Modulation of the Gut Microbiota Alters the Tumour-Suppressive Efficacy of Tim-3 Pathway Blockade in a Bacterial Species- and Host Factor-Dependent Manner
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Construction of Expression Vectors for Tim-3-Blocking Molecules
2.3. Western Blotting for Detection of the Tim3VdIg Protein
2.4. Production and Purification of Tim3VdIg Protein
2.5. Evaluation of Tumour Growth
2.6. Oral Administration of Bacteria to Mice
2.7. Collection of Gut Microbiota Samples and Bacterial DNA Sequencing
2.8. Sequence Processing and Taxonomic Assignment
2.9. Statistics
3. Results
3.1. Tim-3 V Domain—Mouse IgG Fc Fusion Protein Dimer Exerts Tumour-Suppressive Effects
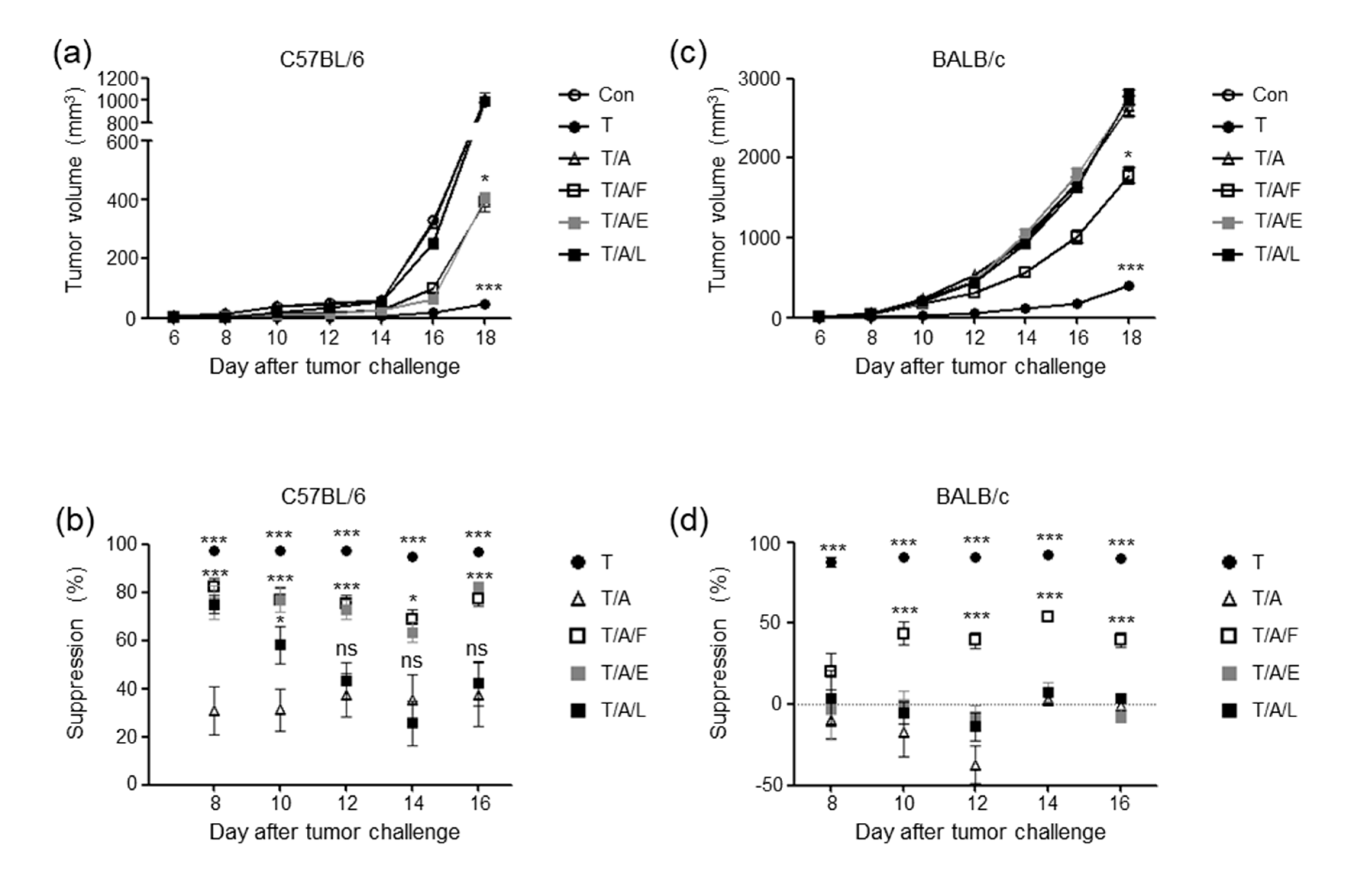
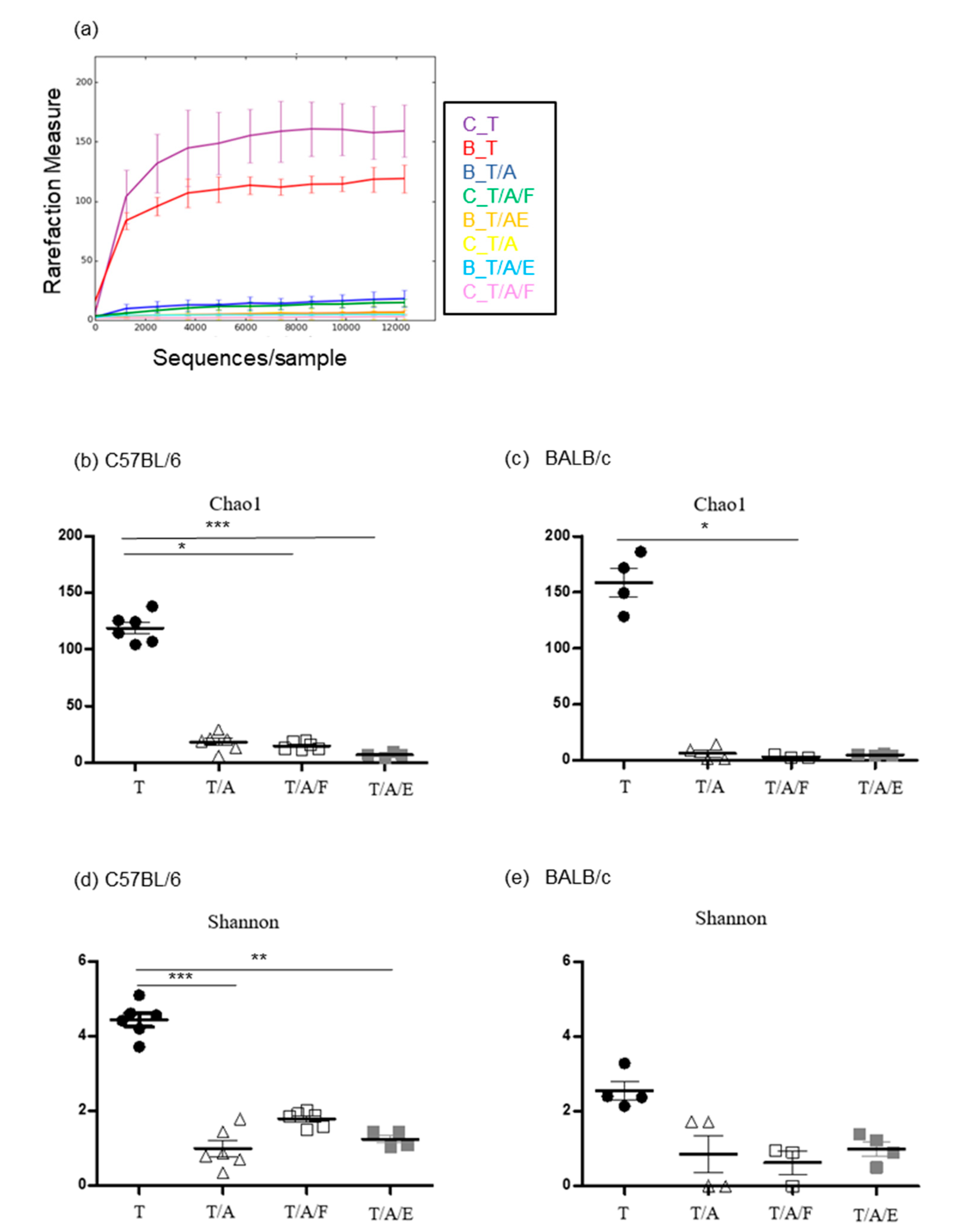
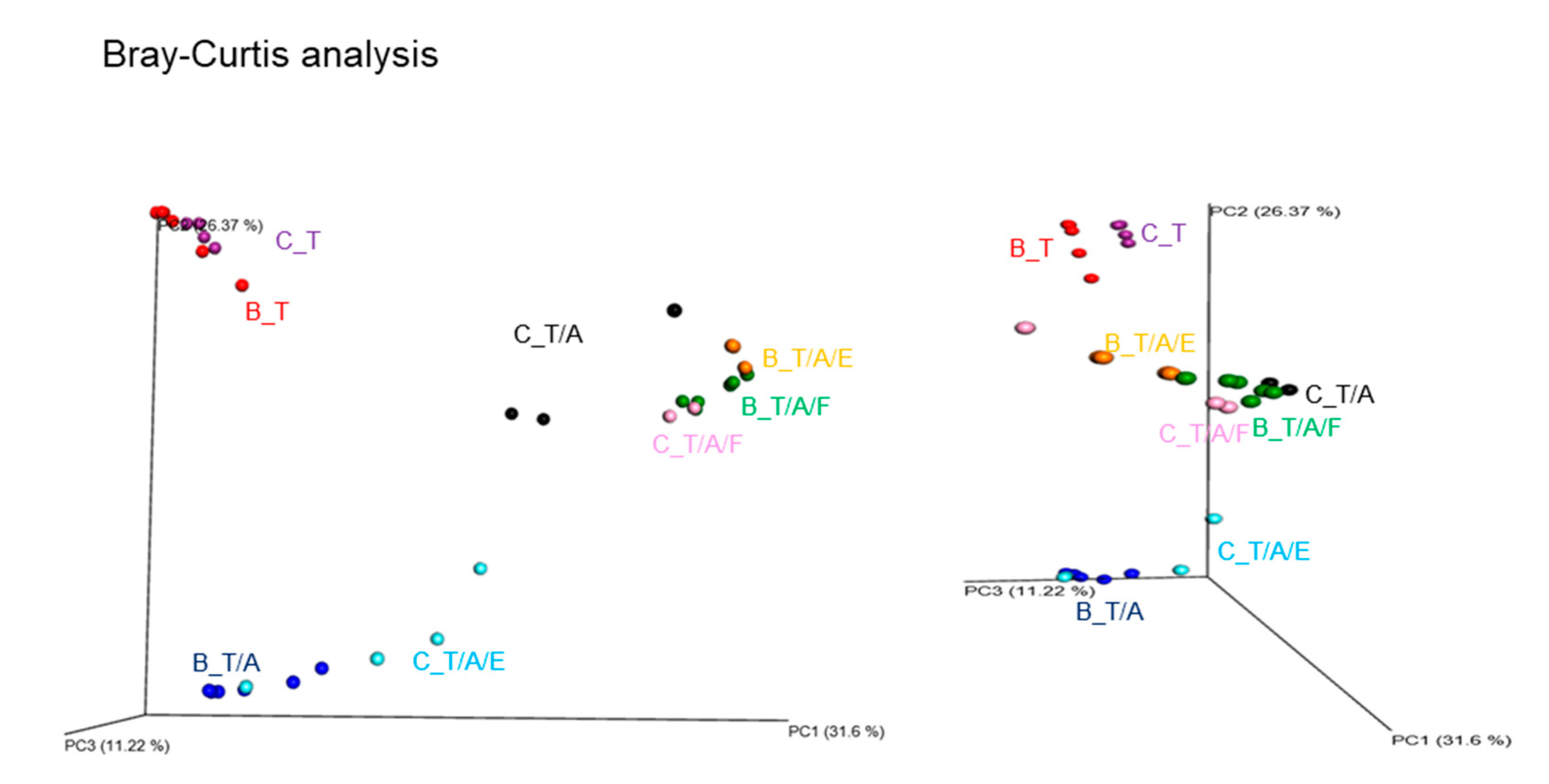
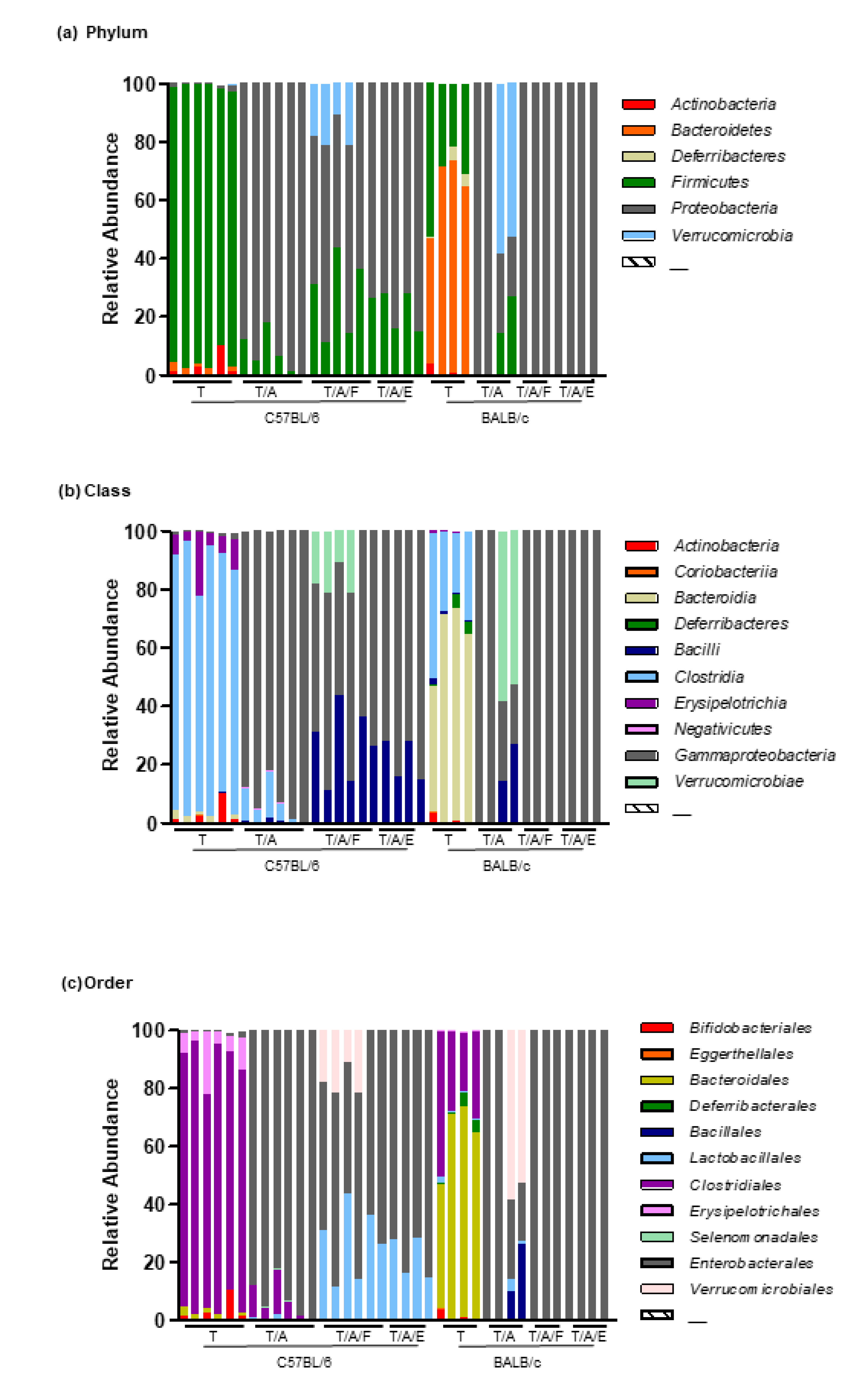
3.2. Oral Administration of Antibiotics to Mice Attenuates the Tumour-Suppressive Effect of Tim-3 Blockade
3.3. Oral Administration of Faecal Bacteria or Enterococcus hirae Restores the Tumour-Suppressive Effect of Tim-3 Blockade in Mice Treated with Antibiotics
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Munger, M.E.; Veenstra, R.G.; Weigel, B.J.; Hirashima, M.; Munn, D.H.; Murphy, W.J.; Azuma, M.; Anderson, A.C.; Kuchroo, V.K.; et al. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood 2011, 117, 4501–4510. [Google Scholar] [CrossRef] [PubMed]
- Gleason, M.K.; Lenvik, T.R.; McCullar, V.; Felices, M.; O’Brien, M.S.; Cooley, S.A.; Verneris, M.R.; Cichocki, F.; Holman, C.J.; Panoskaltsis-Mortari, A.; et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 2012, 119, 3064–3072. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Akiba, H.; Takeda, K.; Kojima, Y.; Hashiguchi, M.; Azuma, M.; Yagita, H.; Okumura, K. Tim-3 mediates phagocytosis of apoptotic cells and cross-presentation. Blood 2009, 113, 3821–3830. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Yin, Y.; Ju, H.; Xu, X.; Liu, W.; Fu, Q.; Hu, J.; Zhang, X.; Sun, B. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 2018, 9, 478. [Google Scholar] [CrossRef]
- Avery, L.; Filderman, J.; Szymczak-Workman, A.L.; Kane, L.P. Tim-3 co-stimulation promotes short-lived effector T cells; restricts memory precursors; and is dispensable for T cell exhaustion. Proc. Natl. Acad. Sci. USA 2018, 115, 2455–2460. [Google Scholar] [CrossRef] [Green Version]
- Gallois, A.; Silva, I.; Osman, I.; Bhardwaj, N. Reversal of natural killer cell exhaustion by TIM-3 blockade. Oncoimmunology 2014, 3, e946365. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef]
