Assessment of Multilocus Sequence Analysis (MLSA) for Identification of Candidatus Liberibacter Solanacearum from Different Host Plants in Spain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Analysed Throughout the Work
2.2. DNA Extraction and Q-PCR Analysis for Detection of CLso
2.3. Haplotype Identification and Characterization
2.4. Phylogenetic Analysis and Molecular Dating of Candidatus Liberibacter Isolates
2.5. Descriptive Analysis of the Sequences
3. Results
3.1. Detection of CLso from Different Samples
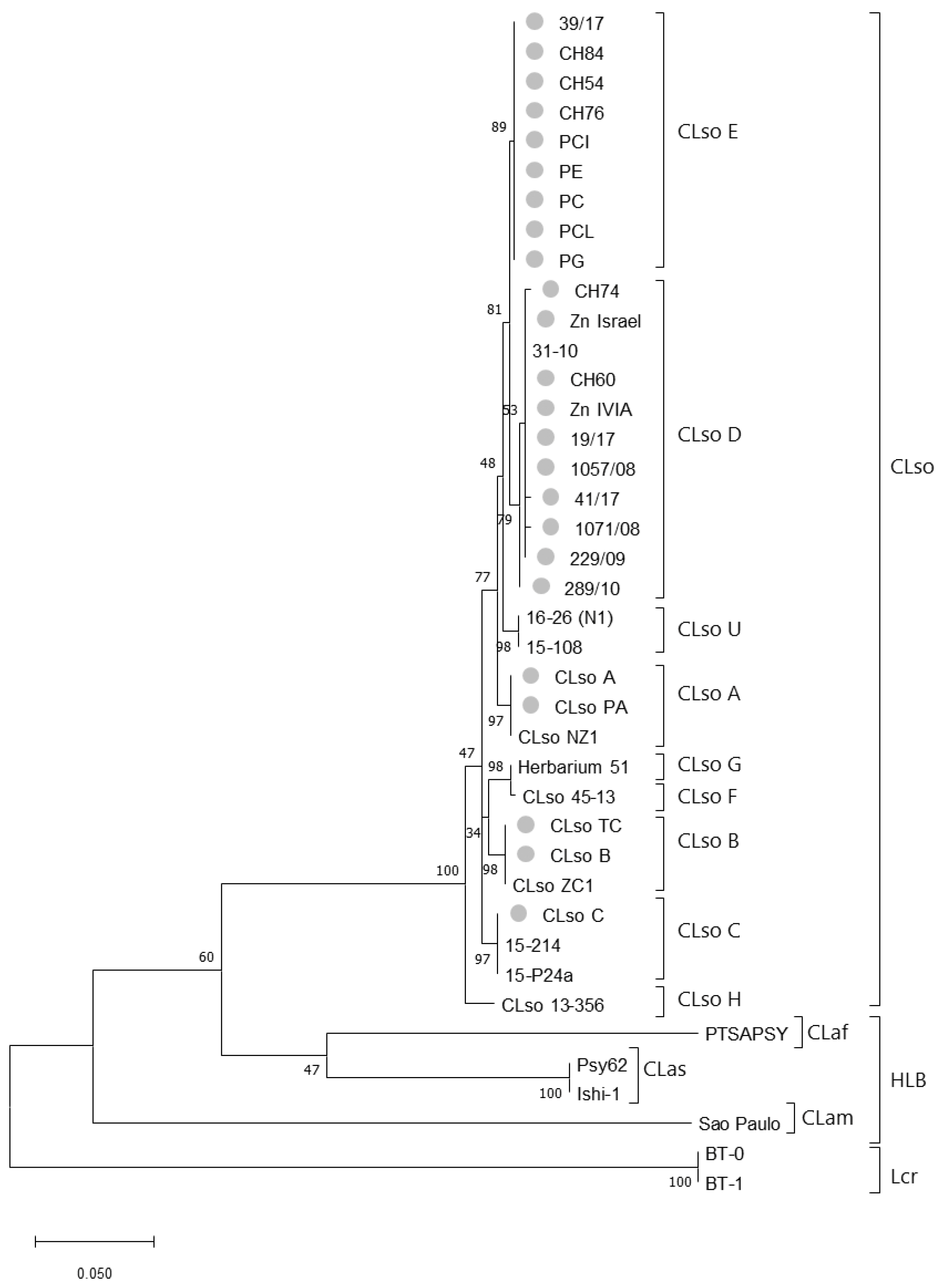
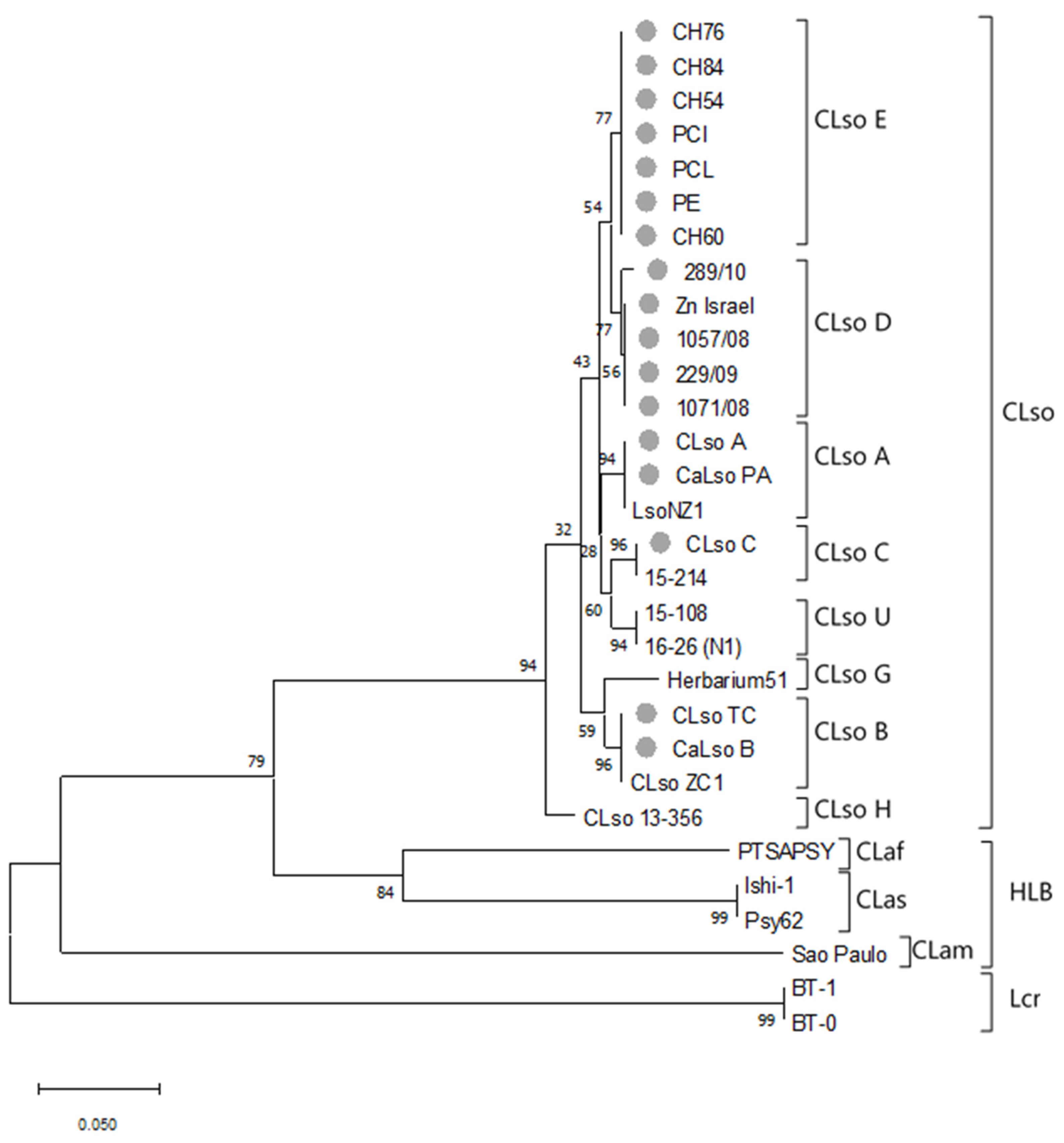
3.2. 50S Ribosomal Subunit Proteins L10/L12 (rplJ/rplL) Genes Sequence Analysis
3.3. MLSA Sequence Analysis
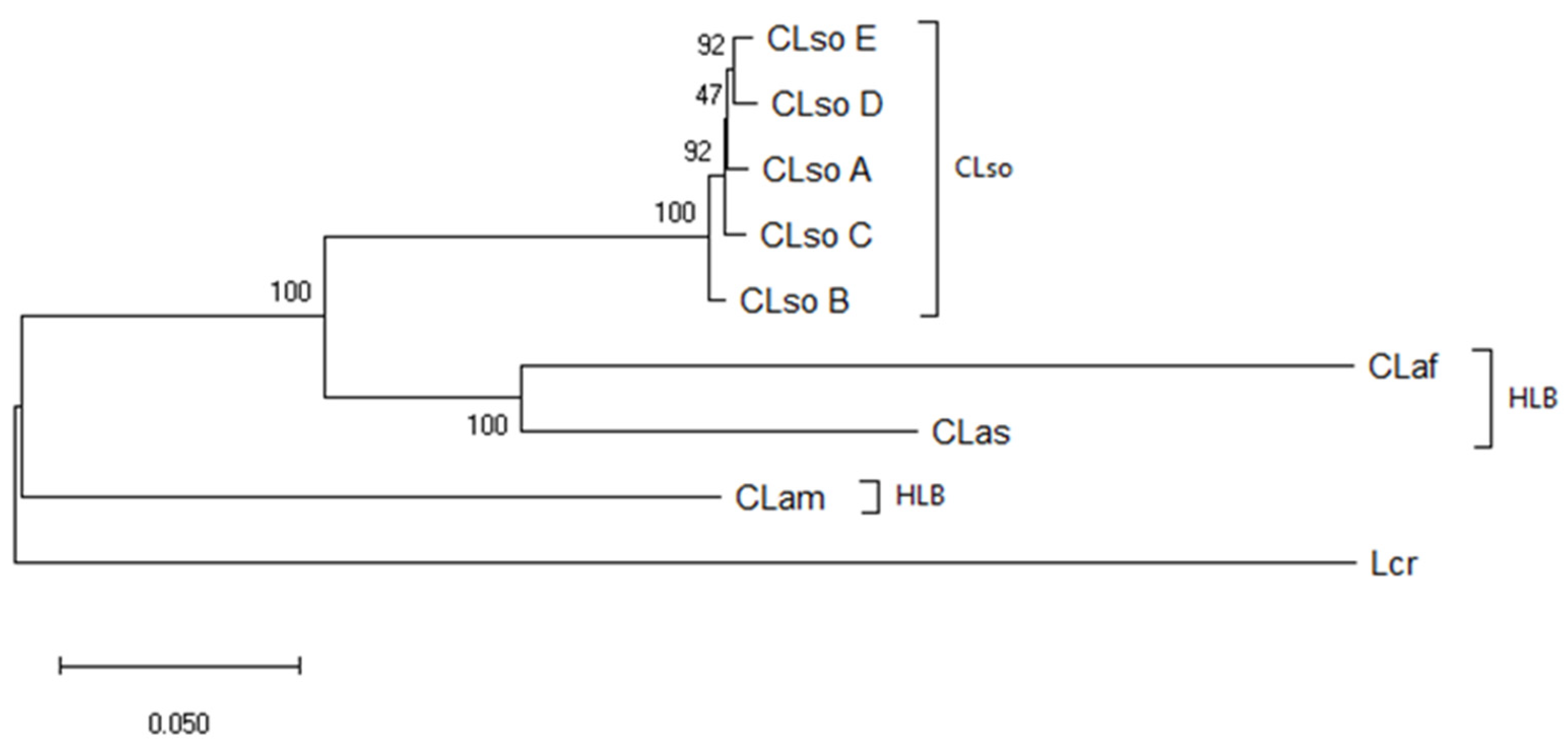
3.4. Concatenated Sequence Analysis
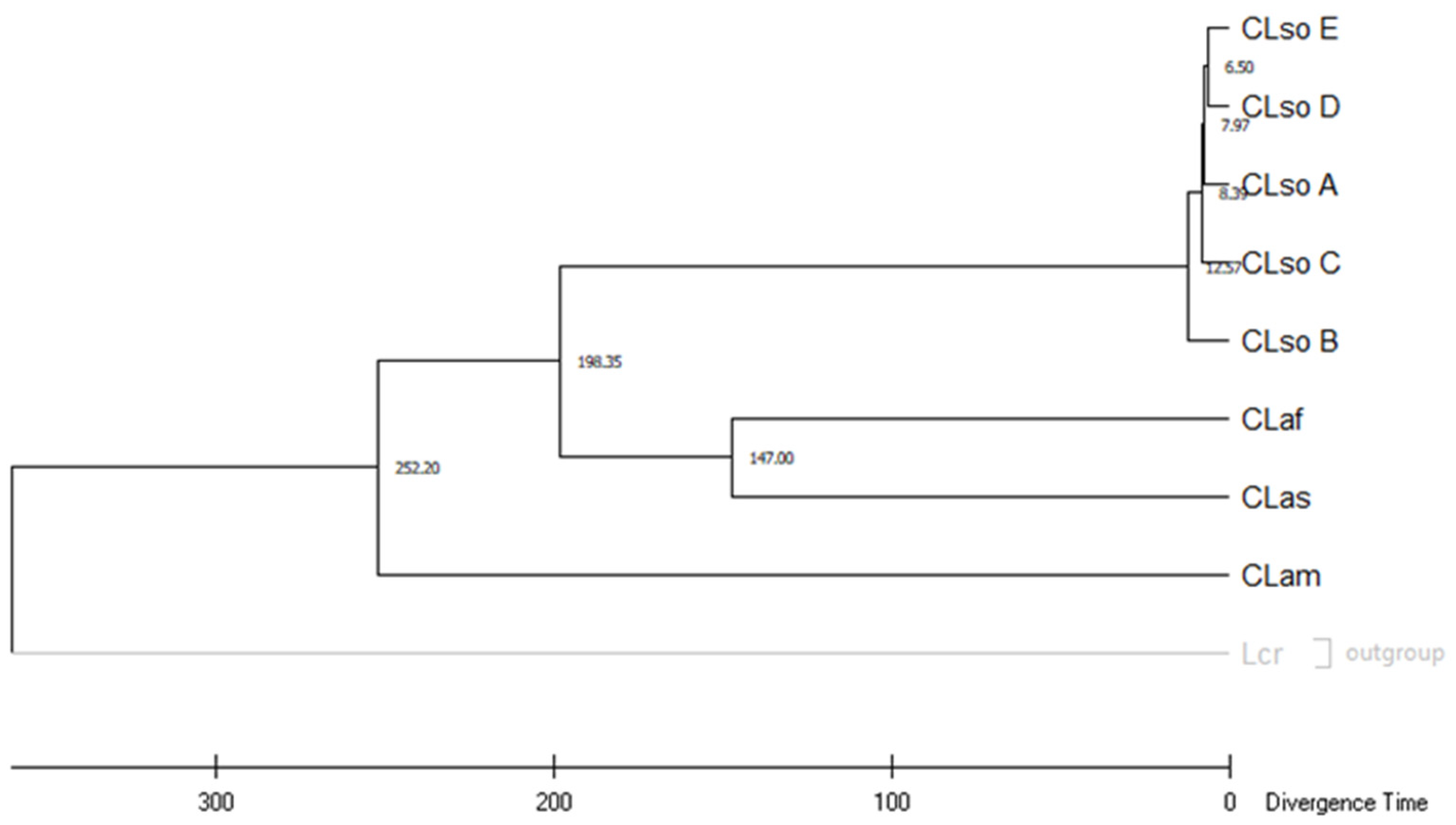
3.5. Molecular Dating on Liberibacter Species
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Haapalainen, M. Biology and epidemics of Candidatus Liberibacter species, psyllid-transmitted plant-pathogenic bacteria. Ann. Appl. Biol. 2014, 165, 172–198. [Google Scholar] [CrossRef]
- Raddadi, N.; Gonella, E.; Camerota, C.; Pizzinat, A.; Tedeschi, R.; Crotti, E.; Mandrioli, M.; Attilio Bianco, P.; Daffonchio, D.; Alma, A. ‘Candidatus Liberibacter europaeus’ sp. nov. that is associated with and transmitted by the psyllid Cacopsylla pyri apparently behaves as an endophyte rather than a pathogen. Environ. Microbiol. 2011, 13, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Pierson, E.A.; Setubal, J.C.; Xu, J.; Levy, J.G.; Zhang, Y.; Li, J.; Rangel, L.T.; Martins, J. The Candidatus Liberibacter–Host Interface: Insights into Pathogenesis Mechanisms and Disease Control. Annu. Rev. Phytopathol. 2017, 55, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.; Shiller, J.; Mann, R.; Smith, G.; Yen, A.; Rodoni, B. Novel ‘Candidatus Liberibacter’ species identified in the Australian eggplant psyllid, Acizzia solanicola. Microb. Biotechnol. 2017, 10, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Alfaro-Fernández, A.; Hernández-Llopis, D.; Font, M.I. Haplotypes of ‘Candidatus Liberibacter solanacearum’ identified in Umbeliferous crops in Spain. Eur. J. Plant Pathol. 2017, 149, 127–131. [Google Scholar] [CrossRef]
- Palomo, J.L.; Bertolini, E.; Martín-Robles, M.J.; Teresiani, G.; López, M.M.; Cambra, M. Detección en patata en España de un haplotipo de “Candidatus Liberibacter solanacearum” no descrito en solanáceas. In Proceedings of the Abstracts of XVII Congress of Spanish Phytopathological Society, Lleida, Spain, 7–10 October 2014; Sociedad Española de Fitopatología: Lleida, Spain, 2014; p. 125. [Google Scholar]
