastQC [37], assembly of the paired-end reads with Pear [38], and assign taxonomy (including pick representative set of sequences with ‘longest’ method) with blast by using the Silva 128 release as reference database (https://www.arb-silva.de/). For analysis purposes, only species with a relative abundance greater than 0.001 were assumed significant. For tables and figures, results of duplicate bottles of the same treatment were averaged. The 16S rRNA amplicon reads (raw data) have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA605700.
2.5. Quantitative PCR 16S rRNA Gene
16S rRNA gene copies were quantified using quantitative PCR (qPCR) with the same primer pair as used for amplicon sequencing (515F, 806RB) on a Rotor-Gene 6000 (Corbett Research, Mortlake, Australia). The qPCR reaction mixture (25 µL) contained 0.5 U of Phusion High-Fidelity DNA Polymerase (Thermo Scientific), 1× Phusion HF Buffer, 0.2 µM of each dNTP, 20 μg of BSA, 0.6 pmol µL−1 of both primers, 0.5× EvaGreen dye (0.625 µM) in aqueous solution (Biotium, Hayward, CA, USA) and AccuGENE Molecular Biology Water (Lonza, Basel, Switzerland). The cycling conditions for the qPCR reaction were as follow: initial denaturation 98 °C for 30 s, 45 cycles of 98 °C for 10 s, 50 °C for 20 s, followed by fluorescence data acquisition, 72 °C for 30 s, and 80 °C for 25 s. Specificity of the reaction was tested with a gradient melting temperature assay, from 55 °C to 95 °C with 0.5 °C increments of 5 s apiece. The qPCR reactions were performed in duplicate with standard curves encompassing a range from 101 to 107 molecules µL−1. qPCR efficiency for the 16S rRNA gene quantification was 100% with R2 = 0.996. For quantification of microbial groups, the assumption that all microorganisms of the microbial community contained a single 16S rRNA copy in their genome was made, which has been confirmed by genome analysis for the ANME-1 group [25].
3. Results
The microbial community of the deep Black Sea water column was studied during incubation experiments, specifically focused on the response to additions of different electron acceptors and their effect on anaerobic methane oxidation. The methanotrophic activity in the incubations was assessed by the addition of 13C-labeled methane, followed by the analysis of 13CO2 concentrations over time.
3.1. Water Column Physicochemical Conditions and In Situ Microbial Community
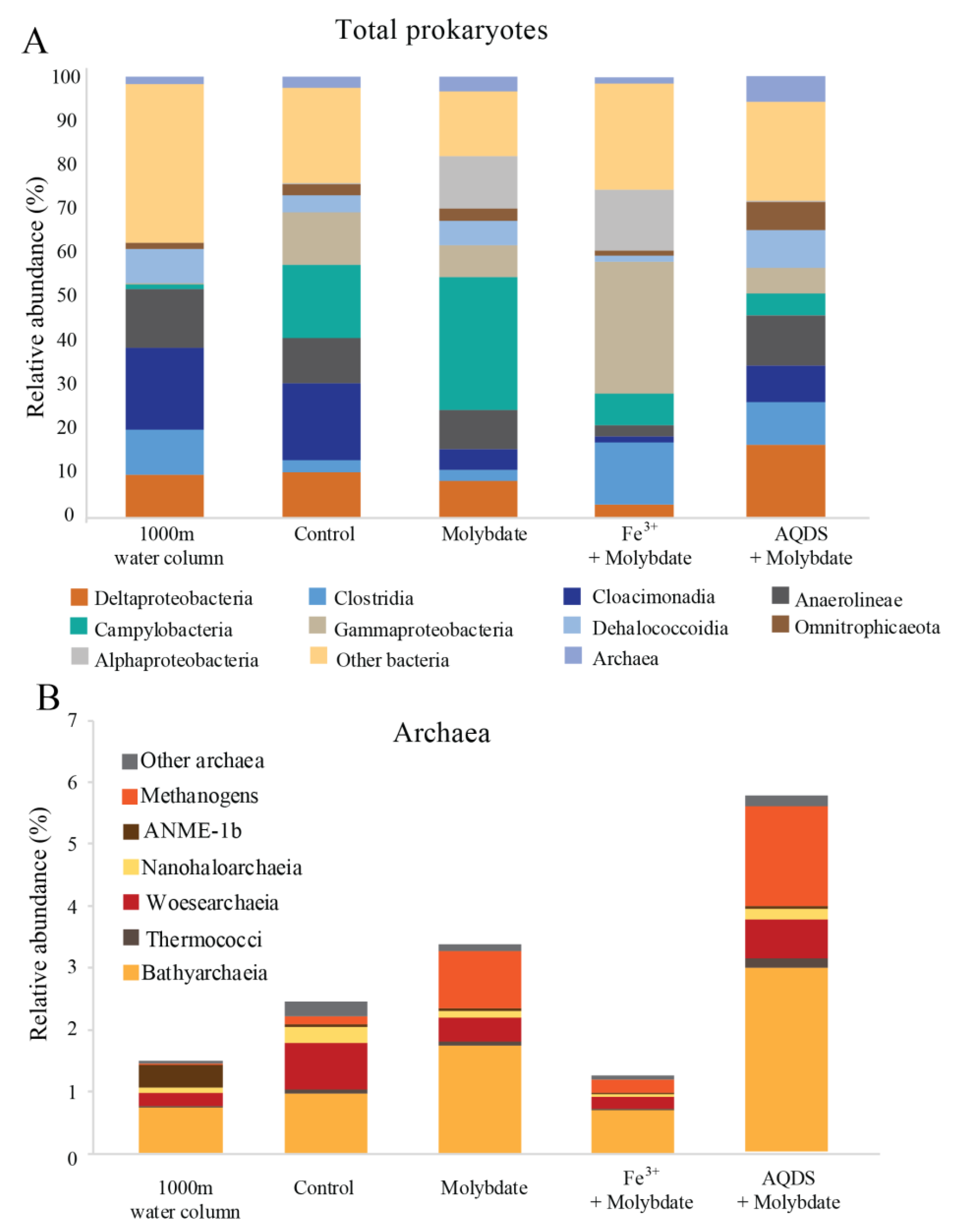
The Black Sea water column is over 2000 m deep and permanently stratified, with, at the studied station, an oxycline around 75 m depth (Figure S1). Water was sampled at 1000 m depth. The sulfate concentration at this depth was 17 mM and the nitrite concentration 21 nM (Figure S1). Diversity estimates based on 16S rRNA gene amplicon sequencing at 1000 m depth showed that the archaeal abundance was 2 × 105 copies per L (i.e., 1.5% of the total 16S rRNA reads; Figure 1). 21% of the archaeal 16S rRNA reads were classified as ANME-1b (0.3% of the total 16S rRNA reads; Table 1 and Table S2; Figure 2), corresponding to 4 × 104 copies per L. No known methanotrophs other than ANME-1b were detected in the library of 16S rRNA reads from 1000 m depth (average 190,000 reads per sample; Table S2). The abundance of methanogenic archaea (defined as 16S rRNA gene reads assigned to the Methanomicrobia, Methanococci, Methanobacteria, Methanomassiliicoccales, and Methanofastidiosales, but excluding ANME) amounted 4 × 103 copies per L (2% of the archaeal 16S rRNA gene reads; Table 1 and Table S2; Figure 1 and Figure 2). 16S rRNA gene reads attributed to bacteria were closely related to Cloacimonadia (2 × 106 copies per L, 19% of total 16S rRNA reads), Dehalococcoidia (9 × 105 copies per L, 7.7%), Deltaproteobacteria (1 × 106 copies per L, 9.8%, of which 5 × 105 copies per L falling into SEEP-SRB cluster and 6 × 105 copies per L to Desulfatiglans spp.), plus many other, less abundant groups (36% ‘other bacteria’; Figure 1).