- Huang, X.; Bai, X.; Cao, Y.; Wu, J.; Huang, M.; Tang, D.; Tao, S.; Zhu, T.; Liu, Y.; Yang, Y.; et al. Lymphoma endothelium preferentially expresses Tim-3 and facilitates the progression of lymphoma by mediating immune evasion. J. Exp. Med. 2010, 207, 505–520. [Google Scholar] [CrossRef]
- Li, H.; Wu, K.; Tao, K.; Chen, L.; Zheng, Q.; Lu, X.; Liu, J.; Shi, L.; Liu, C.; Wang, G.; et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology 2012, 56, 1342–1351. [Google Scholar] [CrossRef]
- Shindo, Y.; Hazama, S.; Suzuki, N.; Iguchi, H.; Uesugi, K.; Tanaka, H.; Aruga, A.; Hatori, T.; Ishizaki, H.; Umeda, Y.; et al. Predictive biomarkers for the efficacy of peptide vaccine treatment: Based on the results of a phase II study on advanced pancreatic cancer. J. Exp. Clin. Cancer Res. 2017, 36, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikushige, Y.; Shima, T.; Takayanagi, S.; Urata, S.; Miyamoto, T.; Iwasaki, H.; Takenaka, K.; Teshima, T.; Tanaka, T.; Inagaki, Y.; et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell 2010, 7, 708–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngiow, S.F.; von Scheidt, B.; Akiba, H.; Yagita, H.; Teng, M.W.; Smyth, M.J. Anti-TIM3 antibody promotes T cell IFN-γ-mediated antitumor immunity and suppresses established tumors. Cancer Res. 2011, 71, 3540–3551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuishi, K.; Apetoh, L.; Sullivan, J.M.; Blazar, B.R.; Kuchroo, V.K.; Anderson, A.C. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 2010, 207, 2187–2194. [Google Scholar] [CrossRef]
- Koyama, S.; Akbay, E.A.; Li, Y.Y.; Herter-Sprie, G.S.; Buczkowski, K.A.; Richards, W.G.; Gandhi, L.; Redig, A.J.; Rodig, S.J.; Asahina, H.; et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 2016, 7, 10501. [Google Scholar] [CrossRef]
- Pfannenstiel, L.W.; Diaz-Montero, C.M.; Tian, Y.F.; Scharpf, J.; Ko, J.S.; Gastman, B.R. Immune-Checkpoint Blockade Opposes CD8(+) T-cell Suppression in Human and Murine Cancer. Cancer Immunol. Res. 2019, 7, 510–525. [Google Scholar] [CrossRef] [Green Version]
- de Mingo Pulido, A.; Gardner, A.; Hiebler, S.; Soliman, H.; Rugo, H.S.; Krummel, M.F.; Coussens, L.M.; Ruffell, B. TIM-3 Regulates CD103(+) Dendritic Cell Function and Response to Chemotherapy in Breast Cancer. Cancer Cell 2018, 33, 60–74.e6. [Google Scholar] [CrossRef] [Green Version]