- Haapalainen, M.; Wang, J.; Latvala, S.; Lehtonen, M.T.; Pirhonen, M.; Nissinen, A.I. Genetic Variation of ‘Candidatus Liberibacter solanacearum’ Haplotype C and Identification of a Novel Haplotype from Trioza urticae and Stinging Nettle. Phytopathology 2018, 108, 925–934. [Google Scholar] [CrossRef] [Green Version]
- Haapalainen, M.; Latvala, S.; Wickström, A.; Wang, J.; Pirhonen, M.; Nissinen, A.I. A novel haplotype of ‘Candidatus Liberibacter solanacearum’ found in Apiaceae and Polygonaceae family plants. Eur. J. Plant Pathol. 2020, 156, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Mauck, K.E.; Sun, P.; Meduri, V.R.; Hansen, A.K. New Ca. Liberibacter psyllaurous haplotype resurrected from a 49-year-old specimen of Solanum umbelliferum: A native host of the psyllid vector. Sci. Rep. 2019, 9, 9530. [Google Scholar] [CrossRef]
- Teixeira, D.C.; Eveillard, S.; Sirand-Pugnet, P.; Wulff, A.; Saillard, C.; Ayres, A.J.; Bove, J.M. The tufB-secE-nusG-rplKAJL-rpoB gene cluster of the liberibacters: Sequence comparisons, phylogeny and speciation. Int. J. Syst. Evol. Microbiol. 2008, 58, 1414–1421. [Google Scholar] [CrossRef] [Green Version]
- Glaeser, S.P.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef]
- Glynn, J.M.; Islam, M.S.; Bai, Y.; Lan, S.; Wen, A.; Gudmestad, N.C.; Civerolo, E.L.; Lin, H. Multilocus sequence typing of “Candidatus Liberibacter solanacearum” isolates from North America and New Zealand. J. Plant Pathol. 2012, 94, 223–228. [Google Scholar] [CrossRef]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; Van de Peer, Y.; Vandamme, P.; Thompson, F.L.; et al. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005, 3, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Swisher Grimm, K.D.; Garczynski, S.F. Identification of a New Haplotype of ‘Candidatus Liberibacter solanacearum’ in Solanum tuberosum. Plant Dis. 2019, 103, 468–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Lou, B.; Glynn, J.M.; Doddapaneni, H.; Civerolo, E.L.; Chen, C.; Duan, Y.; Zhou, L.; Vahling, C.M. The Complete Genome Sequence of ‘Candidatus Liberibacter solanacearum’, the Bacterium Associated with Potato Zebra Chip Disease. PLoS ONE 2011, 6, e19135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, S.M.; Johnson, C.P.; Lu, A.Y.; Frampton, R.A.; Sullivan, K.L.; Fiers, M.W.E.J.; Crowhurst, R.N.; Pitman, A.R.; Scott, I.A.W.; Wen, A.; et al. Genomes of ‘Candidatus Liberibacter solanacearum’ Haplotype A from New Zealand and the United States Suggest Significant Genome Plasticity in the Species. Phytopathology 2015, 105, 863–871. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Pietersen, G.; Han, C.; Read, D.A.; Lou, B.; Gupta, G.; Civerolo, E.L. Complete Genome Sequence of “Candidatus Liberibacter africanus,” a Bacterium Associated with Citrus Huanglongbing. Genome Announc. 2015, 3, e00733-15. [Google Scholar] [CrossRef] [Green Version]
- Wulff, N.A.; Zhang, S.; Setubal, J.C.; Almeida, N.F.; Martins, E.C.; Harakava, R.; Kumar, D.; Rangel, L.T.; Foissac, X.; Bové, J.M.; et al. The Complete Genome Sequence of ‘Candidatus Liberibacter americanus’, Associated with Citrus Huanglongbing. Mol. Plant Microbe Interact. 2014, 27, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Zhou, L.; Hall, D.G.; Li, W.; Doddapaneni, H.; Lin, H.; Liu, L.; Vahling, C.M.; Gabriel, D.W.; Williams, K.P.; et al. Complete Genome Sequence of Citrus Huanglongbing Bacterium, ‘Candidatus Liberibacter asiaticus’ Obtained Through Metagenomics. Mol. Plant-Microbe Interact. 2009, 22, 1011–1020. [Google Scholar] [CrossRef] [Green Version]
- Katoh, H.; Miyata, S.; Inoue, H.; Iwanami, T. Unique Features of a Japanese ‘Candidatus Liberibacter asiaticus’ Strain Revealed by Whole Genome Sequencing. PLoS ONE 2014, 9, e106109. [Google Scholar] [CrossRef] [Green Version]
- Leonard, M.T.; Fagen, J.R.; Davis-Richardson, A.G.; Davis, M.J.; Triplett, E.W. Complete genome sequence of Liberibacter crescens BT-1. Stand. Genom. Sci. 2012, 7, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Teresani, G.R.; Bertolini, E.; Alfaro-Fernández, A.; Martínez, C.; Tanaka, F.A.O.; Kitajima, E.W.; Roselló, M.; Sanjuán, S.; Ferrándiz, J.C.; López, M.M.; et al. Association of ‘Candidatus Liberibacter solanacearum’ with a Vegetative Disorder of Celery in Spain and Development of a Real-Time PCR Method for Its Detection. Phytopathology 2014, 104, 804–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Doddapaneni, H.; Munyaneza, J.E.; Civerolo, E.L.; Sengoda, V.G.; Buchman, J.L.; Stenger, D.C. Molecular characterization and phylogenetic analysis of 16s rRNA from a new “Candidatus liberibacter” strain associated with zebra chip disease of potato (Solanum tuberosum L.) and the potato psyllid (Bactericera cockerelli Sulc). J. Plant Pathol. 2009, 91, 215–219. [Google Scholar] [CrossRef]