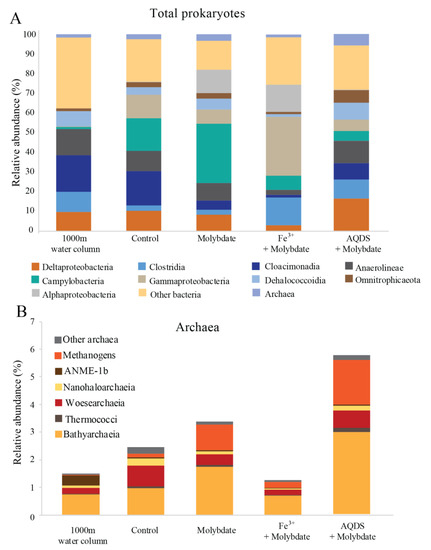
Figure 1.
Total prokaryotic (A) and archaeal (B) diversity in Black Seas water from 1000 m depth and at the end of the various incubation experiments, expressed as percentage of total 16S rRNA gene reads. Methanogens defined here as all species belonging to the Methanomicrobia, Methanococci, Methanobacteria, Methanomassiliicoccales, and Methanofastidiosales, excluding any anaerobic methane oxidizing archaea (ANME). Values for duplicate bottles are averaged, except for the 1000 m sample, for which only one sample was used.

Table 1.
Abundance (16S rRNA copies per L) of major species in the incubation experiments and the Black Sea water column.
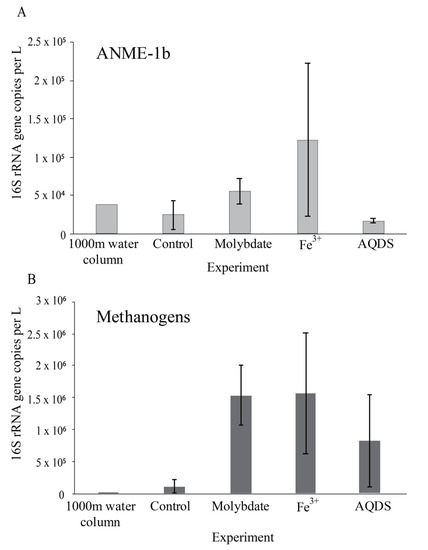
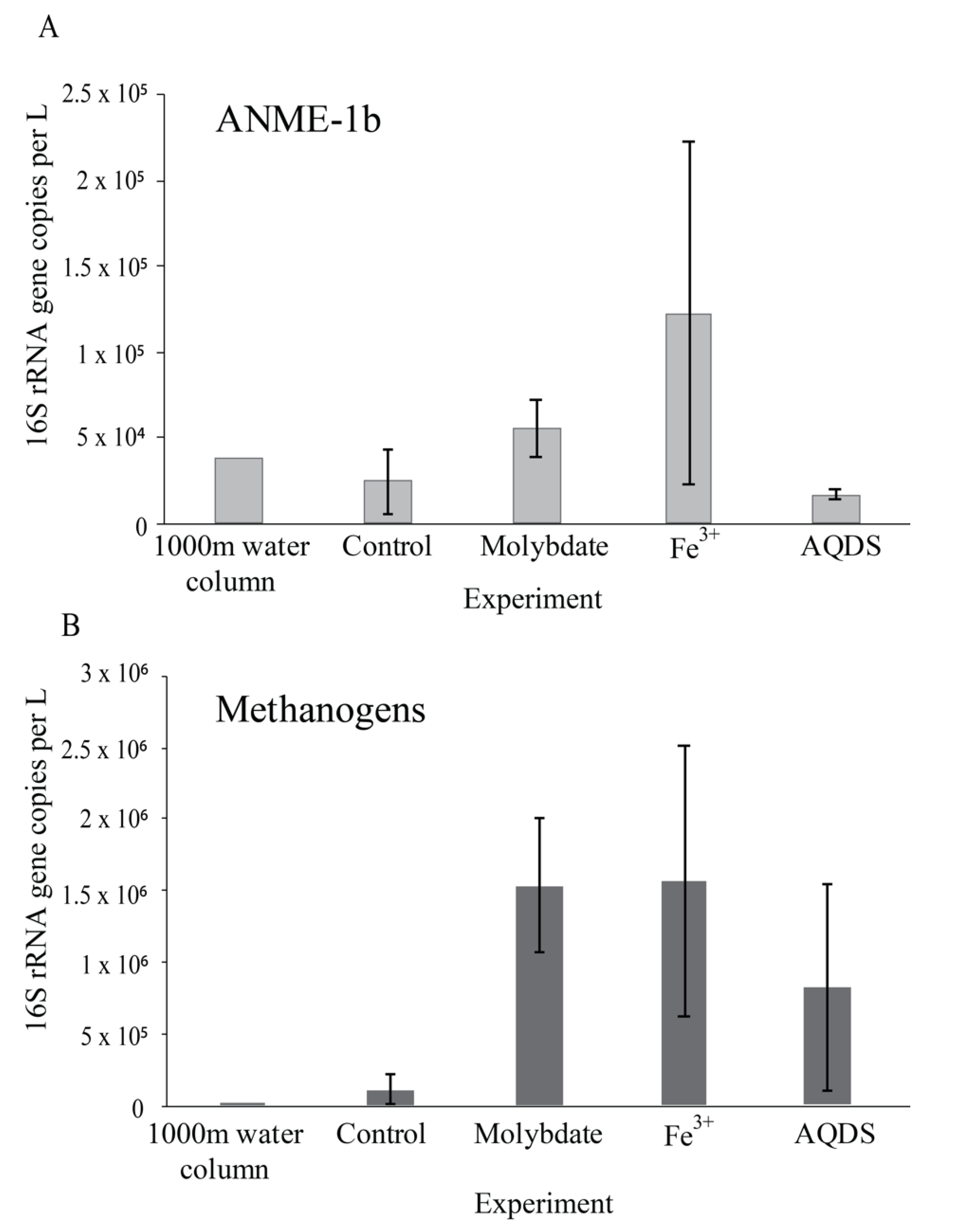
Figure 2.
Abundance of ANME-1b (A) and methanogens (B); defined here as all species belonging to the Methanomicrobia, Methanococci, Methanobacteria, Methanomassiliicoccales, and Methanofastidiosales, (excluding any ANME) in natural Black Sea water from 1000 m depth and at the end of the various incubation experiments, in 16S rRNA gene copies per L. Values for duplicate bottles are averaged, except for the 1000 m sample, for which only one sample was used. Notice the different scale on the y-axis.