- da Silva, I.P.; Gallois, A.; Jimenez-Baranda, S.; Khan, S.; Anderson, A.C.; Kuchroo, V.K.; Osman, I.; Bhardwaj, N. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol. Res. 2014, 2, 410–422. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.J.; Woo, M.Y.; Heo, Y.M.; Kim, J.S.; Kwon, M.H.; Kim, K.; Park, S. The inhibition of the T-cell immunoglobulin and mucin domain 3 (Tim3) pathway enhances the efficacy of tumor vaccine. Biochem. Biophys. Res. Commun. 2010, 402, 88–93. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome; cancer; and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Hong, J.S.; Waud, W.R.; Levasseur, D.N.; Townes, T.M.; Wen, H.; McPherson, S.A.; Moore, B.A.; Bebok, Z.; Allan, P.W.; Secrist, J.A.; et al. Excellent in vivo bystander activity of fludarabine phosphate against human glioma xenografts that express the escherichia coli purine nucleoside phosphorylase gene. Cancer Res. 2004, 64, 6610–6615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viaud, S.; Daillere, R.; Boneca, I.G.; Lepage, P.; Langella, P.; Chamaillard, M.; Pittet, M.J.; Ghiringhelli, F.; Trinchieri, G.; Goldszmid, R.; et al. Gut microbiome and anticancer immune response: Really hot Sh*t! Cell Death Differ. 2015, 22, 199–214. [Google Scholar] [CrossRef] [Green Version]
- Daillere, R.; Vetizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [Green Version]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, V.; Brunetti, O.; Azzariti, A.; Galetta, D.; Nardulli, P.; Leonetti, F.; Silvestris, N. Strategies to Improve Cancer Immune Checkpoint Inhibitors Efficacy; Other Than Abscopal Effect: A Systematic Review. Cancers 2019, 11, 539. [Google Scholar] [CrossRef] [Green Version]
- Brattain, M.G.; Strobel-Stevens, J.; Fine, D.; Webb, M.; Sarrif, A.M. Establishment of mouse colonic carcinoma cell lines with different metastatic properties. Cancer Res. 1980, 40, 2142–2146. [Google Scholar]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Ianiro, G.; Tilg, H.; Gasbarrini, A. Antibiotics as deep modulators of gut microbiota: Between good and evil. Gut 2016, 65, 1906–1915. [Google Scholar] [CrossRef]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [Green Version]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manichanh, C.; Reeder, J.; Gibert, P.; Varela, E.; Llopis, M.; Antolin, M.; Guigo, R.; Knight, R.; Guarner, F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010, 20, 1411–1419. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.L.; Maier, I.; Dang, A.T.; Berry, D.; Liu, J.; Ruegger, P.M.; Yang, J.I.; Soto, P.A.; Presley, L.L.; Reliene, R.; et al. Intestinal bacteria modify lymphoma incidence and latency by affecting systemic inflammatory state; oxidative stress; and leukocyte genotoxicity. Cancer Res. 2013, 73, 4222–4232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcial, G.E.; Ford, A.L.; Haller, M.J.; Gezan, S.A.; Harrison, N.A.; Cai, D.; Meyer, J.L.; Perry, D.J.; Atkinson, M.A.; Wasserfall, C.H.; et al. Lactobacillus johnsonii N6.2 Modulates the Host Immune Responses, A Double-Blind; Randomized Trial in Healthy Adults. Front. Immunol. 