- Li, W.; Hartung, J.S.; Levy, L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 2006, 66, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Munyaneza, J.E.; Sengoda, V.G.; Crosslin, J.M.; De la Rosa-Lozano, G.; Sanchez, A. First Report of ‘Candidatus Liberibacter psyllaurous’ in Potato Tubers with Zebra Chip Disease in Mexico. Plant Dis. 2009, 93, 552. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Liao, J.; Wiedmann, M.; Kovac, J. Genetic Stability and Evolution of the sigB Allele, Used for Listeria Sensu Stricto Subtyping and Phylogenetic Inference. Appl. Environ. Microbiol. 2017, 83, e00306-17. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Tao, Q.; Kumar, S. Theoretical Foundation of the RelTime Method for Estimating Divergence Times from Variable Evolutionary Rates. Mol. Biol. Evol. 2018, 35, 1770–1782. [Google Scholar] [CrossRef]
- López-Hermoso, C.; de la Haba, R.R.; Sánchez-Porro, C.; Papke, R.T.; Ventosa, A. Assessment of MultiLocus Sequence Analysis as a Valuable Tool for the Classification of the Genus Salinivibrio. Front. Microbiol. 2017, 8, 1107. [Google Scholar] [CrossRef] [PubMed]
- Hajri, A.; Loiseau, M.; Cousseau-Suhard, P.; Renaudin, I.; Gentit, P. Genetic Characterization of ‘Candidatus Liberibacter solanacearum’ Haplotypes Associated with Apiaceous Crops in France. Plant Dis. 2017, 101, 1383–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Wang, Y.; Liu, Z.; Dai, H.; Cai, H.; Li, Z.; Du, Z.; Wang, X.; Jing, H.; Wei, Q.; et al. Multilocus sequence analysis, a rapid and accurate tool for taxonomic classification, evolutionary relationship determination, and population biology studies of the genus Shewanella. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konstantinidis, K.T.; Ramette, A.; Tiedje, J.M. Toward a More Robust Assessment of Intraspecies Diversity, Using Fewer Genetic Markers. Appl. Environ. Microbiol. 2006, 72, 7286–7293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajene, I.J.; Khamis, F.; Ballo, S.; Pietersen, G.; van Asch, B.; Seid, N.; Azerefegne, F.; Ekesi, S.; Mohamed, S. Detection of Asian Citrus Psyllid (Hemiptera: Psyllidae) in Ethiopia: A New Haplotype and its Implication to the Proliferation of Huanglongbing. J. Econ. Entomol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Thapa, S.P.; De Francesco, A.; Trinh, J.; Gurung, F.B.; Pang, Z.; Vidalakis, G.; Wang, N.; Ancona, V.; Ma, W.; Coaker, G. Genome-wide analyses of Liberibacter species provides insights into evolution, phylogenetic relationships, and virulence factors. Mol. Plant Pathol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Antolinez, C.A.; Fereres, A.; Moreno, A. Risk assessment of ‘Candidatus Liberibacter solanacearum’ transmission by the psyllids Bactericera trigonica and B. tremblayi from Apiaceae crops to potato. Sci. Rep. 2017, 7, 45534. [Google Scholar] [CrossRef] [Green Version]