3.2. Abiotic and Control Incubations
13CO2 concentrations in the abiotic incubations (artificial seawater with added 13CH4) and control (artificial seawater with added 13CH4 and microbial matter from the SPM) remained constant over the course of the experiment after a small initial decrease (Figure 3). The ANME-1b abundance in the control incubations was 2 × 104 copies per L, corresponding to 1.4% of the archaeal 16S rRNA gene reads. No other microorganisms known to be capable of methane oxidation were detected. Reads assigned to methanogenic archaea made up 1 × 105 copies per L (6.6% of the archaeal 16S rRNA gene reads; Table 1; Figure 2). The bacterial community was similar to that of the water column at 1000 m depth (Figure 1 and Figure S2), except for a higher relative abundance of Campylobacteria (1 × 107 copies per L, 16% of total 16S rRNA reads; Figure 1) and Gammaproteobacteria (9 × 106 copies per L, 12%; Figure 1). The genus Sulfurimonas, belonging to the Campylobacteria, comprised 5 × 106 copies per L, corresponding to 7% of the 16S rRNA reads, the genus Sulfurospirillum (Campylobacteria) 5 × 106 copies per L (6%; Table S2), Desulfatiglans (Deltaproteobacteria) 2 × 106 copies per L (3%), Fusibacter (Clostridia) 1 × 106 copies per L (2%) and SEEP-SRB (Deltaproteobacteria) 4 × 105 copies per L (1%; Table 1 and Table S2). Each of these bacterial groups, except for SEEP-SRB, was more abundant in the control incubations than in the water column (Table 1; Figure S2).
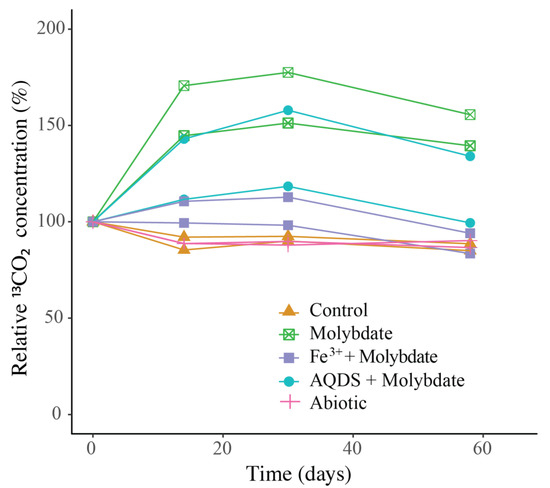
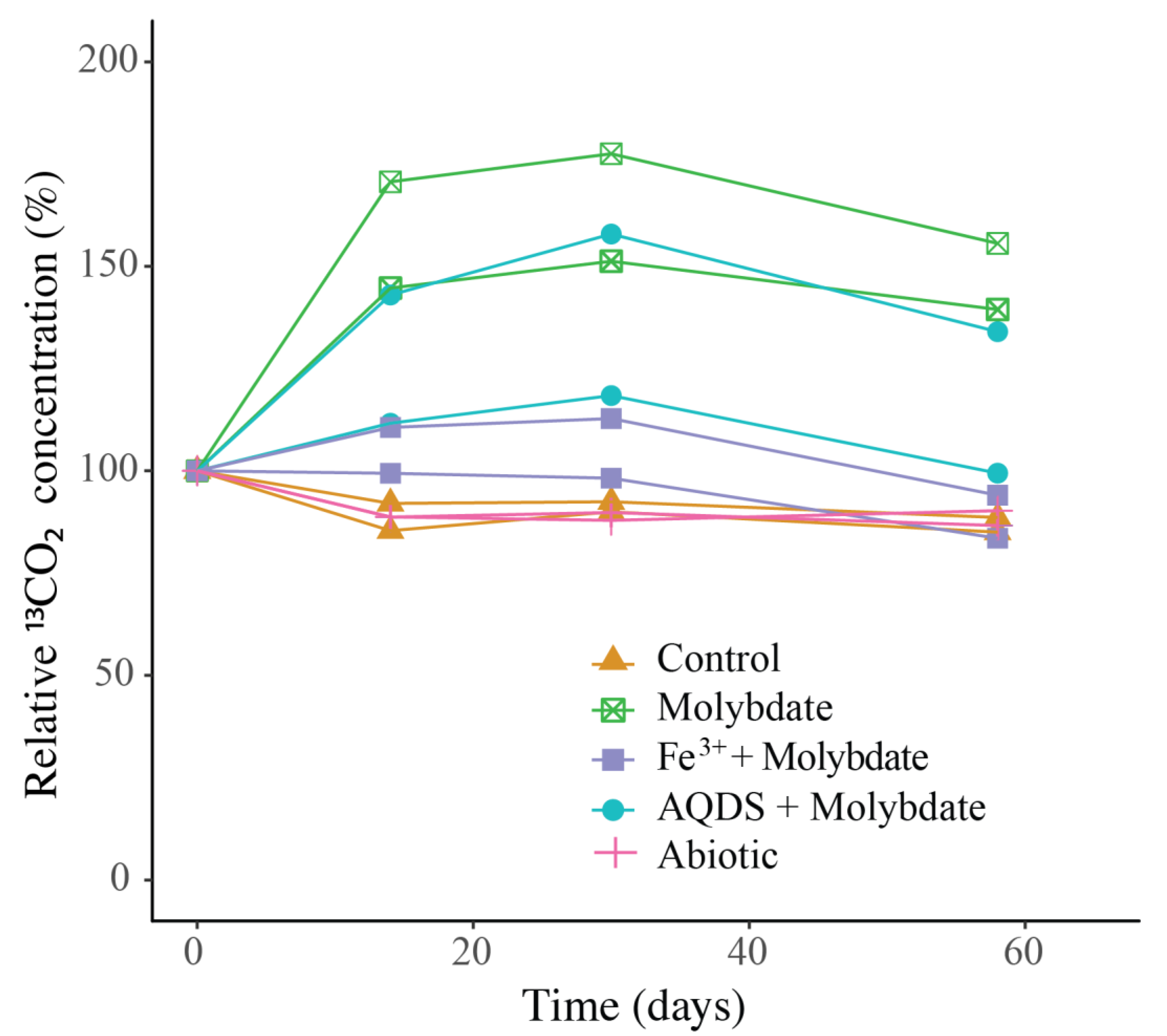
Figure 3.
Change over time of 13CO2 in the incubation experiments, normalized to the concentration at t0 (t0 = 100%). The duplicate incubation bottles of each experiment are both shown individually. Actual concentrations are shown in Figure S3.
3.3. Incubations with Sodium Molybdate
Sodium molybdate was used as an inhibitor of sulfate reduction in a subset of the incubation experiments, both with and without the addition of alternative electron acceptors (overview available in Table S1). In the incubation with only sodium molybdate, an increase of 65% in the 13CO2 concentration was observed from day 0 to day 30, after which the concentration slightly decreased (Figure 3 and Figure S3). The abundance of ANME-1b was 6 × 104 copies per L in the molybdate incubation, which corresponded to a relative abundance of 1% of the archaeal 16S rRNA gene reads (Table 1; Figure 2). The abundance of methanogenic archaea increased substantially to 1.5 × 106 copies per L (27% of the archaeal 16S rRNA reads; Table 1; Figure 2). Archaea of the Bathyarchaeia were relatively abundant compared to other archaea (3 × 106 copies per L, 52% of archaeal 16S rRNA reads; Figure 1). The total 16S rRNA gene reads were strongly dominated by reads attributed to Campylobacteria (5 × 107 copies per L, 31%), specifically of the genus Sulfurospirillum (5 × 107 copies per L, 27%; Table 1 and Table S2). Members of the Deltaproteobacteria Desulfatiglans comprised 1 × 107 copies per L (7%; Table 1 and Table S2), the Sulfurimonas 4 × 106 copies per L (2%) and SEEP-SRB 9 × 105 copies per L in the incubations with molybdate only (1%; Table 1 and Table S2).