2017, 8, 655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haller, D.; Serrant, P.; Granato, D.; Schiffrin, E.J.; Blum, S. Activation of human NK cells by staphylococci and lactobacilli requires cell contact-dependent costimulation by autologous monocytes. Clin. Diagn. Lab. Immunol. 2002, 9, 649–657. [Google Scholar] [CrossRef] [Green Version]
- Takada, H.; Kawabata, Y.; Arakaki, R.; Kusumoto, S.; Fukase, K.; Suda, Y.; Yoshimura, T.; Kokeguchi, S.; Kato, K.; Komuro, T.; et al. Molecular and structural requirements of a lipoteichoic acid from Enterococcus hirae ATCC 9790 for cytokine-inducing; antitumor; and antigenic activities. Infect. Immun. 1995, 63, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Rong, Y.; Dong, Z.; Hong, Z.; Jin, Y.; Zhang, W.; Zhang, B.; Mao, W.; Kong, H.; Wang, C.; Yang, B.; et al. Reactivity toward Bifidobacterium longum and Enterococcus hirae demonstrate robust CD8(+) T cell response and better prognosis in HBV-related hepatocellular carcinoma. Exp. Cell Res. 2017, 358, 352–359. [Google Scholar] [CrossRef]
- Mariman, R.; Tielen, F.; Koning, F.; Nagelkerken, L. The Probiotic Mixture VSL#3 Has Differential Effects on Intestinal Immune Parameters in Healthy Female BALB/c and C57BL/6 Mice. J. Nutr. 2015, 145, 1354–1361. [Google Scholar]
- Yang, F.; Wang, D.; Li, Y.; Sang, L.; Zhu, J.; Wang, J.; Wei, B.; Lu, C.; Sun, X. Th1/Th2 Balance and Th17/Treg-Mediated Immunity in relation to Murine Resistance to Dextran Sulfate-Induced Colitis. J. Immunol. Res. 2017, 2017, 7047201. [Google Scholar] [CrossRef]
- Yasuda, Y.; Shimoda, T.; Uno, K.; Tateishi, N.; Furuya, S.; Yagi, K.; Suzuki, K.; Fujita, S. The effects of MPTP on the activation of microglia/astrocytes and cytokine/chemokine levels in different mice strains. J. Neuroimmunol. 2008, 204, 43–51. [Google Scholar] [CrossRef]

| p Value 2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Species | T vs T/A | T vs T/A/F | T vs T/A/E | T/A vs T/A/F | T/A vs T/A/E | T/A/F vs T/A/E | |
| Actinobacteria | Asaccharobacter celatus | 0.0011 | 0.0052 | 0.0052 | 0.0116 | 1.0000 | 1.0000 | 1.0000 |