- Antolínez, C.A.; Moreno, A.; Ontiveros, I.; Pla, S.; Plaza, M. Spain Seasonal abundance of psyllid species associated with carrots and potato fields in Spain. Insects 2019, 10, 287. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Haapalainen, M.; Schott, T.; Thompson, S.M.; Smith, G.R.; Nissinen, A.I.; Pirhonen, M. Genomic sequence of “Candidatus Liberibacter solanacearum” haplotype C and its comparison with haplotype A and B genomes. PLoS ONE 2017, 12, e0171531. [Google Scholar] [CrossRef] [Green Version]
- Katsir, L.; Zhepu, R.; Santos Garcia, D.; Piasezky, A.; Jiang, J.; Sela, N.; Freilich, S.; Bahar, O. Genome Analysis of Haplotype D of Candidatus Liberibacter Solanacearum. Front. Microbiol. 2018, 9, 2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Chaves, M.Q.-G.; Teresani, G.R.; Hernández-Suárez, E.; Bertolini, E.; Moreno, A.; Fereres, A.; Cambra, M.; Siverio, F. ‘Candidatus Liberibacter Solanacearum’ Is Unlikely to Be Transmitted Spontaneously from Infected Carrot Plants to Citrus Plants by Trioza Erytreae. Insects 2020, 11, 514. [Google Scholar] [CrossRef] [PubMed]



| Sample | Source | Origin | Type | Reference |
|---|---|---|---|---|
| 39/17 1 | Carrot/seeds | Castilla y León/Spain | CLso E | This work |
| CH84 1 | Parsnip/seeds | Valencia/Spain | CLso E | This work |
| CH54 1 | Parsnip/seeds | Valencia/Spain | CLso E | This work |
| CH76 1 | Parsnip/seeds | Valencia/Spain | CLso E | This work |
| PCI 1 | Potato/tubers | Canary/Spain | CLso E | This work |
| PE 1 | Potato/tubers | Euskadi/Spain | CLso E | This work |
| PC 1 | Potato/tubers | Cantabria/Spain | CLso E | This work |
| PCL 1 | Potato/tubers | Castilla y León/Spain | CLso E | This work |
| PG 1 | Potato/tubers | Galicia/Spain | CLso E | This work |
| Zn IVIA 1 | Carrot/seeds | Valencia/Spain | CLso D | This work |
| 19/17 1 | Carrot/seeds | Castilla y León/Spain | CLso D | This work |
| 41/17 1 | Carrot/seeds | Castilla y León/Spain | CLso D | This work |
| CH60 1 | Parsnip/seeds | Valencia/Spain | CLso D | This work |
| CH74 1 | Parsnip/seeds | Valencia/Spain | CLso D | This work |
| CLso TC 1 | Tomato/seeds | Mexico | CLso B | This work |
| CLso PA 1 | Pepper/seeds | Mexico | CLso A | This work |
| CLso C 2 | Unknown | Unknown | CLso C | IVIA control |
| CLso B 2 | Unknown | Unknown | CLso B | IVIA control |
| CLso A 2 | Unknown | Unknown | CLso A | IVIA control |
| Zn Israel 3 | Carrot/leaf | Israel | CLso D | Volcani C |
| 31–10 4 | Carrot/leaf | Canary/Spain | CLso D | [5] |
| 289/10 4 | Carrot/leaf | Valencia/Spain | CLso D | [5] |