3.4. Incubations with Sodium Molybdate and Soluble Fe3+ Complexes
The 13CO2 concentration in the Fe3+ amended incubations showed a slight increase during the experiment (on average 5%; Figure 3 and Figure S3) although the difference with the starting concentration was small. The abundance of ANME-1b was 1 × 105 copies per L (1.3% of the archaeal reads; Table 1; Figure 2). The abundance of methanogenic archaea was 2 × 106 copies per L (17% of the archaeal 16S rRNA gene reads; Table 1; Figure 2). Gammaproteobacteria dominated the Fe3+-amended incubations (2 × 108 copies per L, 30% of total 16S rRNA reads, dominated by Vibrionales; Figure 1). The abundance of Sulfurospirillum was 4 × 107 copies per L (5%, Table 1 and Table S2), the abundances of Desulfatiglans, Fusibacter and Sulfurimonas were 1–2 × 107 copies per L for all three genera (2% of the 16S rRNA gene reads; Table 1 and Table S2).
3.5. Incubations with Sodium Molybdate and AQDS
The addition of AQDS to the incubations resulted in an increase of 38% in 13CO2 concentrations over the course of the experiment (Figure 3 and Figure S3). The abundance of ANME-1b at the end of the incubation was 2 × 104 copies per L (1% of archaeal 16S rRNA reads; Table 1; Figure 2). The abundance of methanogenic archaea was 8 × 105 copies per L (30% of archaeal reads; Table 1; Figure 2). The Bathyarchaeia made up just over half of the archaeal 16S rRNA reads (1 × 106 copies per L, 52%; Figure 1). The 16S rRNA gene reads of bacteria were assigned to several major groups, each making up 5–18% of the community, with the Deltaproteobacteria being the most abundant (18%; Figure 1). The Deltaproteobacteria were dominated by the genus Desulfatiglans (6 × 106 copies per L, 13% of 16S rRNA reads; Table 1 and Table S2). The abundance of the genus Fusibacter was 3 × 106 copies per L (7%; Table 1 and Table S2). The abundance of the SEEP-SRB cluster was 8 × 105 copies per L (representing 2% of 16S rRNA reads; Table 1 and Table S2). Sulfurimonas, Sulfurospirillum, and Sulfurovum abundances were 2 × 106, 3 × 105, and 2 × 105 copies per L, respectively (representing 3, 0.6 and 0.4% of 16S rRNA reads, respectively; Table 1 and Table S2).
4. Discussion
Most commonly, marine AOM is coupled to sulfate reduction via a syntrophic relationship between ANME and SRB, based on the exchange of electrons or reaction intermediates [4]. In the past decade, several alternative electron acceptors for marine methane oxidation were proposed to be used by ANME, such as nitrate [39], iron and manganese [40], and humic substances [7,10,41]. ANME-1 are often found in the vicinity of SRB, but unlike ANME-2, they are found to be only loosely associated, not tightly aggregated [42,43], which raises the question if ANME-1 performs anaerobic methane oxidation independent of SRB and sulfate reduction. Here, this question was addressed by performing incubation experiments with alternative electron acceptors, using suspended particulate matter collected from the anoxic Black Sea water column which is naturally relatively rich in sulfate (17 mM, Figure S1) and where ANME-1b is present in the deep waters (4 × 104 ANME-1b 16S rRNA gene copies per L at 1000 m; Figure 2; Table 1). No other organisms known to perform methane oxidation were detected at 1000 m depth. Therefore, we assume that AOM at this depth in the water column and in the incubation experiments is performed by ANME-1b, allowing us to test the electron donor preferences of ANME-1b in an incubation setup.
4.1. Enhanced Methane Oxidation by ANME-1b in Molybdate and in AQDS Incubations
In the control incubations, which contained the same sulfate-containing artificial seawater, SPM, and 13CH4 as the other experiments, no increase in the 13CO2 concentration over time was observed. Possibly, methane oxidation did not occur, or at too low rates to observe over the 60-day experiment. Methane oxidation rates by ANME in the Black Sea were previously reported to be 0.5–7 nmol L−1 day−1 at 1000 m [29]. The highest 13CO2 production rates observed in our experiments were much lower than would be expected at those methane oxidation rates (Figure S3). Potentially, the change in conditions from the water column to the incubation bottles affected the activity and methane turnover rate of ANME-1b. A change in pressure is known to affect both ANME and SRB abundance and activity [44]. Another possibility would be that 13CO2 is produced by ANME-1b but is simultaneously consumed by methanogens, SRB, or other microbial groups, and therefore no increase is detectable in the gas headspace. As 13C incorporation in the biomass was not measured, the fate of the labeled substrate cannot be fully determined.
The addition of only molybdate, and of molybdate plus AQDS, increased methane oxidation as seen by the detected increase of 13CO2 (Figure 3). We believe that the results found in these experiments are interesting to discuss in more detail, despite the low number of replicates per treatment. As sulfate reduction was assumed to be inhibited by the addition of sodium molybdate [45], it seemed surprising that methane oxidation could occur in the incubations with the addition of molybdate only and no alternative electron acceptor such as humic substances or Fe3+. Even if the inhibition of sulfate reduction was incomplete, and some sulfate reduction-coupled AOM would still have occurred, the methane oxidation rate in the molybdate-only experiments would not be expected to be higher than in the control incubations, that also contain sulfate and methane, but no inhibiting molybdate. It is therefore considered unlikely that sulfate-mediated AOM was responsible for the observed 13CO2 production, in both the molybdate only and the molybdate + AQDS incubation. More likely, an alternative form of AOM occurred when sulfate-mediated AOM was inhibited, possibly leading to more favorable energetic conditions for ANME (ΔG°′ sulfate-mediated AOM −17 kJ mol−1, ΔG°′ AQDS-mediated AOM −41 kJ mol−1 [10]). Although it is considered likely that AQDS was involved in AOM in the AQDS incubations, the question arises what was the role of molybdate, and whether it was molybdate rather than AQDS causing the enhanced 13CO2 production that was observed in those incubations, given the high 13CO2 production in the molybdate only incubation. Possibly, molybdate could have played a role as electron acceptor, as several studies have found specific bacteria are capable of the reduction of molybdate to molybdenum blue [46,47,48]. However, if it was purely the molybdate that caused the increase in methane oxidation rates in both the molybdate and in the AQDS incubations, the incubations with Fe3+ (containing molybdate) would have also been expected to show increased 13CO2 production, which was not the case (Figure 3). No substantial 13CO2 production was observed in the Fe3+ incubations, despite the relatively high abundance of ANME-1b compared to the AQDS incubations (Figure 2). The ANME-1b in the Fe3+ incubations may have been inactive or involved in methanogenesis rather than in methanotrophic pathways [49,50]. Possibly, the total 16S rRNA gene copies in the AQDS amended incubations, and thus also the ANME-1b abundance, was underestimated due to an inhibitory effect of humic substances on qPCR reactions, which is widely recognized [51,52] and could complicate the comparison between incubations with and without humic substances.