| Actinobacteria | Bifidobacterium pseudolongum | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Bacteroidetes | Bacteroides xylanolyticus | 0.0003 | 0.0045 | 0.0012 | 0.0045 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Acetatifactor muris | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Anaerocolumna cellulosilytica | 0.0065 | 0.0219 | 0.0219 | 0.0373 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Anaerocolumna jejuensis | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Anaerotaenia torta | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Beduini massiliensis | 0.0011 | 0.0052 | 1.0000 | 1.0000 | 0.0052 | 0.0116 | 1.0000 |
| Firmicutes | Blautia producta | 0.0007 | 0.0578 | 0.0013 | 0.0048 | 0.6363 | 0.6363 | 1.0000 |
| Firmicutes | Caecibacterium sporoformans | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Christensenella massiliensis | 0.0006 | 1.0000 | 0.0114 | 0.0218 | 0.0114 | 0.0218 | 1.0000 |
| Firmicutes | Clostridium aerotolerans | 0.0011 | 0.0052 | 0.0052 | 0.0116 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Clostridium asparagiforme | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Clostridium Cocleatum | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Clostridium indolis | 0.0013 | 0.8387 | 0.054 | 0.0814 | 0.0063 | 0.0169 | 1.0000 |
| Firmicutes | Clostridium methylpentosum | 0.0065 | 0.0219 | 0.0219 | 0.0373 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Clostridium saccharolyticum | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Desulfitobacterium metallireducens | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Eubacterium coprostanoligenes | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Falcatimonas natans | 0.0065 | 0.0219 | 0.0219 | 0.0373 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Flintibacter butyricus | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Hungateiclostridium thermocellum | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Hydrogenoanaerobacterium saccharovorans | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Kineothri alysoides | 0.0003 | 0.0012 | 0.0045 | 0.0045 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Lachnoclostridium pacaense | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Lactobacillus animalis | 0.0004 | 0.3507 | 0.0011 | 0.0081 | 0.0666 | 0.1573 | 0.8399 |
| Firmicutes | Muricomes intestini | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Murimonas intestini | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Neglecta timonensis | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Oscillibacter ruminantium | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Oscillibacter valericigenes | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Phocea massiliensis | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Propionispira paucivorans | 0.0065 | 0.0219 | 1.0000 | 1.0000 | 0.0219 | 0.0373 | 1.0000 |
