Molecular Methods for the Detection of Toxoplasma gondii Oocysts in Fresh Produce: An Extensive Review
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dubey, J.P. Toxoplasmosis of Animals and Humans, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Torgerson, P.R.; Devleesschauwer, B.; Praet, N.; Speybroeck, N.; Willingham, A.L.; Kasuga, F.; Rokni, M.B.; Zhou, X.N.; Fèvre, E.M.; Sripa, B.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001920. [Google Scholar] [CrossRef] [PubMed]
- ECDC. European Centre for Disease Prevention and Control. Congenital toxoplasmosis. In Annual Epidemiological Report for 2015; ECDC: Stockholm, Sweden, 2018. [Google Scholar]
- Hald, T.; Aspinall, W.; Devleesschauwer, B.; Cooke, R.; Corrigan, T.; Havelaar, A.H.; Gibb, H.J.; Torgerson, P.R.; Kirk, M.D.; Angulo, F.J.; et al. World Health Organization Estimates of the Relative Contributions of Food to the Burden of Disease Due to Selected Foodborne Hazards: A Structured Expert Elicitation. PLoS ONE 2016, 11, e0145839. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; et al. Public health risks associated with food-borne parasites. Efsa J. 2018, 16, e05495. [Google Scholar] [CrossRef] [PubMed]
- Bouwknegt, M.; Devleesschauwer, B.; Graham, H.; Robertson, L.J.; van der Giessen, J.W. The Euro-Fbp Workshop Participants. Prioritisation of food-borne parasites in Europe, 2016. Eurosurveillance 2018, 23, 17-00161. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations/World Health Organization. Multicriteria Based Ranking for Risk Management of Food Borne Parasites; FAO, World Health Organization: Rome, Italy, 2014; 287p. [Google Scholar]
- Castro-Ibáñez, I.; Gil, M.I.; Allende, A. Ready-to-eat vegetables: Current problems and potential solutions to reduce microbial risk in the production chain. LWT Food Sci. Technol. 2017, 85, 284–292. [Google Scholar] [CrossRef]
- Murphy, H.R.; Almeria, S.; da Silva, A.J. BAM Chapter 19b: Molecular Detection of Cyclospora cayetanensis in Fresh Produce Using Real-Time PCR. U.S: Food and Drug Administration. 2019. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-19b-molecular-detection-cyclospora-cayetanensis-fresh-produce-using-real-time-pcr (accessed on 10 July 2020).
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Lalle, M.; Slana, I.; Bier, N.; Mayer-Scholl, A.; Jokelainen, P. Deliverable D-JRP-TOXOSOURCES-WP3.1 Report on Available Analytical Procedures for Detection of Toxoplasma Gondii in Fresh Produce and List of Promising Analytical Procedures. April 2020. Available online: https://zenodo.org/record/3778719#.X8ENmbPSLcs (accessed on 10 July 2020).
- Chandra, V.; Torres, M.; Ortega, Y.R. Efficacy of wash solutions in recovering Cyclospora cayetanensis, Cryptosporidium parvum, and Toxoplasma gondii from basil. J. Food Prot. 2014, 77, 1348–1354. [Google Scholar] [CrossRef]
- Hohweyer, J.; Cazeaux, C.; Travaillé, E.; Languet, E.; Dumètre, A.; Aubert, D.; Terryn, C.; Dubey, J.P.; Azas, N.; Houssin, M.; et al. Simultaneous detection of the protozoan parasites Toxoplasma, Cryptosporidium and Giardia in food matrices and their persistence on basil leaves. Food Microbiol. 2016, 57, 36–44. [Google Scholar] [CrossRef]
- Lalle, M.; Possenti, A.; Dubey, J.P.; Pozio, E. Loop-Mediated Isothermal Amplification-Lateral-Flow Dipstick (LAMP-LFD) to detect Toxoplasma gondii oocyst in ready-to-eat salad. Food Microbiol. 2018, 70, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, K.; Kim, M.; Rajal, V.B.; Arrowood, M.J.; Packham, A.; Aguilar, B.; Wuertz, S. Simultaneous detection of four protozoan parasites on leafy greens using a novel multiplex PCR assay. Food Microbiol. 2019, 84, 103252. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.Z.; Rafael, K.; Sanders, A.P.; Tiyo, B.T.; Marchioro, A.A.; Colli, C.M.; Gomes, M.L.; Falavigna-Guilherme, A.L. An alternative method to recover Toxoplasma gondii from greenery and fruits. Int. J. Environ. Health Res. 2016, 26, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, L.F.; Gajadhar, A.A. Optimization and validation of methods for isolation and real-time PCR identification of protozoan oocysts on leafy green vegetables and berry fruits. Food Waterborne Parasitol. 2016, 2, 1–7. [Google Scholar] [CrossRef]
- Marchioro, A.A.; Tiyo, B.T.; Colli, C.M.; de Souza, C.Z.; Garcia, J.L.; Gomes, M.L.; Falavigna-Guilherme, A.L. First Detection of Toxoplasma gondii DNA in the Fresh Leafs of Vegetables in South America. Vector Borne Zoonotic Dis. 2016, 16, 624–626. [Google Scholar] [CrossRef] [PubMed]
- Lass, A.; Pietkiewicz, H.; Szostakowska, B.; Myjak, P. The first detection of Toxoplasma gondii DNA in environmental fruits and vegetables samples. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1101–1108. [Google Scholar] [CrossRef]
- Temesgen, T.T.; Robertson, L.J.; Tysnes, K.R. A novel multiplex real-time PCR for the detection of Echinococcus multilocularis, Toxoplasma gondii, and Cyclospora cayetanensis on berries. Food Res. Int. 2019, 125, 108636. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, L.F.; Gajadhar, A.A. Detection of Cyclospora cayetanensis, Cryptosporidium spp., and Toxoplasma gondii on imported leafy green vegetables in Canadian survey. Food Waterborne Parasitol. 2016, 2, 8–14. [Google Scholar] [CrossRef]