4.2. Potential Role of Sulfur Cycling Organisms
Several different sulfur processes have been linked to marine AOM, i.e., the reduction of sulfate [4], zero-equivalent sulfur [19], or polysulfides [53]. To explore the role of sulfur cycling in the incubation experiments, the organisms that were present and are known to be involved in sulfur cycling were studied. The only sulfur compounds that were added to the incubation experiments were sulfate, as part of the artificial seawater salts mixture, organic sulfur-containing microbial biomass of the SPM, and possibly small amounts of sulfur compounds that were present in the natural water column and that were transferred with the SPM that was used as the inoculum. We chose to add sulfate to all experiments, including those with molybdate, Fe3+, and AQDS, to retain similar conditions in all experiments, varying only the molybdate and alternative electron acceptor availability. Several microbial groups that are known to be involved in the sulfur cycle were abundant in the incubations. SRB were assumed to be inhibited by the addition of molybdate to all but the control incubations, but as sulfate consumption or sulfide production within the incubation experiments were not measured, it was not possible to determine whether sulfate reduction was completely inhibited.
The genera Sulfurimonas, Sulfurospirillum, and Sulfurovum increased drastically in abundance in the control incubations when compared to the water column (Table 1 and Table S2; Figure S2). This change could potentially be attributed to a bottle effect, in which the incubation conditions favor specific bacterial groups. These microorganisms have all been described to use sulfur (elemental S or polysulfides), thiosulfate, or sulfite as electron acceptor, and, for some strains, also as electron donor [54,55,56,57]. Sulfurimonas, and likely also Sulfurovum and Sulfurospirillum, are capable of oxidizing sulfide to produce sulfate as an end product and elemental sulfur and polysulfide as intermediate products [58,59,60]. The ability to autotrophically fix CO2 is likely widely present in different Sulfurimonas, Sulfurospirillum, and Sulfurovum species [56,58]. Sulfurovum and some Sulfurimonas strains are also capable of oxidizing sulfide and of using H2 as an electron donor [56,61]. It is unclear which compounds were cycled in the incubation experiments, and which affect the addition of molybdate had on these processes. In the control incubation, where no inhibitor of sulfate reduction was present, active sulfate reduction and sulfur cycling are expected to occur. As the named species are capable of CO2 fixation, they may have decreased the 13CO2 concentration in the control experiments, possibly diminishing the increase in headspace 13CO2 that was expected to occur, and that was taken as a measure of methane oxidation.
In the incubation with only molybdate, Sulfurospirillum sp. dominated the community (5 × 107 copies per L, 27% of 16S rRNA reads; Table 1 and Table S2). As this was the main distinguishing factor between the molybdate and the other incubations, it could potentially be related to the enhanced AOM that was observed in these incubations (Figure 3). Recent research has suggested partner-independent AOM coupled to polysulfide reduction as a novel pathway of methane oxidation by ANME-1 [53], and potentially, Sulfurospirillum could produce these polysulfides. The production of polysulfides, however, requires the oxidation of sulfide, which was not added to our incubation experiments. It could be produced by the sulfur cycling organisms present (Sulfurimonas, Sulfurospirillum, Sulfurovum, SEEP-SRB) but it is unclear whether this could occur in the presence of molybdate, and why then specifically Sulfurospirillum became highly abundant in the molybdate incubations. In the AQDS incubations, Sulfurospirillum abundance was two orders of magnitude lower than in the molybdate only incubation (3 × 105 copies per L, 0.6%; Table 1 and Table S2). In the Fe3+ amended incubation, the Sulfurospirillum abundance (4 × 107 copies per L; Table 1) was comparable to the molybdate incubation, although the relative abundance was much lower (5%; Table S2).
Bacteria of the genera Desulfatiglans and Fusibacter became relatively more abundant in the incubations with AQDS (Desulfatiglans 13% of the 16S rRNA gene reads AQDS incubations, versus 3% in control incubations; Fusibacter 7% in AQDS incubations, vs. 2% in control incubations; Table S2), although this is not reflected in the absolute abundances (Table 1). Desulfatiglans sp. and Fusibacter sp. are known as strict anaerobes that can reduce sulfate, thiosulfate or sulfur, while oxidizing carbohydrates or other organic electron donors, such as AQDS [62,63,64]. Recently, Desulfatiglans sp. have been found to cooccur with ANME-1 and SEEP-SRB in estuarine sediments [65]. It is, however, unknown whether the sulfur compound reduction by Desulfatiglans sp. and Fusibacter sp. here could be coupled to AOM, and which sulfur compound they use here, as sulfate reduction is expected to be inhibited by the molybdate that was present in the incubations with AQDS.
4.3. Potential Methanogenesis by Methanogens, ANME and Bathyarchaeota
The abundance of methanogens in the molybdate and Fe3+ incubations was 10-fold higher than in control incubations (Table 1; Figure 2). Possibly, part of the produced CO2 in the incubations is converted back to CH4 by these methanogens, which would mean that the net produced 13CO2 is higher than was measured. It remains unclear what caused this increase in the methanogenic abundance.
Besides, ANME-1b has been shown to also be capable of methanogenesis, specifically under high H2 concentrations [65]. Although the concentration of H2 in the incubations was not measured, H2 may have built up as the consumption of H2 by SRB was likely inhibited by molybdate. Therefore, it is possible that ANME-1b switched to a methanogenic metabolism, producing methane rather than consuming it. In the molybdate and AQDS + molybdate incubations, a decrease in the 13CO2 concentration is observed after day 30 (Figure 3 and Figure S2). Possibly, this could be linked to such a metabolic switch.
The phylum Bathyarchaeota (i.e., the former Miscellaneous Crenarchaeotal Group) increased in abundance in all incubations supplemented with molybdate (i.e., from 8 × 105 copies per L in the control to 3 × 106, 5 × 106 and 1 × 106 copies per L in molybdate only, Fe3+ and AQDS, respectively; Figure 1). The Bathyarchaeota are known to be metabolically diverse, with subgroups potentially capable of methanogenesis [66], although organisms performing organic matter degradation and dissimilatory nitrogen and sulfur reduction are also present within this phylum [67,68,69,70,71]. The co-occurrence of Bathyarchaeota and ANME-1b in methane cold seeps has been suggested to be based on an indirect trophic relationship rather than a direct interaction [72] and the cause of the increase of the abundance of this phylum in the molybdate, Fe3+ and AQDS amended incubations remains unclear.