| 1057/08 4 | Carrot/leaf | Valencia/Spain | CLso D | [5] |
| 1071/08 4 | Carrot/leaf | Valencia/Spain | CLso D | [5] |
| 229/09 4 | Celery/leaf | Valencia/Spain | CLso D | [5] |
| 15-214 5 | Trioza anthisci | Finland | CLso C | [8] |
| 15-P24a 5 | Trioza apicalis | Finland | CLso C | [8] |
| CLso 13-356 5 | Carrot/leaf | Finland | CLso H | [8] |
| 16-26 (N1) 5 | Nettle plants | Finland | CLso U | [7] |
| 15-108 5 | Trioza urticae | Finland | CLso U | [7] |
| Herbarium 51 5 | Solanum umbelliferum | USA | CLso G | [9] |
| CLso 45-13 5 | Potato/tuber | USA | CLso F | [14] |
| CLso ZC1 5 | Bactericera cockerelli | USA | CLso B | [15] |
| CLso NZ1 5 | Bactericera cockerelli | New Zealand | CLso A | [16] |
| PTSAPSY 5 | Citrus sp. | South Africa | CLaf | [17] |
| São Paulo 5 | Citrus sp. | Brazil | CLam | [18] |
| Psy62 5 | Citrus sp. | USA | CLas | [19] |
| Ishi-1 5 | Citrus sp. | Japan | CLas | [20] |
| BT-0 5 | Mountain Papaya | Puerto Rico | Lcr | [21] |
| BT-1 5 | Mountain Papaya | Puerto Rico | Lcr | [21] |
| Oligos | Sequence (5′-3′) | Target | Ref. |
|---|---|---|---|
| CaLsppF | GCAGGCCTAACACATGCAAGT | 16S rRNA | [22] |
| CaLsppR | GCACACGTTTCCATGCGTTAT | ||
| CaLspl 1 | FAM-AGCGCTTATTTTTAATAGGAGCGGCAGACG-TAMRA | ||
| LsoF | GTCGAGCGCTTATTTTTAATAGGA | 16S rRNA | [23] |
| HLBr | GCGTTATCCCGTAGAAAAAGGTAG | ||
| HLBp 1 | FAM-AGACGGGTGAGTAACGCG-BHQ-1 | ||
| COXf | GTATGCCACGTCGCATTCCAGA | Cytochrome oxidase (COX) | [24] |
| COXr | GCCAAAACTGCTAAGGGCATTC | ||
| COXp 1 | TET-ATCCAGATGCTTACGCTGG-BHQ-2 |
| Oligos | Sequence (5′-3′) | Target | Ref. |
|---|---|---|---|
| CL514F | CTCTAAGATTTCGGTTGGTT | 50S proteins L10/L12 genes (rplJ/rplL) | [25] |
| CL514R | TATATCTATCGTTGCACCAG | ||
| ADK | ATGAGAATTATATTTCTAGGCCCTCC | adenylate kinase (adk) | [12] |
| CKC_0526 0 | ATCATATTTATCATCTGATCGCACAG | ||
| DnaG | TTGCTATTGACTTTGATTAATCATCC | DNA primase (dnaG) | [12] |
| CKC_05195 | CAAAGCCTTCTATTATGGCTTCTTG | ||
| FumC | TTCCTTTAGTCGTCTGGCAAACAGG | fumarate hydratase (fumC) | [12] |
| CKC_050 75 | ACTTGTGCAGCGTATCCTGAAAATTC | ||
| GrpE | TAGAAATACCAACTAAAGCGGGGCG | heat shock protein (grpE) | [12] |
| CKC_00585 | GGAAATCCCCTAACGGAACCATTCG | ||
| IcdA | TAGAAATACCAACTAAAGCGGGGCG | isocitrate dehydrogenase (icdA) | [12] |
| CKC_043 65 | GGAAATCCCCTAACGGAACCATTCG | ||
| MetG | TCGTACGATGATTTTATTCGCACAACGG | methionyl-tRNA synthetase (metG) | [12] |
| CKC_02965 | GGATCGTTAGGAATTTTTATTCCCCAATC | ||
| MutS | CCAACAGATTCTAATTATCTCATGG | DNA mismatch repair protein (mutS) | [12] |
| CKC_008 15 | TCTAAATTGGAACGAGCGGCGGA | ||