- Caradonna, T.; Marangi, M.; Del Chierico, F.; Ferrari, N.; Reddel, S.; Bracaglia, G.; Normanno, G.; Putignani, L.; Giangaspero, A. Detection and prevalence of protozoan parasites in ready-to-eat packaged salads on sale in Italy. Food Microbiol. 2017, 67, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Lass, A.; Ma, L.; Kontogeorgos, I.; Zhang, X.; Li, X.; Karanis, P. First molecular detection of Toxoplasma gondii in vegetable samples in China using qualitative, quantitative real-time PCR and multilocus genotyping. Sci. Rep. 2019, 26, 17581. [Google Scholar] [CrossRef] [PubMed]
- Slany, M.; Dziedzinska, R.; Babak, V.; Kralik, P.; Moravkova, M.; Slana, I. Toxoplasma gondii in vegetables from fields and farm storage facilities in the Czech Republic. FEMS Microbiol. Lett. 2019, 366, fnz170. [Google Scholar] [CrossRef]
- Herrmann, D.C.; Maksimov, A.; Pantchev, N.; Vrhovec, M.G.; Conraths, F.J.; Schares, G. Comparison of different commercial DNA extraction kits to detect Toxoplasma gondii oocysts in cat faeces. Berl Munch Tierarztl. Wochenschr. 2011, 124, 497–502. [Google Scholar] [PubMed]
- Staggs, S.E.; Keely, S.P.; Ware, M.W.; Schable, N.; See, M.J.; Gregorio, D.; Zou, X.; Su, C.; Dubey, J.P.; Villegas, E.N. The development and implementation of a method using blue mussels (Mytilus spp.) as biosentinels of Cryptosporidium spp. and Toxoplasma gondii contamination in marine aquatic environments. Parasitol. Res. 2015, 114, 4655–4667. [Google Scholar] [CrossRef] [PubMed]
- Manore, A.J.W.; Harper, S.L.; Aguilar, B.; Weese, J.S.; Shapiro, K. Comparison of freeze-thaw cycles for nucleic acid extraction and molecular detection of Cryptosporidium parvum and Toxoplasma gondii oocysts in environmental matrices. J. Microbiol. Methods 2019, 156, 1–4. [Google Scholar] [CrossRef]
- Durand, L.; La Carbona, S.; Geffard, A.; Possenti, A.; Dubey, J.P.; Lalle, M. Comparative evaluation of loop-mediated isothermal amplification (LAMP) vs qPCR for detection of Toxoplasma gondii oocysts DNA in mussels. Exp. Parasitol. 2020, 208, 107809. [Google Scholar] [CrossRef] [PubMed]
- Géba, E.; Aubert, D.; Durand, L.; Escotte, S.; La Carbona, S.; Cazeaux, C.; Bonnard, I.; Bastien, F.; Palos Ladeiro, M.; Dubey, J.P.; et al. Use of the bivalve Dreissena polymorpha as a biomonitoring tool to reflect the protozoan load in freshwater bodies. Water Res. 2020, 170, 115297. [Google Scholar] [CrossRef] [PubMed]
- Escotte-Binet, S.; Da Silva, A.M.; Cancès, B.; Aubert, D.; Dubey, J.; La Carbona, S.; Villena, I.; Poulle, M.L. A rapid and sensitive method to detect Toxoplasma gondii oocysts in soil samples. Vet. Parasitol. 2019, 274, 108904. [Google Scholar] [CrossRef]
- Galvani, A.T.; Christ, A.P.G.; Padula, J.A.; Barbosa, M.R.F.; de Araújo, R.S.; Sato, M.I.Z.; de Araújo, R.S.; Sato, M.I.Z.; Razzolini, M.T.P. Real-time PCR detection of Toxoplasma gondii in surface water samples in São Paulo, Brazil. Parasitol. Res. 2019, 118, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, L.A.; Santos, L.K.; Brito, P.A., Jr.; Maciel, B.M.; Da Silva, A.V.; Albuquerque, G.R. Detection of Toxoplasma gondii DNA in Brazilian oysters (Crassostrea rhizophorae). Genet. Mol. Res. 2015, 14, 4658–4665. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lindquist, H.D.; Cama, V.; Schaefer, F.W., 3rd; Villegas, E.; Fayer, R.; Lewis, E.J.; Feng, Y.; Xiao, L. Detection of Toxoplasma gondii oocysts in water sample concentrates by real-time PCR. Appl. Environ. Microbiol. 2009, 75, 3477–3483. [Google Scholar] [CrossRef] [PubMed]
- Lass, A.; Pietkiewicz, H.; Modzelewska, E.; Dumètre, A.; Szostakowska, B.; Myjak, P. Detection of Toxoplasma gondii oocysts in environmental soil samples using molecular methods. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Salant, H.; Markovics, A.; Spira, D.T.; Hamburger, J. The development of a molecular approach for coprodiagnosis of Toxoplasma gondii. Vet. Parasitol. 2007, 146, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Tavalla, M.; Oormazdi, H.; Akhlaghi, L.; Shojaee, S.; Razmjou, E.; Hadighi, R.; Meamar, A. Genotyping of Toxoplasma gondii Isolates from Soil Samples in Tehran, Iran. Iran. J. Parasitol. 2013, 8, 227–233. [Google Scholar]
- Du, F.; Feng, H.L.; Nie, H.; Tu, P.; Zhang, Q.L.; Hu, M.; Zhou, Y.Q.; Zhao, J.L. Survey on the contamination of Toxoplasma gondii oocysts in the soil of public parks of Wuhan, China. Vet. Parasitol. 2012, 184, 141–146. [Google Scholar] [CrossRef]
- Schares, G.; Vrhovec, M.G.; Pantchev, N.; Herrmann, D.C.; Conraths, F.J. Occurrence of Toxoplasma gondii and Hammondia hammondi oocysts in the faeces of cats from Germany and other European countries. Vet. Parasitol. 2008, 152, 34–45. [Google Scholar] [CrossRef]
- Matsuo, J.; Kimura, D.; Rai, S.K.; Uga, S. Detection of Toxoplasma oocysts from soil by modified sucrose flotation and PCR methods. Southeast. Asian J. Trop Med. Public Health 2004, 35, 270–274. [Google Scholar] [PubMed]