5. Conclusions
Overall, these results show that both molybdate and AQDS can stimulate methane oxidation in incubations with material from the Black Sea water column, containing ANME-1b. Enhanced methane oxidation by AQDS has been shown for ANME-2 in marine sediments [10] and also Valenzuela et al. (2017) observed an increase in 13CO2 production in sediment incubations with AQDS [73], but only when sulfate reduction was not inhibited and with barely detectable ANME-1 and ANME-3 abundances, making it difficult to assess which organisms were involved. To our knowledge, this is the first study showing that additions of molybdate, and of molybdate plus AQDS, are stimulating methane oxidation in incubations with water column SPM. We also believe this is the first study that suggests that ANME-1b is involved in an AQDS-stimulated AOM pathway. The mechanism behind the stimulating effects of molybdate and AQDS remains unclear. More research, including detailed measurements of the sulfur compounds in solution and gene expression analysis, is needed to reveal whether ANME-1b is indeed involved in AQDS-dependent AOM, whether sulfur compounds and a partner organism are involved, and what could be the role of this process in marine environments.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/9/1288/s1, Figure S1. Environmental conditions in the Black Sea water column at the time of sampling, Figure S2. Abundance of major groups in the Black Sea water column (1000 m depth) and the control incubation experiment, in 16S rRNA copies per L-1, Figure S3. 13CO2 concentration in the headspace of the incubations with different electron acceptors, Table S1. Overview of the incubation experiments, Table S2. Total number of 16S rRNA reads per incubation, and the relative abundance (as % of the total 16S rRNA gene reads) of major species in the incubation experiments and the Black Sea water column. Values for duplicate bottles are averaged.
Author Contributions
Conceptualization, S.v.G. and L.V.; methodology, S.v.G. and L.V.; software, L.V.; writing—original draft preparation S.v.G., J.S.S.D. and L.V.; writing—review and editing, S.v.G., J.S.S.D. and L.V.; funding acquisition, J.S.S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by the Soehngen Institute of Anaerobic Microbiology (SIAM) Gravitation grant (024.002.002) to JSSD and LV of the Netherlands Ministry of Education, Culture and Science (OCW) and the Netherlands Organisation for Scientific Research (NWO).
Acknowledgments
The authors thank the captain and crew of the R/V Pelagia, Dina Castillo Boukhchtaber and Saara Suominen for help with sampling. Marianne Baas, Marcel van der Meer, Maartje Brouwer and Sanne Vreugdenhil are thanked for help with laboratory procedures. We thank two anonymous reviewers for their helpful comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stocker, T.F.; Qin, D.; Plattner, G.-K.; Tignor, M.M.B.; Allen, S.K.; Boschung, J.; Nauels, A.; Xia, Y.; Bex, V.; Midgley, P.M. IPCC, 2013: Summary for policymakers. In Climate Change 2013: The Physical Science Basis. Contribution of Working group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge Univ. Press: Cambridge, UK, 2013. [Google Scholar]
- Conrad, R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 2009, 1, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Reeburgh, W.S. Oceanic methane biogeochemistry. Chem. Rev. 2007, 107, 486–513. [Google Scholar] [CrossRef] [PubMed]
- Boetius, A.; Ravenschlag, K.; Schubert, C.J.; Rickert, D.; Widdel, F.; Gieseke, A.; Amann, R.; Jùrgensen, B.B.; Witte, U.; Pfannkuche, O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Orphan, V.J.; Turk, K.A.; Green, A.M.; House, C.H. Patterns of 15N assimilation and growth of methanotrophic ANME-2 archaea and sulfate-reducing bacteria within structured syntrophic consortia revealed by FISH-SIMS. Environ. Microbiol. 2009, 11, 1777–1791. [Google Scholar] [CrossRef]
- Nauhaus, K.; Albrecht, M.; Elvert, M.; Boetius, A.; Widdel, F. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ. Microbiol. 2007, 9, 187–196. [Google Scholar] [CrossRef]
- Segarra, K.E.A.; Comerford, C.; Slaughter, J.; Joye, S.B. Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments. Geochim. Cosmochim. Acta 2013, 115, 15–30. [Google Scholar] [CrossRef]
- Ettwig, K.F.; Zhu, B.; Speth, D.; Keltjens, J.T.; Jetten, M.S.M.; Kartal, B. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. USA 2016, 113, 12792–12796. [Google Scholar] [CrossRef]
- Egger, M.; Rasigraf, O.; Sapart, C.J.; Jilbert, T.; Jetten, M.S.M.; Röckmann, T.; Van Der Veen, C.; Banda, N.; Kartal, B.; Ettwig, K.F.; et al. Iron-mediated anaerobic oxidation of methane in brackish coastal sediments. Environ. Sci. Technol. 2014, 49, 277–283. [Google Scholar] [CrossRef]
- Scheller, S.; Yu, H.; Chadwick, G.L.; Mcglynn, S.E. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 2016, 351, 703–707. [Google Scholar] [CrossRef]
- Valenzuela, E.I.; Avendaño, K.A.; Balagurusamy, N.; Arriaga, S.; Nieto-Delgado, C.; Thalasso, F.; Cervantes, F.J. Electron shuttling mediated by humic substances fuels anaerobic methane oxidation and carbon burial in wetland sediments. Sci. Total Environ. 2019, 650, 2674–2684. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, X.; Wu, J.; Lu, Y.; Fu, L.; Zhang, F.; Lau, T.; Zeng, R.J. Humic substances as electron acceptors for anaerobic oxidation of methane driven by ANME-2d. Water Res. 2019, 164, 114935. [Google Scholar] [CrossRef] [PubMed]
- Hinrichs, K.U.; Hayes, J.M.; Sylva, S.P.; Brewert, P.G.; DeLong, E.F. Methane-consuming archaebacteria in marine sediments. Nature 1999, 398, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Hallam, S.J.; Putnam, N.; Preston, C.M.; Detter, J.C.; Rokhsar, D.; Richardson, P.M.; DeLong, E.F. Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science 2004, 305, 1457–1462. [Google Scholar] [CrossRef] [PubMed]
- Orphan, V.J.; House, C.H.; Hinrichs, K. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 2001, 293, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Niemann, H.; Lösekann, T.; De Beer, D.; Elvert, M.; Nadalig, T.; Knittel, K.; Amann, R.; Sauter, E.J.; Schlüter, M.; Klages, M.; et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 2006, 443, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Knittel, K.; Losekann, T.; Boetius, A.; Kort, R.; Amann, R. Diversity and distribution of methantrophic acrhaea at cold seeps. Appl. Environ. Microbiol. 2005, 71, 467–479. [Google Scholar] [CrossRef]