| PurA | TGTAGTTGTGGTCGGCTTACAATGG | adenylosuccinate synthetase (purA) | [12] |
| CKC_003 15 | TATCTTCATAAGCTGGGCCAATACC | ||
| RecA | TTGGAAATAACAGATATGCTGGTGCG | recombinase A (recA) | [12] |
| CKC_050 85 | AACCACGCTCCTGATTTATCAACGAT | ||
| JG-gyrB1 | AACGCTAGCCGTCTTGTGAA | DNA topoisomerase IV sub B (gyrB) | This work |
| JG-gyrB2 | TTGCCACGCAAGGGAAGTAT |
| CLso A | CLso B | CLso C | CLso D | Clso E | CLso F | CLso G | CLso H | CLso U | |
|---|---|---|---|---|---|---|---|---|---|
| CLso A | 0.65 1 | 0.59 | 0.58 | 0.50 | 0.68 | 0.64 | 0.79 | 0.56 | |
| CLso B | 97.91 | 0.53 | 0.77 | 0.71 | 0.63 | 0.58 | 0.77 | 0.67 | |
| CLso C | 98.38 | 98.61 | 0.68 | 0.61 | 0.68 | 0.62 | 0.73 | 0.58 | |
| CLso D | 98.38 | 97.22 | 97.69 | 0.43 | 0.78 | 0.73 | 0.79 | 0.59 | |
| CLso E | 98.84 | 97.68 | 98.15 | 99.07 | 0.71 | 0.67 | 0.81 | 0.52 | |
| CLso F | 97.90 | 98.13 | 97.91 | 97.20 | 97.66 | 0.23 | 0.78 | 0.69 | |
| CLso G | 98.14 | 98.37 | 98.15 | 97.44 | 97.91 | 99.77 | 0.73 | 0.64 | |
| CLso H | 96.98 | 97.22 | 97.47 | 96.71 | 96.75 | 97.22 | 97.46 | 0.78 | |
| CLso U | 98.61 | 97.91 | 98.38 | 98.38 | 98.84 | 97.90 | 98.14 | 96.98 |
| Gene | No. of Mutations | No. of Polymorphic Sites | ds 1 | k 2 | Tajima’s D Value |
|---|---|---|---|---|---|
| rplJ/rplL3 | 291 | 223 | 0.09040 | 35.97841 | −1.78652 |
| rplJ/rplL4 | 19 | 19 | 0.01076 | 4.63768 | −0.32106 |
| adk3 | 173 | 120 | 0.22971 | 46.86111 | −1.36942 |
| adk4 | 13 | 13 | 0.02621 | 5.40000 | −0.97762 |
| dnaG3 | 286 | 223 | 0.19587 | 80.30556 | −1.23317 |
| dnaG4 | 11 | 11 | 0.01073 | 4.40000 | −1.19955 |
| fumC3 | 148 | 114 | 0.14153 | 41.75000 | −1.20958 |
| fumC4 | 11 | 11 | 0.01549 | 4.60000 | −0.95426 |
| grpE3 | 225 | 167 | 0.17818 | 64.50000 | −1.14858 |
| grpE4 | 7 | 7 | 0.00826 | 3.00000 | −0.74682 |
| icdA3 | 203 | 161 | 0.14866 | 58.72222 | −1.11107 |
| icdA4 | 5 | 5 | 0.00557 | 2.40000 | −0.56199 |
| metG3 | 194 | 163 | 0.16769 | 54.50000 | −1.22856 |
| metG4 | 11 | 11 | 0.01354 | 4.40000 | −1.19955 |
| mutS3 | 223 | 281 | 0.19543 | 82.66667 | −1.11084 |
| mutS4 | 14 | 14 | 0.01389 | 6.00000 | −0.78089 |
| purA3 | 160 | 125 | 0.15681 | 43.75000 | −1.33243 |
| purA4 | 5 | 5 | 0.00717 | 2.00000 | −1.12397 |
| recA3 | 195 | 150 | 0.12953 | 60.36111 | −0.82453 |
| recA4 | 10 | 10 | 0.00944 | 4.40000 | −0.59633 |
| gyrB3 | 611 | 465 | 0.23816 | 163.58333 | −1.42139 |
| gyrB4 | 18 | 18 | 0.01178 | 8.40000 | −0.20459 |
| Gene | No. of Mutations | No. of Polymorphic Sites | ds 1 | k 2 | Tajima’s D Value |
|---|---|---|---|---|---|
| A3 | 657 | 504 | 0.18420 | 187.11111 | −1.17949 |
| A4 | 32 | 32 | 0.01317 | 13.60000 | −0.85795 |