- Chemoh, W.; Sawangjaroen, N.; Nissapatorn, V.; Sermwittayawong, N. Molecular investigation on the occurrence of Toxoplasma gondii oocysts in cat feces using TOX-element and ITS-1 region targets. Vet. J. 2016, 215, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, D.C.; Pantchev, N.; Vrhovec, M.G.; Barutzki, D.; Wilking, H.; Fröhlich, A.; Lüder, C.G.; Conraths, F.J.; Schares, G. Atypical Toxoplasma gondii genotypes identified in oocysts shed by cats in Germany. Int. J. Parasitol. 2010, 40, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Lélu, M.; Gilot-Fromont, E.; Aubert, D.; Richaume, A.; Afonso, E.; Dupuis, E.; Gotteland, C.; Marnef, F.; Poulle, M.L.; Dumètre, A.; et al. Development of a sensitive method for Toxoplasma gondii oocyst extraction in soil. Vet. Parasitol. 2011, 183, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Villena, I.; Aubert, D.; Gomis, P.; Ferté, H.; Inglard, J.C.; Denis-Bisiaux, H.; Dondon, J.M.; Pisano, E.; Ortis, N.; Pinon, J.M. Evaluation of a strategy for Toxoplasma gondii oocyst detection in water. Appl. Environ. Microbiol. 2004, 70, 4035–4039. [Google Scholar] [CrossRef]
- de Wit, L.A.; Kilpatrick, A.M.; VanWormer, E.; Croll, D.A.; Tershy, B.R.; Kim, M.; Shapiro, K. Seasonal and spatial variation in Toxoplasma gondii contamination in soil in urban public spaces in California, United States. Zoonoses Public Health 2020, 67, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Wells, B.; Shaw, H.; Innocent, G.; Guido, S.; Hotchkiss, E.; Parigi, M.; Opsteegh, M.; Green, J.; Gillespie, S.; Innes, E.A.; et al. Molecular detection of Toxoplasma gondii in water samples from Scotland and a comparison between the 529bp real-time PCR and ITS1 nested PCR. Water Res. 2015, 87, 175–181. [Google Scholar] [CrossRef]
- Poulle, M.L.; Bastien, M.; Richard, Y.; Josse-Dupuis, É.; Aubert, D.; Villena, I.; Knapp, J. Detection of Echinococcus multilocularis and other foodborne parasites in fox, cat and dog faeces collected in kitchen gardens in a highly endemic area for alveolar echinococcosis. Parasite 2017, 24, 29. [Google Scholar] [CrossRef] [PubMed]
- Bigot-Clivot, A.; Palos Ladeiro, M.; Lepoutre, A.; Bastien, F.; Bonnard, I.; Dubey, J.P.; Villena, I.; Aubert, D.; Geffard, O.; François, A.; et al. Bioaccumulation of Toxoplasma and Cryptosporidium by the freshwater crustacean Gammarus fossarum: Involvement in biomonitoring surveys and trophic transfer. Ecotoxicol. Environ. Saf. 2016, 133, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, H.; Wang, H.; Qin, H.; Xiao, J. Land use and soil contamination with Toxoplasma gondii oocysts in urban areas. Sci. Total Environ. 2016, 568, 1086–1091. [Google Scholar] [CrossRef]
- Poulle, M.L.; Forin-Wiart, M.A.; Josse-Dupuis, É.; Villena, I.; Aubert, D. Detection of Toxoplasma gondii DNA by qPCR in the feces of a cat that recently ingested infected prey does not necessarily imply oocyst shedding. Parasite 2016, 23, 29. [Google Scholar] [CrossRef] [PubMed]
- Palos Ladeiro, M.; Bigot-Clivot, A.; Aubert, D.; Villena, I.; Geffard, A. Assessment of Toxoplasma gondii levels in zebra mussel (Dreissena polymorpha) by real-time PCR: An organotropism study. Environ. Sci. Pollut. Res. Int. 2015, 22, 13693–13701. [Google Scholar]
- Bier, N.S.; Schares, G.; Johne, A.; Martin, A.; Nöckler, K.; Mayer-Scholl, A. Performance of three molecular methods for detection of Toxoplasma gondii in pork. Food Waterborne Parasitol. 2019, 14, e00038. [Google Scholar] [CrossRef] [PubMed]
- Gotteland, C.; Gilot-Fromont, E.; Aubert, D.; Poulle, M.L.; Dupuis, E.; Dardé, M.L.; Forin-Wiart, M.A.; Rabilloud, M.; Riche, B.; Villena, I. Spatial distribution of Toxoplasma gondii oocysts in soil in a rural area: Influence of cats and land use. Vet. Parasitol. 2014, 205, 629–637. [Google Scholar] [CrossRef]
- Afonso, E.; Lemoine, M.; Poulle, M.L.; Ravat, M.C.; Romand, S.; Thulliez, P.; Villena, I.; Aubert, D.; Rabilloud, M.; Riche, B.; et al. Spatial distribution of soil contamination by Toxoplasma gondii in relation to cat defecation behaviour in an urban area. Int. J. Parasitol. 2008, 38, 1017–1023. [Google Scholar] [CrossRef]
- Aubert, D.; Villena, I. Detection of Toxoplasma gondii oocysts in water: Proposition of a strategy and evaluation in Champagne-Ardenne Region, France. Mem Inst. Oswaldo Cruz. 2009, 104, 290–295. [Google Scholar] [CrossRef]
- Sroka, J.; Karamon, J.; Dutkiewicz, J.; Wójcik-Fatla, A.; Cencek, T. Optimization of flotation, DNA extraction and PCR methods for detection of Toxoplasma gondii oocysts in cat faeces. Ann. Agric. Environ. Med. 2018, 25, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Reischl, U.; Bretagne, S.; Krüger, D.; Ernault, P.; Costa, J.M. Comparison of two DNA targets for the diagnosis of Toxoplasmosis by real-time PCR using fluorescence resonance energy transfer hybridization probes. BMC Infect. Dis. 2003, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, K.; Mazet, J.A.; Schriewer, A.; Wuertz, S.; Fritz, H.; Miller, W.A.; Largier, J.; Conrad, P.A. Detection of Toxoplasma gondii oocysts and surrogate microspheres in water using ultrafiltration and capsule filtration. Water Res. 2010, 44, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Arkush, K.D.; Miller, M.A.; Leutenegger, C.M.; Gardner, I.A.; Packham, A.E.; Heckeroth, A.R.; Tenter, A.M.; Barr, B.C.; Conrad, P.A. Molecular and bioassay-based detection of Toxoplasma gondii oocyst uptake by mussels (Mytilus galloprovincialis). Int. J. Parasitol. 2003, 33, 1087–1097. [Google Scholar] [CrossRef]