- Cui, M.; Ma, A.; Qi, H.; Zhuang, X.; Zhuang, G. Anaerobic oxidation of methane: An “active” microbial process. Microbiologyopen 2015, 4, 1–11. [Google Scholar] [CrossRef]
- Milucka, J.; Ferdelman, T.G.; Polerecky, L.; Franzke, D.; Wegener, G.; Schmid, M.; Lieberwirth, I.; Wagner, M.; Widdel, F.; Kuypers, M.M.M. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 2012, 491, 541–546. [Google Scholar] [CrossRef]
- Moran, J.J.; Beal, E.J.; Vrentas, J.M.; Orphan, V.J.; Freeman, K.H.; House, C.H. Methyl sulfides as intermediates in the anaerobic oxidation of methane. Environ. Microbiol. 2008, 10, 162–173. [Google Scholar] [CrossRef]
- Valentine, D.L.; Reeburgh, W.S. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2000, 2, 477–484. [Google Scholar] [CrossRef]
- Wegener, G.; Krukenberg, V.; Riedel, D.; Tegetmeyer, H.E.; Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 2015, 526, 587–590. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, S.E.; Chadwick, G.L.; Kempes, C.P.; Orphan, V.J. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 2015, 526, 531–535. [Google Scholar] [CrossRef]
- Krukenberg, V.; Riedel, D.; Gruber-Vodicka, H.R.; Buttigieg, P.L.; Tegetmeyer, H.E.; Boetius, A.; Wegener, G. Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia. Environ. Microbiol. 2018, 20, 1651–1666. [Google Scholar] [CrossRef] [PubMed]
- Meyerdierks, A.; Kube, M.; Kostadinov, I.; Teeling, H.; Glöckner, F.O.; Reinhardt, R.; Amann, R. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 2010, 12, 422–439. [Google Scholar] [CrossRef] [PubMed]
- Reeburgh, W.S.; Ward, B.B.; Whalen, S.C.; Sandbeck, K.A.; Kilpatrickt, K.A.; Kerkhof, L.J. Black Sea methane geochemistry. Deep Sea Res. Part A Oceanogr. Res. Pap. 1991, 38, S1189–S1210. [Google Scholar] [CrossRef]
- Schubert, C.J.; Coolen, M.J.L.; Neretin, L.N.; Schippers, A.; Abbas, B.; Durisch-Kaiser, E.; Wehrli, B.; Hopmans, E.C.; Damsté, J.S.S.; Wakeham, S.; et al. Aerobic and anaerobic methanotrophs in the Black Sea water column. Environ. Microbiol. 2006, 8, 1844–1856. [Google Scholar] [CrossRef]
- Durisch-Kaiser, E.; Klauser, L.; Wehrli, B.; Schubert, C. Evidence of Intense Archaeal and Bacterial Methanotrophic Activity in the Black Sea Water Column Evidence of Intense Archaeal and Bacterial Methanotrophic Activity in the Black Sea Water Column. Appl. Environ. Microbiol. 2005, 71, 8099–8106. [Google Scholar] [CrossRef]
- Wakeham, S.G.; Lewis, C.M.; Hopmans, E.C.; Schouten, S.; Damsté, J.S.S. Archaea mediate anaerobic oxidation of methane in deep euxinic waters of the Black Sea. Geochim. Cosmochim. Acta 2003, 67, 1359–1374. [Google Scholar] [CrossRef]
- Sollai, M.; Villanueva, L.; Hopmans, E.C.; Reichart, G.J.; Damsté, J.S.S. A combined lipidomic and 16S rRNA gene amplicon sequencing approach reveals archaeal sources of intact polar lipids in the stratified Black Sea water column. Geobiology 2019, 17, 91–109. [Google Scholar] [CrossRef]
- Schubert, C.J.; Durisch-Kaiser, E.; Holzner, C.P.; Klauser, L.; Wehrli, B.; Schmale, O.; Greinert, J.; McGinnis, D.F.; De Batist, M.; Kipfer, R. Methanotrophic microbial communities associated with bubble plumes above gas seeps in the Black Sea. Geochem. Geophys. Geosyst. 2006, 7. [Google Scholar] [CrossRef]
- Yanagawa, K.; Sunamura, M.; Lever, M.A.; Morono, Y.; Hiruta, A.; Ishizaki, O.; Matsumoto, R.; Urabe, T.; Inagaki, F. Niche separation of methanotrophic archaea (ANME-1 and -2) in methane-seep sediments of the Eastern Japan Sea offshore Joetsu. Geomicrobiol. J. 2011, 28, 118–129. [Google Scholar] [CrossRef]
- Suominen, S.; Dombrowski, N.; Damsté, J.S.S.; Villanueva, L. A diverse uncultivated microbial community is responsible for organic matter degradation in the Black Sea sulphidic zone. Environ. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Besseling, M.A.; Hopmans, E.C.; Christine Boschman, R.; Damsté, J.S.S.; Villanueva, L. Benthic archaea as potential sources of tetraether membrane lipids in sediments across an oxygen minimum zone. Biogeosciences 2018, 15, 4047–4064. [Google Scholar] [CrossRef]
- Asbun, A.A.; Besseling, M.A.; Balzano, S.; van Bleijswijk, J.; Witte, H.; Villanueva, L.; Engelmann, J.C. Cascabel: A flexible, scalable and easy-to-use amplicon sequence data analysis pipeline. BioRxiv 2019, 809384. [Google Scholar]
- Andrews, S.; Krueger, F.; Seconds-Pichon, A.; Biggins, F.; Wingett, S. FastQC. A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 July 2020).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Haroon, M.F.; Hu, S.; Shi, Y.; Imelfort, M.; Keller, J.; Hugenholtz, P.; Yuan, Z.; Tyson, G.W. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 2013, 500, 567–570. [Google Scholar] [CrossRef]
- Beal, E.J.; House, C.H.; Orphan, V.J. Manganese- and iron-dependent marine methane oxidation. Science 2009, 325, 184–187. [Google Scholar] [CrossRef]
- Sivan, O.; Adler, M.; Pearson, A.; Gelman, F.; Bar-Or, I.; John, S.G.; Eckert, W. Geochemical evidence for iron-mediated anaerobic oxidation of methane. Limnol. Oceanogr. 2011, 56, 1536–1544. [Google Scholar] [CrossRef]
- Reitner, J.; Peckmann, J.; Reimer, A.; Schumann, G.; Thiel, V. Methane-derived carbonate build-ups and associated microbial communities at cold seeps on the lower Crimean shelf (Black Sea). Facies 2005, 51, 66–79. [Google Scholar] [CrossRef]
- Gründger, F.; Carrier, V.; Svenning, M.M.; Panieri, G.; Vonnahme, T.R.; Klasek, S.; Niemann, H. Methane-fuelled biofilms predominantly composed of methanotrophic ANME-1 in Arctic gas hydrate-related sediments. Sci. Rep. 2019, 9, 9725. [Google Scholar] [CrossRef] [PubMed]
- Cassarini, C.; Zhang, Y.; Lens, P.N.L. Pressure Selects Dominant Anaerobic Methanotrophic Phylotype and Sulfate Reducing Bacteria in Coastal Marine Lake Grevelingen Sediment. Front. Environ. Sci. 2019, 6, 162. [Google Scholar] [CrossRef]
- Wilson, L.G.; Bandurski, R.S. Enzymatic reactions involving sulfate, sulfite, selenate, and molybdate. J. Biol. Chem. 1958, 233, 975–981. [Google Scholar] [PubMed]
- Campbell, A.M.; Del Campillo-Campbell, A.; Villaret, D.B. Molybdate reduction by Escherichia coli K-12 and its chl mutants. Proc. Natl. Acad. Sci. USA 1985. [Google Scholar] [CrossRef]