| Firmicutes | Pseudobutyrivibrio ruminis | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Pseudoflavonifractor capillosus | 0.0065 | 0.0219 | 0.0219 | 0.0373 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Pseudoflavonifractor phocaeensis | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Roseburia hominis | 0.0001 | 0.0011 | 0.0011 | 0.0033 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Roseburia intestinalis | 0.0011 | 0.0052 | 0.0052 | 0.0116 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Ruminococcus gnavus | 0.0003 | 0.0045 | 0.0012 | 0.0045 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Ruthenibacterium lactatiformans | 0.0003 | 0.0045 | 0.0012 | 0.0045 | 1.0000 | 1.0000 | 1.0000 |
| Firmicutes | Turicibacter sanguinis | 0.0008 | 0.2083 | 0.0016 | 0.0056 | 0.2675 | 0.2675 | 1.0000 |
| Proteobacteria | Escherichia fergusonii | 0.0003 | 0.2236 | 0.0001 | 0.0669 | 0.0627 | 0.4252 | 0.3023 |
| Proteobacteria | Klebsiella variicola | 0.0002 | 0.1948 | 0.1367 | 0.1367 | 0.001 | 0.0017 | 0.8369 |
| Proteobacteria | Proteus alimentorum | 0.0002 | 0.3285 | 0.0069 | 0.0003 | 0.3285 | 0.0347 | 0.3285 |
| Verrucomicrobia | Akkermansia muciniphila | 0.0014 | 0.9065 | 0.0531 | 0.9065 | 0.0027 | 1.0000 | 0.0085 |
| p Value 2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Species | T vs T/A | T vs T/A/F | T vs T/A/E | T/A vs T/A/F | T/A vs T/A/E | T/A/F vs T/A/E | |
| Actinobacteria | Bifidobacterium pseudolongum | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Bacteroidetes | Bacteroides caccae | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Bacteroidetes | Bacteroides paurosaccharolyticus | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Bacteroidetes | Bacteroides vulgatus | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Bacteroidetes | Parabacteroides goldsteinii | 0.007 | 0.051 | 0.027 | 0.016 | 1.000 | 1.000 | 1.000 |
| Deferribacteres | Mucispirillum schaedleri | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Absiella dolichum | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Acetatifactor muris | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Anaerotaenia torta | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Christensenella massiliensis | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Christensenella minuta | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Clostridium aerotolerans | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Clostridium asparagiforme | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Clostridium cocleatum | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Clostridium populeti | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Clostridium saccharolyticum | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Clostridium scindens | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Desulfitobacterium metallireducens | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Eisenbergiella massiliensis | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Eubacterium coprostanoligenes | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Eubacterium siraeum | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Falcatimonas natans | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Flintibacter butyricus | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Hungateiclostridium thermocellum | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Hydrogenoanaerobacterium saccharovorans | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Intestinimonas butyriciproducens | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Kineothri alysoides | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Lachnoclostridium pacaense | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Lactobacillus animalis | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Lactobacillus rogosae | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Muricomes intestini | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Neglecta timonensis | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Oscillibacter ruminantium | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Oscillibacter valericigenes | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Peptococcus simiae | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Pseudoflavonifractor phocaeensis | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Roseburia faecis | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Ruminococcus gnavus | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Firmicutes | Vallitalea pronyensis | 0.004 | 0.014 | 0.019 | 0.014 | 1.000 | 1.000 | 1.000 |
| Proteobacteria | Hafnia alvei | 0.004 | 1.000 | 1.000 | 0.014 | 1.000 | 0.014 | 0.019 |
| Proteobacteria | Klebsiella variicola | 0.004 | 1.000 | 1.000 | 0.014 | 1.000 | 0.014 | 0.019 |
| Phylum | Class | Species | p Value 2 | More in |
|---|---|---|---|---|
| Bacteroidetes | Bacteroidia | Bacteroides xylanolyticus | 0.01 | C57BL/6 |
| Firmicutes | Clostridia | [Clostridium] indolis | 0.01 | |