- Marquis, N.D.; Bishop, T.J.; Record, N.R.; Countway, P.D.; Fernández Robledo, J.A. Molecular Epizootiology of Toxoplasma gondii and Cryptosporidium parvum in the Eastern Oyster (Crassostrea virginica) from Maine (USA). Pathogens 2019, 8, 125. [Google Scholar] [CrossRef]
- Coupe, A.; Howe, L.; Shapiro, K.; Roe, W.D. Comparison of PCR assays to detect Toxoplasma gondii oocysts in green-lipped mussels (Perna canaliculus). Parasitol. Res. 2019, 118, 2389–2398. [Google Scholar] [CrossRef] [PubMed]
- Opsteegh, M.; Langelaar, M.; Sprong, H.; den Hartog, L.; De Craeye, S.; Bokken, G.; Ajzenberg, D.; Kijlstra, A.; van der Giessen, J. Direct detection and genotyping of Toxoplasma gondii in meat samples using magnetic capture and PCR. Int. J. Food Microbiol. 2010, 139, 193–201. [Google Scholar] [CrossRef]
- Aksoy, U.; Marangi, M.; Papini, R.; Ozkoc, S.; Bayram Delibas, S.; Giangaspero, A. Detection of Toxoplasma gondii and Cyclospora cayetanensis in Mytilus galloprovincialis from Izmir Province coast (Turkey) by Real Time PCR/High-Resolution Melting analysis (HRM). Food Microbiol. 2014, 44, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Marangi, M.; Giangaspero, A.; Lacasella, V.; Lonigro, A.; Gasser, R.B. Multiplex PCR for the detection and quantification of zoonotic taxa of Giardia, Cryptosporidium and Toxoplasma in wastewater and mussels. Mol. Cell Probes. 2015, 29, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, R.; Robertson, L.; Dorny, P.; Suzanne, J.; Kärssin, A.; Katzer, F.; La Carbona, S.; Lalle, M.; Lassen, B.; Mladineo, I.; et al. Parasite detection in food: Current status and future needs for validation. Trends Food Sci. Technol. 2020, 99, 337–350. [Google Scholar] [CrossRef]
- Kourenti, C.; Karanis, P. Development of a sensitive polymerase chain reaction method for the detection of Toxoplasma gondii in water. Water Sci. Technol. 2004, 50, 287–291. [Google Scholar] [CrossRef][Green Version]
- Cook, N.; Paton, C.A.; Wilkinson, N.; Nichols, R.A.; Barker, K.; Smith, H.V. Towards standard methods for the detection of Cryptosporidium parvum on lettuce and raspberries. Part 1: Development and optimization of methods. Int. J. Food Microbiol. 2006, 109, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- International Standards Organisation. Microbiology of Food and Animal Feeding Stuffs–Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens—General Requirements and Definitions (ISO 22174:2005); International Standards Organisation: Geneva, Switzerland, 2005. [Google Scholar]
- Hoorfar, J.; Malorny, B.; Abdulmawjood, A.; Cook, N.; Wagner, M.; Fach, P. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 2004, 42, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Why the need for qPCR publication guidelines?—The case for MIQE. Methods 2010, 50, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- International Standards Organisation. Microbiology of Food and Animal Feeding Stuffs—Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens (ISO 22174:2005); International Standards Organisation: Geneva, Switzerland, 2005. [Google Scholar]
- International Standards Organisation. Microbiology of Food and Animal Feeding Stuffs—Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens—Performance Testing for Thermal Cyclers (ISO/TS 20836:2005); International Standards Organisation: Geneva, Switzerland, 2005. [Google Scholar]
- International Standards Organisation. Microbiology of Food and Animal Feeding Stuffs—Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens—Requirements for Sample Preparation for Qualitative Detection (ISO 20837:2006); International Standards Organisation: Geneva, Switzerland, 2006. [Google Scholar]
- International Standards Organisation. Microbiology of Food and Animal Feeding Stuffs—Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens—Requirements for Amplification and Detection for Qualitative Methods (ISO 20838:2006); International Standards Organisation: Geneva, Switzerland, 2006. [Google Scholar]
- International Standards Organisation. Microbiology of Food and Animal Feeding Stuffs—General Requirements and Guidance for Microbiological Examinations (ISO 7218:2007 and Amendments 2013); International Standards Organisation: Geneva, Switzerland, 2013. [Google Scholar]
- International Standards Organisation. Microbiology of the Food Chain—Method Validation—Part. 2: Protocol for the Validation of Alternative (Proprietary) Methods against a Reference Method (ISO 16140-2:2016); International Standards Organisation: Geneva, Switzerland, 2016. [Google Scholar]
- Food and Drug Administration (US); Foods Program Regulatory Science Steering Committee (RSSC). Guidelines for the Validation of Analytical Methods for the Detection of Microbial Pathogens in Foods and Feeds, 3rd ed.; U.S. FDA: Silver Spring, MD, USA, 2019.