- Shukor, M.Y.; Rahman, M.F.; Shamaan, N.A.; Syed, M.S. Reduction of molybdate to molybdenum blue by Enterobacter sp. strain Dr.Y13. J. Basic Microbiol. 2009. [Google Scholar] [CrossRef]
- Shukor, M.Y.; Ahmad, S.A.; Nadzir, M.M.M.; Abdullah, M.P.; Shamaan, N.A.; Syed, M.A. Molybdate reduction by Pseudomonas sp. strain DRY2. J. Appl. Microbiol. 2010. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Alperin, M.J.; Teske, A. Environmental evidence for net methane production and oxidation in putative ANaerobic MEthanotrophic (ANME) archaea. Environ. Microbiol. 2011, 13, 2548–2564. [Google Scholar] [CrossRef]
- Bertram, S.; Blumenberg, M.; Michaelis, W.; Siegert, M.; Krüger, M.; Seifert, R. Methanogenic capabilities of ANME-archaea deduced from 13C-labelling approaches. Environ. Microbiol. 2013, 15, 2384–2393. [Google Scholar] [CrossRef]
- Albers, C.N.; Jensen, A.; Bælum, J.; Jacobsen, C.S. Inhibition of DNA Polymerases Used in Q-PCR by Structurally Different Soil-Derived Humic Substances. Geomicrobiol. J. 2013, 30, 675–681. [Google Scholar] [CrossRef]
- Sidstedt, M.; Jansson, L.; Nilsson, E.; Noppa, L.; Forsman, M.; Rådström, P.; Hedman, J. Humic substances cause fluorescence inhibition in real-time polymerase chain reaction. Anal. Biochem. 2015, 487, 30–37. [Google Scholar] [CrossRef]
- Vigneron, A.; Alsop, E.B.; Cruaud, P.; Philibert, G.; King, B.; Baksmaty, L. Contrasting Pathways for Anaerobic Methane Oxidation in Gulf of Mexico Cold Seep Sediments. Appl. Environ. Sci. 2019, 4, e00091-18. [Google Scholar] [CrossRef] [PubMed]
- Straub, K.L.; Schink, B. Ferrihydrite-Dependent Growth of Sulfurospirillum deleyianum through Electron Transfer via Sulfur Cycling. Society 2004, 70, 5744–5749. [Google Scholar] [CrossRef] [PubMed]
- Goris, T.; Diekert, G. The genus Sulfurospirillum. In Organohalide-Respiring Bacteria; Springer: Berlin/Heidelberg, Germany, 2016; pp. 209–234. ISBN 9783662498750. [Google Scholar]
- Mino, S.; Kudo, H.; Arai, T.; Sawabe, T.; Takai, K.; Nakagawa, S. Sulfurovum aggregans sp. nov., A hydrogenoxidizing, Thiosulfate-reducing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent chimney, And an emended description of the genus Sulfurovum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3195–3201. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Takai, K.; Inagaki, F.; Hirayama, H.; Nunoura, T.; Horikoshi, K.; Sako, Y. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 2005, 7, 1619–1632. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Perner, M. The globally widespread genus Sulfurimonas: Versatile energy metabolisms and adaptations to redox clines. Front. Microbiol. 2015, 6, 1–17. [Google Scholar] [CrossRef]
- Goris, T.; Schubert, T.; Gadkari, J.; Wubet, T.; Tarkka, M.; Buscot, F.; Adrian, L.; Diekert, G. Insights into organohalide respiration and the versatile catabolism of Sulfurospirillum multivorans gained from comparative genomics and physiological studies. Environ. Microbiol. 2014, 16, 3562–3580. [Google Scholar] [CrossRef]
- Ross, D.E.; Norman, R.; Marshall, C.W.; May, H.D. Comparative genomic analysis of sulfurospirillum cavolei MES reconstructed from the metagenome of an electrosynthetic microbiome. PLoS ONE 2016, 11, e0151214. [Google Scholar] [CrossRef]
- Takai, K.; Suzuki, M.; Nakagawa, S.; Miyazaki, M.; Suzuki, Y.; Inagaki, F.; Horikoshi, K. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteo-bacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as S. Int. J. Syst. Evol. Microbiol. 2006, 56, 1725–1733. [Google Scholar] [CrossRef]
- Fadhlaoui, K.; Hania, W.B.; Postec, A.; Fauque, G.; Hamdi, M.; Ollivier, B.; Fardeau, M.L. Fusibacter fontis sp. Nov., a sulfur-reducing, anaerobic bacterium isolated from a mesothermic Tunisian spring. Int. J. Syst. Evol. Microbiol. 2015, 65, 3501–3506. [Google Scholar] [CrossRef]
- Ravot, G.; Magot, M.; Fardeau, M.L.; Patel, B.K.C.; Thomas, P.; Garcia, J.L.; Ollivier, B. Fusibacter paucivorans gen. nov., sp. nov., an anaerobic, thiosulfate- reducing bacterium from an oil-producing well. Int. J. Syst. Bacteriol. 1999, 49, 1141–1147. [Google Scholar] [CrossRef]
- Suzuki, D.; Li, Z.; Cui, X.; Zhang, C.; Katayama, A. Reclassification of Desulfobacterium anilini as Desulfatiglans anilini comb. nov. within Desulfatiglans gen. nov., And description of a 4-chlorophenol-degrading sulfate-reducing bacterium, Desulfatiglans parachlorophenolica sp. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 3081–3086. [Google Scholar] [CrossRef] [PubMed]
- Kevorkian, R.; Callahan, S.; Winstead, R.; Lloyd, K.G. ANME-1 archaea drive methane accumulation and removal in estuarine sediments. BioRxiv Prepr. 2020. [Google Scholar] [CrossRef]
- Evans, P.N.; Parks, D.H.; Chadwick, G.L.; Robbins, S.J.; Orphan, V.J.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Webster, G.; Rinna, J.; Roussel, E.G.; Fry, J.C.; Weightman, A.J.; Parkes, R.J. Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol. Ecol. 2010, 72, 179–197. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.G.; Schreiber, L.; Petersen, D.G.; Kjeldsen, K.U.; Lever, M.A.; Steen, A.D.; Stepanauskas, R.; Richter, M.; Kleindienst, S.; Lenk, S.; et al. Predominant archaea in marine sediments degrade detrital proteins. Nature 2013, 496, 215–218. [Google Scholar] [CrossRef]
- Seyler, L.M.; McGuinness, L.M.; Kerkhof, L.J. Crenarchaeal heterotrophy in salt marsh sediments. ISME J. 2014, 8, 1534–1543. [Google Scholar] [CrossRef]
- Zhou, Z.; Pan, J.; Wang, F.; Gu, J.D.; Li, M. Bathyarchaeota: Globally distributed metabolic generalists in anoxic environments. FEMS Microbiol. Rev. 2018, 42, 639–655. [Google Scholar] [CrossRef]
- Zhang, W.; Ding, W.; Yang, B.; Tian, R.; Gu, S.; Luo, H.; Qian, P.Y. Genomic and transcriptomic evidence for carbohydrate consumption among microorganisms in a cold seep brine pool. Front. Microbiol. 2016, 7, 1825. [Google Scholar] [CrossRef]
- Niu, M.; Fan, X.; Zhuang, G.; Liang, Q.; Wang, F. Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea. FEMS Microbiol. Ecol. 2017, 93, 1–13. [Google Scholar] [CrossRef]
- Valenzuela, E.I.; Prieto-davó, A.; López-lozano, N.E.; García-gonzález, A.S.; López, M.G.; Cervantes, J. Anaerobic Methane Oxidation Driven by Microbial Reduction of Natural Organic Matter in a Tropical Wetland. Appl. Environ. Microbiol. 2017, 83, 1–15. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).