| Firmicutes | Clostridia | Blautia producta | 0.011 | |
| Firmicutes | Clostridia | Caecibacterium sporoformans | 0.014 | |
| Firmicutes | Clostridia | Hungateiclostridium thermocellum | 0.01 | |
| Firmicutes | Clostridia | Pseudoflavonifractor phocaeensis | 0.01 | |
| Firmicutes | Erysipelotrichia | [Clostridium] cocleatum | 0.014 | |
| Firmicutes | Erysipelotrichia | Turicibacter sanguinis | 0.011 | |
| Proteobacteria | Gammaproteobacteria | Klebsiella variicola | 0.011 | |
| Proteobacteria | Gammaproteobacteria | Proteus alimentorum | 0.013 | |
| Bacteroidetes | Bacteroidia | Bacteroides caccae | 0.006 | BALB/c |
| Bacteroidetes | Bacteroidia | Bacteroides paurosaccharolyticus | 0.006 | |
| Bacteroidetes | Bacteroidia | Bacteroides vulgatus | 0.006 | |
| Bacteroidetes | Bacteroidia | Parabacteroides goldsteinii | 0.006 | |
| Deferribacteres | Deferribacteres | Mucispirillum schaedleri | 0.006 | |
| Firmicutes | Bacilli | Lactobacillus animalis | 0.006 | |
| Firmicutes | Bacilli | Lactobacillus rogosae | 0.006 | |
| Firmicutes | Clostridia | [Clostridium] viride | 0.006 | |
| Firmicutes | Clostridia | Anaerovorax odorimutans | 0.006 | |
| Firmicutes | Clostridia | Blautia faecis | 0.005 | |
| Firmicutes | Clostridia | Christensenella minuta | 0.006 | |
| Firmicutes | Clostridia | Flavonifractor plautii | 0.006 | |
| Firmicutes | Clostridia | Intestinimonas butyriciproducens | 0.009 | |
| Firmicutes | Clostridia | Peptococcus simiae | 0.006 | |
| Firmicutes | Clostridia | Vallitalea pronyensis | 0.006 | |
| Firmicutes | Erysipelotrichia | Absiella dolichum | 0.006 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, B.; Lee, J.; Woo, M.-Y.; Lee, M.J.; Shin, H.-J.; Kim, K.; Park, S. Modulation of the Gut Microbiota Alters the Tumour-Suppressive Efficacy of Tim-3 Pathway Blockade in a Bacterial Species- and Host Factor-Dependent Manner. Microorganisms 2020, 8, 1395. https://doi.org/10.3390/microorganisms8091395
Lee B, Lee J, Woo M-Y, Lee MJ, Shin H-J, Kim K, Park S. Modulation of the Gut Microbiota Alters the Tumour-Suppressive Efficacy of Tim-3 Pathway Blockade in a Bacterial Species- and Host Factor-Dependent Manner. Microorganisms. 2020; 8(9):1395. https://doi.org/10.3390/microorganisms8091395
Chicago/Turabian StyleLee, Bokyoung, Jieun Lee, Min-Yeong Woo, Mi Jin Lee, Ho-Joon Shin, Kyongmin Kim, and Sun Park. 2020. "Modulation of the Gut Microbiota Alters the Tumour-Suppressive Efficacy of Tim-3 Pathway Blockade in a Bacterial Species- and Host Factor-Dependent Manner" Microorganisms 8, no. 9: 1395. https://doi.org/10.3390/microorganisms8091395