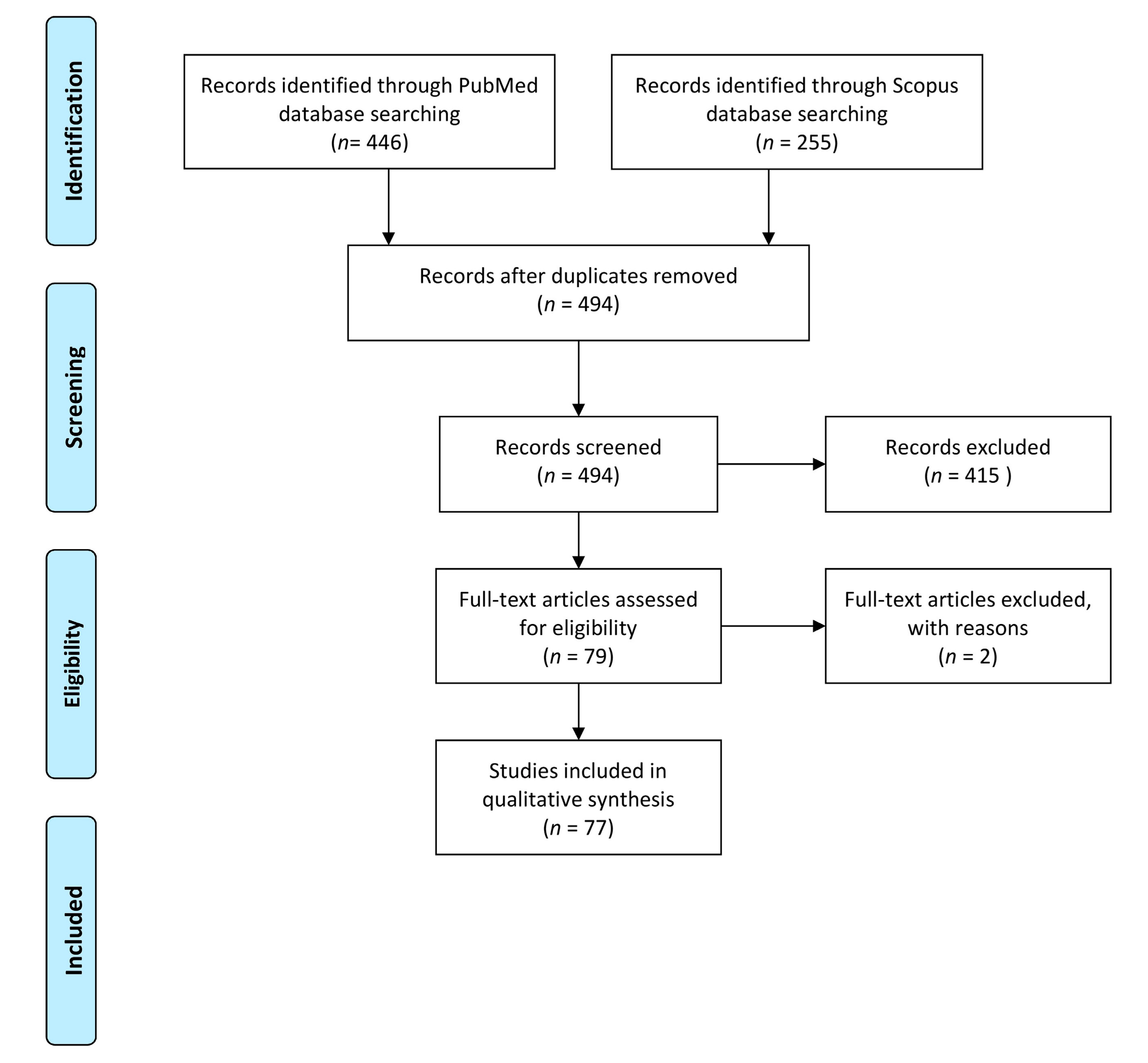

| Type of Spiking | Matrix (Grams) | Spiking Level (oo)Cysts | Time after Spiking | Processing Method | Washing Buffer (mL) | Recovery (%) (Quantitative Evaluation) ### | Pre-Treatment before DNA Extract | DNA Extraction | Detection Method (Target Gene) | Amplicon Size (bp) | LoD (Oocysts) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dripping | basil (25) | 102 | ON at 4 °C | wash by hand shaking for 15 s, hand rubbing for 30 s, shaking vigorously for 15 s and centrifugtion | six different buffers (200 mL) # | NR | BB (5.5 m/s for 30 s) | Fast DNA Spin for Soil kit | PCR (529 RE) | 529 | depend on the washing buffer ## | [12] |
| basil (30) raspberries (30) | 5 to 104 | 2 h at RT | wash by automatic shaker (80 rpm 10 min), centrifugation, IMS Toxo | 1 M glycine pH 5.5 (200 mL) | basil: 0.2% microscopy; 35% qPCR raspberry: 2% microscopy; 29% qPCR | FT 6× (−80 °C for 5 min/95 °C for 5 min) and US (1 min at 37 Hz) | InstaGene Matrix | qPCR Taqman (529 RE) | 81 | Basil: <33/g Raspberries: <33/g | [13] | |
| wash by automatic shaker (80 rpm 10 min), centrifugation | basil: 35% qPCR raspberries: 2.5% qPCR | Basil: <1/g Raspberries: <1/g | ||||||||||
| baby lettuce (50) | 25, 50, 100 for 50 g | ON at 4 °C | wash by stomaching b and centrifugation | 1 M glycine pH 5.5 (200 mL) | NR | BB 2× (6.0 m/s for 40 s) | FastPrep for soil kit | LAMP (529 RE) | NA | 0.5/g or 5/mL | [14] | |
| 5, 10, 50 in 800 µL (pellet) | ON at −20 °C | qPCR_Taqman (529 RE) | 163 | |||||||||
| spinach (10) | 101 to 104 | 2 h at RT | wash by stomaching and centrifugation | 0.1% Tween 80 (100 mL) | ≤30% by microscopy (IMS/membrane filtration) | FT 1× (4 min in N2 and 4 min 100°C) | DNeasy Blood and Tissue Kit | nPCR (rDNA 18S) | 715 | 0.1–1/g | [15] | |
| manual wash and centrifugation | ≥40% by microscopy (IMS/membrane filtration) | qPCR_Taqman (529 RE) | 163 | 1/g | ||||||||
| strawberry (50) lettuce (50) | 101 to 104, in 100 mL ddH2O for 250 g sample | 30 min at RT | wash by manual stirring, filtration through a cellulose ester membrane and centrifugation | 1% Tween 80 (100 mL) | NR | US 5× (45 s at 20 Hz, at 2 min intervals | Axy Prep Blood Genomic DNA kit | PCR (B1 gene) | 115 | 1000/250 g by immersion using any pre-treatment | [16] | |
| 10/250 g by drip using FT or US | ||||||||||||
| blackberries, blueberries, cranberries, raspberries, strawberries (60) herbs a (35) green onions (35) | 5 × 103 Eimeria papillata | ON at 4 °C | wash by orbital shaking in bottles for 1 min, centrifugation and Sheather’s solution in the flotation | 1 M glycine pH 5.5 (200 mL) | higher for blueberries, leafy herbs and thyme | FT 8× (1 min in N2/1 min at 95 °C) and proteinase K | spin columns (QIAamp DNA Mini Kit | qPCR_HRM (18S rDNA) | 312 | For all berry types, 3/g For herbs and green onions, 5–9/g | [17] * | |
| ON at RT | wash by stomaching in stomacher filter bag, centrifugation and Sheather’s solution in the flotation | elution solution 0.563 mM H2Na2P2O7/42.8 mM NaCl (200 mL) | higher for blackberries, cranberries, raspberries and green onions | |||||||||
| wash by shaking in stomacher filter bag (30 min per side), centrifugation and Sheather’s solution in the flotation | ||||||||||||
| immersion | lettuce (50) | 10 to 104, in 2000 mL ddH2O | NR | wash by manual stirring, filtration through a cellulose ester membrane and centrifugation | 1% Tween 80 (100 mL) | NR | FT 5× (5 min in N2/5 min at 65 °C) | spin columns (Axy Prep Blood Genomic DNA | PCR (B1 gene) | 115 | ≥10/ µL | [18] |
| PCR (529 RE) | 529 | ≥100/ µL | ||||||||||
| strawberry (50) lettuce (50) | 101–104, in 2000 mL ddH2O for 250 g sample | stirred 15 s followed by 30 min incubation at RT | wash by manual stirring, filtration through a cellulose ester membrane and centrifugation | 1% Tween 80 (100 mL) | NR | NO | Axy Prep Blood Genomic DNA kit | PCR (529 RE) | 529 | 1000/250 g by immersion using FT or US | [16] | |
| FT 5× (5 min in N2/5 min at 65 °C) | 100/250 g by drip using FT or US | |||||||||||