| B3 | 662 | 504 | 0.18423 | 185.88889 | −1.23622 |
| B4 | 29 | 29 | 0.01187 | 12.00000 | −1.03047 |
| C3 | 654 | 493 | 0.17364 | 187.52778 | −1.15191 |
| C4 | 34 | 34 | 0.01312 | 14.20000 | −0.97388 |
| D3 | 493 | 649 | 0.17175 | 188.75000 | −1.09384 |
| D4 | 37 | 37 | 0.01431 | 15.80000 | −0.82874 |
| E3 | 376 | 281 | 0.17627 | 105.58333 | −1.23426 |
| E4 | 18 | 18 | 0.01265 | 7.60000 | −0.88654 |
| F3 | 368 | 270 | 0.16003 | 107.22222 | −1.08462 |
| F4 | 23 | 23 | 0.01458 | 9.80000 | −0.83399 |
| G3 | 484 | 384 | 0.17021 | 140.25000 | −1.10811 |
| G4 | 19 | 19 | 0.00992 | 8.20000 | −0.74443 |
| H3 | 476 | 373 | 0.15854 | 141.88889 | −0.99021 |
| H4 | 24 | 24 | 0.01283 | 10.40000 | −0.72278 |
| I3 | 2476 | 1911 | 0.18088 | 695.86111 | −1.23455 |
| I4 | 105 | 105 | 0.01153 | 44.80000 | −0.84470 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Padilla, A.; Redondo, C.; Asensio, A.; Garita-Cambronero, J.; Martínez, C.; Pérez-Padilla, V.; Marquínez, R.; Collar, J.; García-Méndez, E.; Alfaro-Fernández, A.; et al. Assessment of Multilocus Sequence Analysis (MLSA) for Identification of Candidatus Liberibacter Solanacearum from Different Host Plants in Spain. Microorganisms 2020, 8, 1446. https://doi.org/10.3390/microorganisms8091446
Ruiz-Padilla A, Redondo C, Asensio A, Garita-Cambronero J, Martínez C, Pérez-Padilla V, Marquínez R, Collar J, García-Méndez E, Alfaro-Fernández A, et al. Assessment of Multilocus Sequence Analysis (MLSA) for Identification of Candidatus Liberibacter Solanacearum from Different Host Plants in Spain. Microorganisms. 2020; 8(9):1446. https://doi.org/10.3390/microorganisms8091446
Chicago/Turabian StyleRuiz-Padilla, Ana, Cristina Redondo, Adrián Asensio, Jerson Garita-Cambronero, Carmen Martínez, Verónica Pérez-Padilla, Raquel Marquínez, Jesús Collar, Eva García-Méndez, Ana Alfaro-Fernández, and et al. 2020. "Assessment of Multilocus Sequence Analysis (MLSA) for Identification of Candidatus Liberibacter Solanacearum from Different Host Plants in Spain" Microorganisms 8, no. 9: 1446. https://doi.org/10.3390/microorganisms8091446
APA StyleRuiz-Padilla, A., Redondo, C., Asensio, A., Garita-Cambronero, J., Martínez, C., Pérez-Padilla, V., Marquínez, R., Collar, J., García-Méndez, E., Alfaro-Fernández, A., Asensio-S.-Manzanera, C., Palomo, J. L., Siverio, F., León, L. D., & Cubero, J. (2020). Assessment of Multilocus Sequence Analysis (MLSA) for Identification of Candidatus Liberibacter Solanacearum from Different Host Plants in Spain. Microorganisms, 8(9), 1446. https://doi.org/10.3390/microorganisms8091446