| NR | radish (NR) strawberries (NR) | 101 to 104 | NR | wash in glass beaker by automatic shaker (100 rpm 2 h), flocculation method using CaCO3 solution ON, centrifugation | 1% Tween 80 (2000 mL) | NR | BB (6.5 m/s for 2 min), chloroform treatment, and proteinase K incubation | FAST-Prep for soil matrix E combined with Genomic Mini Kit | qPCR_Taqman (B1 gene) | 128 | 100/single radish 10,000/single strawberries | [19] |
| NR | raspberries (30) blueberries (30) | 10 or 50 in 250 µL (sediment) | NA | wash by automatic shaker (300-600 rpm 10 min) and centrifugation | 1% Alconox (200 mL) | NA | BB 2× (4 m/s for 60 s) | DNeasy Power Soil | multiplex qPCR_Taqman (529 RE) | 163 | 10/250 µL (sediment from 30 g berries) | [20] |
| Matrix (Number) | Source (Number) | Amount of Sample Tested | Processing Method | Washing Buffer (mL) | Pre-Treatment before DNA Extraction | DNA Extraction | Detection Method (Target Gene) | Amplicon Size (bp) | LoD (Oocysts) | Sample Positive (%) | Reference |
| strawberries (60) carrot (46) radish (60) lettuce (50) | retail shops and marketplaces (175) kitchen-gardens and allotments (41) | 1 kg strawberries, 0,5 kg carrot, 20 radishes, 1 lettuce | wash in glass beaker? by automatic shaker (100 rpm 2 h), flocculation method using CaCO3 solution ON, centrifugation | 1% Tween 80 (2000 mL) | BB (6.5 m/s for 2 min) | Chloroform, proteinase K incubation and Genomic Mini Kit | qPCR_Taqman (B1) | 128 | 1 | 21/216 (10% ) 3 radish 9 carrot 9 lettuce | [19] * |
| arugula/baby arugula (107) kale (44) spinach/baby spinach (387) romaine (113) chard (39) leaf lettuces1 (226) spring mix (124) leafy green mixes (91) other mixes (3) | retail outlets (1171) | 35 g | wash by orbital shaker or stomacher (romaine, red or green leafy lettuces only), centrifugation and flotation procedure using Sheather’s solution | 1 M glycine pH 5.5 (200 mL) | FT 8× (1 min N2/1 min. 95 °C) and proteinase K | QIAamp DNA Micro Kit or Dneasy Blood and Tissue Kit | Multiplex qPCR_HRM (18S rDNA) | 312 | 10 | 3/387 (0.78%) baby spinach | [21] |
| crisp lettuce (106) regular lettuce (62) chicory (40) rocket (7) parsley (5) | open fairs (77) from producers’ fairs (81), from community fairs (80) | 50 g | wash by manual stirring, filtration through a cellulose ester membrane and centrifugation | 1% Tween 80 (100 mL) | FT 5× (5 min N2/5 min. 65 °C) | Axy Prep Blood Genomic DNA | PCR (B1) | 115 | NR | 9/238 (3.8 %) 4 crisp lettuce 1 chicory 1 rocket 1 parsley | [18]* |
| PCR (529 RE) | 529 | NR | 1 chicory 1 regular lettuce | ||||||||
| RTE mixed salad (curly and escarole lettuce, red radish, rocket salad and carrots) (648) | retail shops (648) | 100 g (9 salad packages, 72 pools) | wash by orbital shaker for 15 min at 120 rpm and centrifugation | 10X PBS, 0.1% Tween-80, 0.1% SDS and 0.05% antifoam B (200 mL) | FT 15× (1 min N2/1 min. 95 °C) | QiAmp Plant Mini Kit | qPCR_HRM (B1) | 128 | NR | 0.8% | [22] |
| lettuce (71) spinach (50) pak choi (34) chinese cabbage (26) rape (22) asparagus (18) chrysanthemum coronarium (16) endive (14) chinese chives (11) cabbage (9) red cabbage (8) | open markets | NR | sample rinsing and Al2(SO4)3 flocculation of washing suspensions | NR (NR) | FT 10× (N2/water bath) | TIANamp Micro DNA Ki | qPCR_Taqman (B1) | 129 | 1 | 10/279 (3.6%) 5 lettuce 2 spinach, 1 pak choi 1 red cabbage 1 rape | [23] |
| carrots (93) slicing cucumbers (109) salads (90) (butterhead lettuce, iceberg lettuce, little gem and lollo lettuce) | 9 farms (292) | 100 g | wash by automatic shaker for 20 min and centrifugation | Tris–glycine beef extract pH 9.5 (230 mL) | BB (6400 rpm for 60 s) | Power-Soil DNA isolation kit | Triplex qPCR_Taqman (B1 + 529RE) | 129 (B1) 163 (529 RE) 157 (IAC) | NR | 28/292 (9.6%) 7 Carrots 13 cucumbers 8 salads | [24] |
| Matrix (Amount) | Spiking Level | Pre-Treatment before DNA Extract | DNA Extraction | Detection (Target Gene) | Limit of Detection (oo)cysts | Reference |
|---|---|---|---|---|---|---|
| faeces (200 mg) | 101–104 | BB | NucleoSpin Soil using Buffer SL2 + Enhancer SLX | PCR (529 RE) | 10 | [25] |
| BB | ZR fecal DNA Kit | 10 | ||||
| FT 3× (10 min at −20 °C/2 min at RT) | phenol/chloroform extraction (in-house) | 100 | ||||
| water or mussels tissues (NR) | 100–103 | FT 5× (liquid N2/70 °C) +BB (glass beads, 30 s at 4200 rpm) + proteinase K 1 h at 56 °C | spin column | PCR (529 RE) | 1 (100%) in water and hemolymph | [26] |
| vortexing (PowerSoil beads max speed for 10 min) + BB (30 s at 4200 rpm) | PowerSoil™ DNA Isolation Kit | 10 (100%) in water and hemolymph, (50%) in dig. glands; 100 (100%) in gills and dig. glands | ||||
| water or mussels tissue homogenate (100 µl) | 100–103 | FT 1× (4 min liquid N2/4 min 100 °C) | spin column | nested PCR (B1) | 100 (100%) with 1, 3 or 6 cycles; 10 (60%) with 1 or 6 cycles; 1 (30%) with 1 cycles | [27] |
| FT 3× (4 min liquid N2/4 min 100 °C | ||||||
| FT 6× (4 min liquid N2/4 min 100 °C |
| Target Gene | Amplicon Size (bp) | LoD (Number of Spiked Oocysts That Provide Positive Amplification) | Primer Pairs | Matrix | Reference |
|---|---|---|---|---|---|
| B1 | 115 | ≥10 oocysts in 250 g of strawberry or 1 lettuce head | B22: 5′-AACGGGCGAGTAGCACCTGAGGAGA-3′ B23: 5′-TGGGTCTACGTCGATGGCATGACAAC-3′ | fresh produce | [16] |
| 10 oocysts | water | [16] | |||
| 10 oocysts/µL spiking level | fresh produce | [18] | |||
| 194 | NR | Oligo1: 5′-GGAACTGCATCCGTTCATGAG-3′ Oligo4: 5′-TCTTTAAAGCGTTCGTGGTC-3′ | soil | [36] | |
| 50 tachyzoites /0.5 g soil | soil | [37] | |||
| NR | soil | [34] | |||
| 100 oocysts | faeces | [38] | |||
| 25 oocysts/30 g soil or ≤1 oocyst/1g soil | soil | [39] | |||
| 529 RE | 529 | ≥100 oocyst in 250 g of strawberry or 1 lettuce head | TOX4: 5′-CTGCAGGGAGGAAGACGAAAGTTG-3′ TOX5: 5′-CTGCAGACAGAGTGCATCTGGATT-3′ | Fresh produce | [16] |
| 10 oocysts | water | [16] | |||
| ≥100 oocysts/ µl spiking level | fresh produce | [18] | |||
| NR | faeces | [40] | |||
| 1 oocyst in water and mussel hemolymph | mussels | [26] | |||
| 100 oocysts in mussel gills and dig. Glands | mussels | [26] | |||
| NR | oysters | [32] | |||
| NR | food | [12] | |||
| 5 tachyzoites /0.5 g soil | soil | [37] | |||
| 10 oocysts | faeces | [38] | |||
| 1–2 oocysts per 200 µL | faeces | [35] | |||
| 450 | 1 oocyst | TOX-8: 5′-CCCAGCTGCGTCTGTCGGGAT-3′ TOX-5: 5′-GACGTCTGTGTCACGTAGACCTAAG-3′ | faeces | [38] | |
| 10 oocysts/200 mg feces | faeces | [25] | |||
| 529 and 450 | NR | TOX4/TOX5 and TOX-8/TOX-5 | faeces | [41] | |
| 134 | NR | TOXO-F: 5′ AGGCGAGGGTGAGGATGA-3′ TOXO-R: 5′-TCGTCTCGTCTGGATCGCAT-3’ | soil | [34] | |
| NR | soil | [36] |
| Type of qPCR | N° of Studies | Target Gene | N° of Studies |
|---|---|---|---|
| MC/HRMC | 9 | B1 | 4 |
| 529RE | 1 | ||
| 18SrDNA | 3 | ||
| B1 and 529RE | 1 | ||
| Taqman | 26 | B1 | 7 |
| 529RE | 16 | ||
| ITS1 | 1 | ||
| 18SrDNA | 1 | ||
| B1 and 18SrDNA | 1 | ||
| FRET | 1 | 529RE | 1 |
| HRM and FRET | 1 | B1 and 529RE | 1 |
| MC and Taqman | 1 | B1 | 1 |
| Total | 38 | 38 |
| Target | Amplicon Size (bp) | IAC (Y/N) | Primer Sequence | Analytical Specificity | Analytical Sensitivity | LoD (Oocysts/g) | LoQ | Matrix | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 529 RE | 169 | N | Tox-9F: 5′-AGGAGAGATATCAGGACTGTAG-3′ Tox-11R: 5′-GCGTCGTCTCGTCTAGATCG-3′ Tox-TP1: 5′-6-FAM-CCGGCTTGGCTGCTTTTCCT-BHQ1-3′ | [14] | 90–100% | 5 | NR | mussel hemolymph | [28] |
| 100 oocysts (90%) 10 oocyst (30%) | 10–100 | mussel tissue | |||||||
| Y | Tox-9F; Tox-11R and Tox-TP1 | NR | 100% 2000 oocysts/g | 200 | NR | soil | [44] | ||
| 164 | Y a | Tox-9F; Tox-11R and Tox-TP1 | 1 with 95% CI of 0.69–1.00 | 100% (Crypto or Neospora) | 50 fg/µL | NR | water | [45] | |
| N | Tox-9F; Tox-11R and Tox-TP1 | NR | NR | 1–10 in water and hemolymph | NR | hemolymph and mussel tissue | [26] | ||
| 163 | Y b | Tox-9F; Tox-11R and Tox-TP1 5’-HEX | NR | NR | NR | NR | fresh produce | [24] f | |
| NR | NR | NR | NR | water | |||||
| 162 | N | Tox-9F; Tox-11R and Tox-TP1 5’-Cy5 | In silico 100% | NR | 0.3 | NR | raspberries blueberries | [20] | |
| N | Tox-9F; Tox-11R and Tox-TP1 | 100% (test on parasites, mammalian and plant DNA) | 50–100 oocysts (100%) 25 oocysts (83%) | 0.5 | baby lettuce | [14] | |||
| 81 | Y | [42] Toxo-P 5’-Cy3-ACGCTTTCCTCGTGGTGATGGCG-BHQ2-3’ | NR | NR | 0.1–1 | 5 oocyst/5 g | mussel | [29] | |
| NR | NR | 10–50 | 100 oocysts | haemolymph | |||||
| N | [42] | NR | NR | NR | NR | faeces | [46] | ||
| N | [42]; Toxo-P5′ | NR | NR | NR | NR | mussels | [47] | ||
| NR | NR | NR | NR | water | |||||
| N | [42] | NR | NR | NR | NR | soil | [48] | ||
| N | [42]; Toxo-P5′ | NR | NR | NR | NR | faeces | [49] | ||
| N | [42]; Toxo-P5′ | NR | NR | <1 | NR | basil leaves raspberries | [13] | ||
| N N | [42]; Toxo-P5′ | NR | NR | NR | NR | hemolymph and mussel tissue | [50] | ||
| NR | NR | NR | NR | water | |||||
| N | [42] | NR [51] | Reported in [42] | 10-100 | NR | soil | [52] | ||
| N | FOR_5′-AGAGACACCGGAATGCGATCT-3′ REV_5′-CCCTCTTCTCCACTCTTCAATTCT-3′ Probe 5′-6FAM-ACGCTTTCCTCGTGGTGATGGGG-3´TAMRA | NR | NR | 10-100 | NR | soil | [42] | ||
| N | [42] Probe Tox-HP-1 GAGTCGGAGAGGGAGAAGATGTT-FL Probe Tox-HP-2 Red 640-CCGGCTTGGCTGCTTTTCCTG-Ph | NR | NR | NR | NR | faeces | [53] | ||
| B1 | 129 | Y c | [54] | NR | NR | NR | NR | fresh produce | [23] |
| Y b | [54] ToxB-69p 5′-FAM-ACCGCGAGATGCACCCGCA- BHQ -3′ | NR | NR | NR | NR | fresh produce | [24] f | ||
| NR | NR | NR | NR | water | |||||
| ToxB-41: 5′′-TCGAAGCTGAGATGCTCAAAGTC-3′ ToxB-169 5′′-AATCCACGTCTGGGAAGAACTC-3′ 5′′-FAM-ACCGCGAGATGCACCCGCA TAMRA-3′ | tested by sequencing | 10 molecules of plasmid | 100 | NR | fruits and vegetables | [19] | |||
| 98 | Y d | Toxo-F 5′-TCCCCTCTGCTGGCGAAAAGT-3′ Toxo-R 5′-AGCGTTCGTGGTCAACTATCGATTG-3′ probe V5′-FAM-TCTGTGCAACTTTGGTGTATTCGCAG-3′ TAMRA | NR | NR | NR | NR | water | [43] | |
| N | [43] Toxo-F; Toxo-R and probe V5′ | NR | NR | NR | 1 | water | [55] | ||
| NR | NR | NR | 250 | faeces | |||||
| Y (N.S.) | [43] Toxo-F; Toxo-R and probe V5′ | NR | NR | NR | NR | water | [54] | ||
| 62 | N | TX2-F 5″-CTAGTATCGTGCGGCAATGTG-3′ TX2-R 5′-GGCAGCGTCTCTTCCTCTTTT-3′ TX2M1 5″-(6-FAM)-CCACCTCGCCTCTTGG-(NFQ-MGB)-3′ | NR | NR | 5 genomic copies/µL | 50 genomic copies/µL | water | [31] | |
| 18S rRNA | NR | N | [56] Tox18-213f 5′-CCGGTGGTCCTCAGGTGAT-3′ Tox18-332r 5′-TGCCACGGTAGTCCAATACAGTA-3′ Tox18-249p 5′-FAM-ATCGCGTTGACTTCGGTCTGCGAC-TAMRA-3′ | NR | NR | NR | NR | water | [57] |
| 120 | Y e | Tox18-213f; Tox18-332r and Tox18-249p | 100% (various parasites tested) | NR | 10 molecules | NR | hemolymph and mussels | [58] | |
| ITS1 | NR | N | For 5′-GATTTGCATTCAAGAAGCGTGATAGTA-3′ Rev 5′-AGTTTAGGAAGCAATCTGAAAGCACATC-3′ Probe 5′-/-TET/-CTGCGCTGC/ZEN/TTCCAATATTGG-/-IABkFQ-/-3′ | NR | NR | NR | NR | oysters | [59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slana, I.; Bier, N.; Bartosova, B.; Marucci, G.; Possenti, A.; Mayer-Scholl, A.; Jokelainen, P.; Lalle, M. Molecular Methods for the Detection of Toxoplasma gondii Oocysts in Fresh Produce: An Extensive Review. Microorganisms 2021, 9, 167. https://doi.org/10.3390/microorganisms9010167
Slana I, Bier N, Bartosova B, Marucci G, Possenti A, Mayer-Scholl A, Jokelainen P, Lalle M. Molecular Methods for the Detection of Toxoplasma gondii Oocysts in Fresh Produce: An Extensive Review. Microorganisms. 2021; 9(1):167. https://doi.org/10.3390/microorganisms9010167
Chicago/Turabian StyleSlana, Iva, Nadja Bier, Barbora Bartosova, Gianluca Marucci, Alessia Possenti, Anne Mayer-Scholl, Pikka Jokelainen, and Marco Lalle. 2021. "Molecular Methods for the Detection of Toxoplasma gondii Oocysts in Fresh Produce: An Extensive Review" Microorganisms 9, no. 1: 167. https://doi.org/10.3390/microorganisms9010167
APA StyleSlana, I., Bier, N., Bartosova, B., Marucci, G., Possenti, A., Mayer-Scholl, A., Jokelainen, P., & Lalle, M. (2021). Molecular Methods for the Detection of Toxoplasma gondii Oocysts in Fresh Produce: An Extensive Review. Microorganisms, 9(1), 167. https://doi.org/10.3390/microorganisms9010167





