A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha’il Hospitals, Saudi Arabia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ha’il City and All Its Socio-Economic Strata
2.2. Study Designs, Data Sources, and Statistical Analysis (From September to December 2020)
2.3. Classification as Multi-Resistant, Extremely-Resistant, and Pan-Resistant Types (MDR, XDR, PDR)
3. Results
3.1. Kleb. pneumoniae (MDR)
3.2. E. coli (MDR)
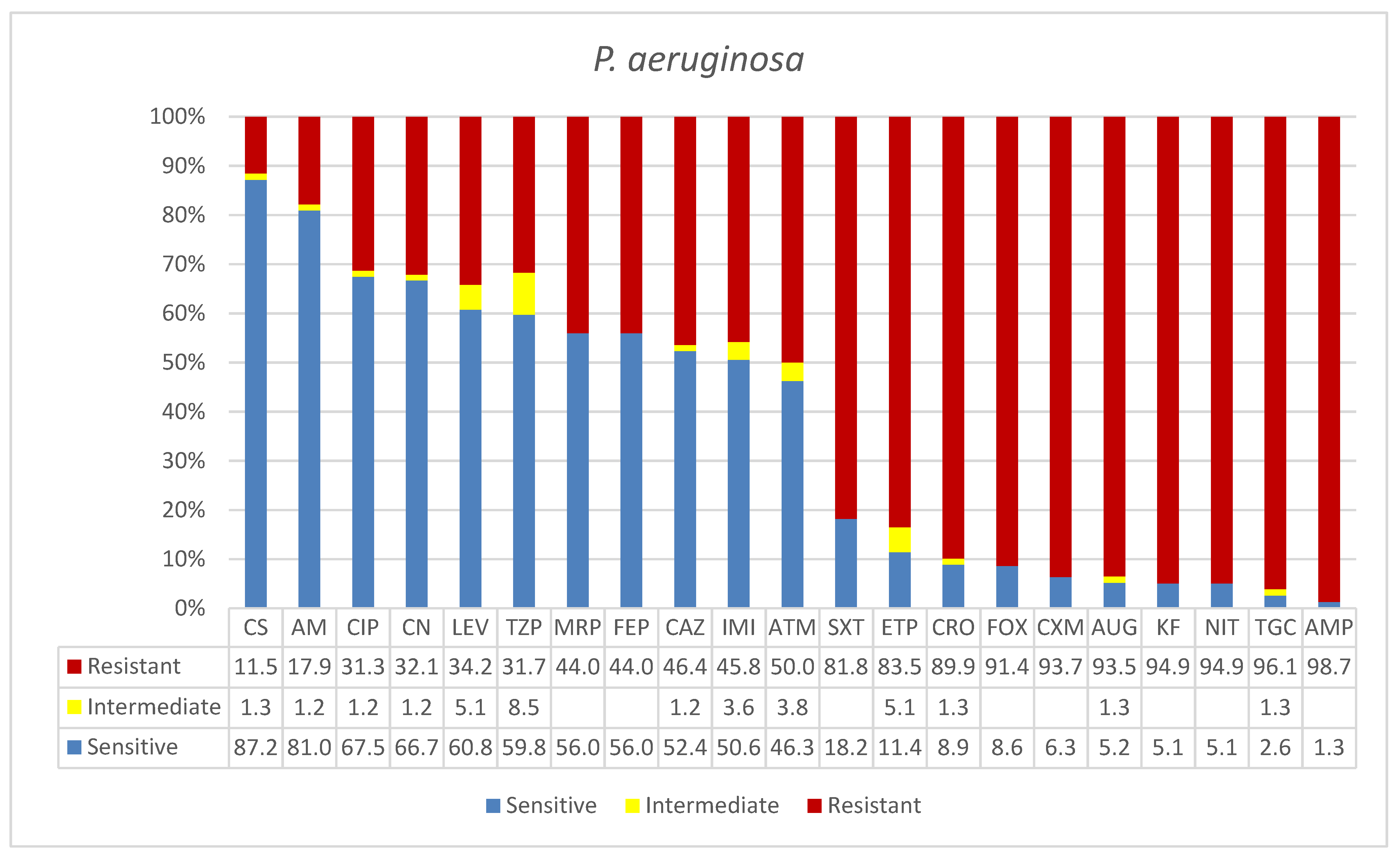
3.3. Pseudomonas aeruginosa (XDR)
3.4. Acinetobacter baumannii (PDR)
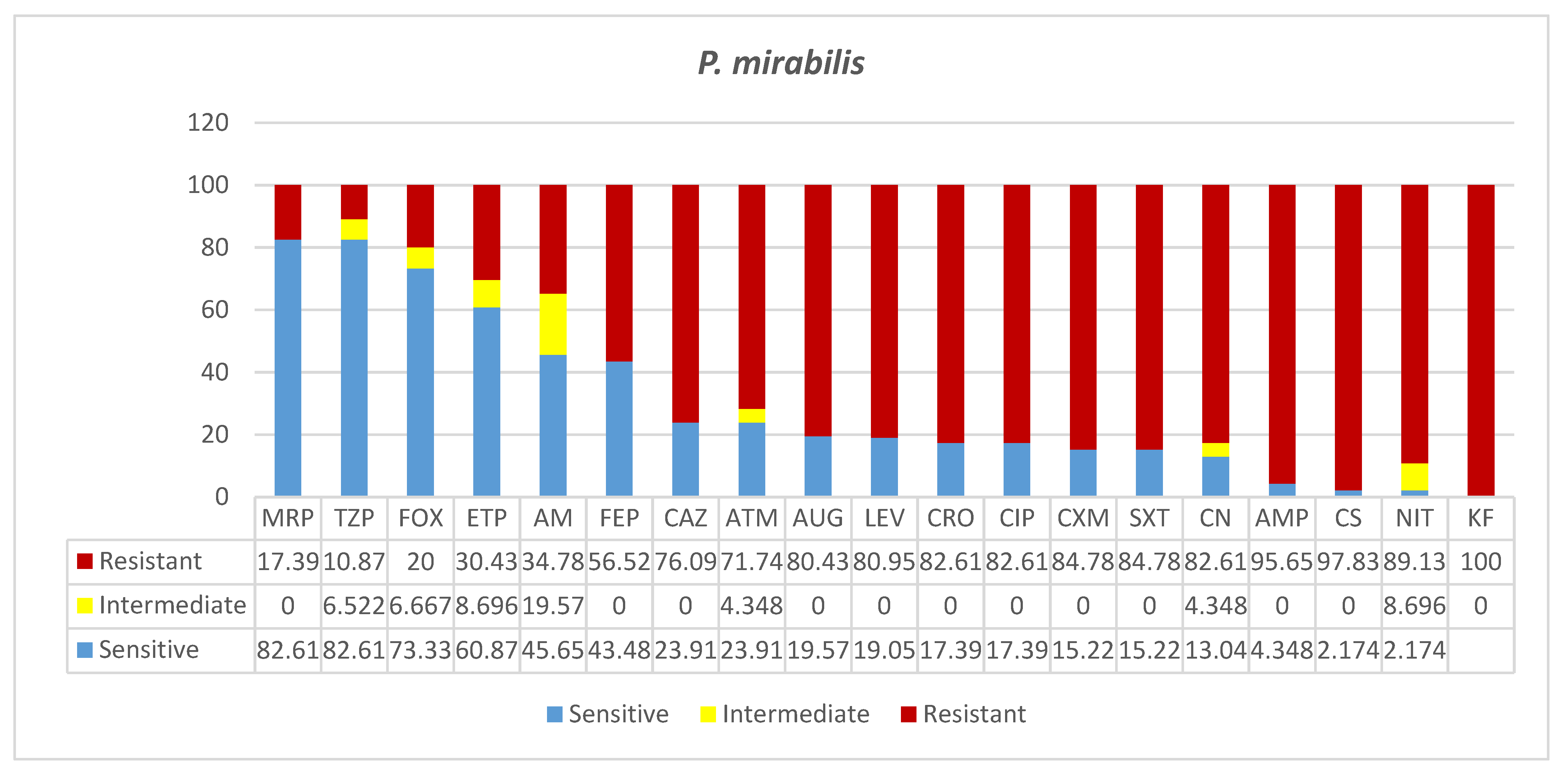
3.5. Proteus mirabilis (MDR)
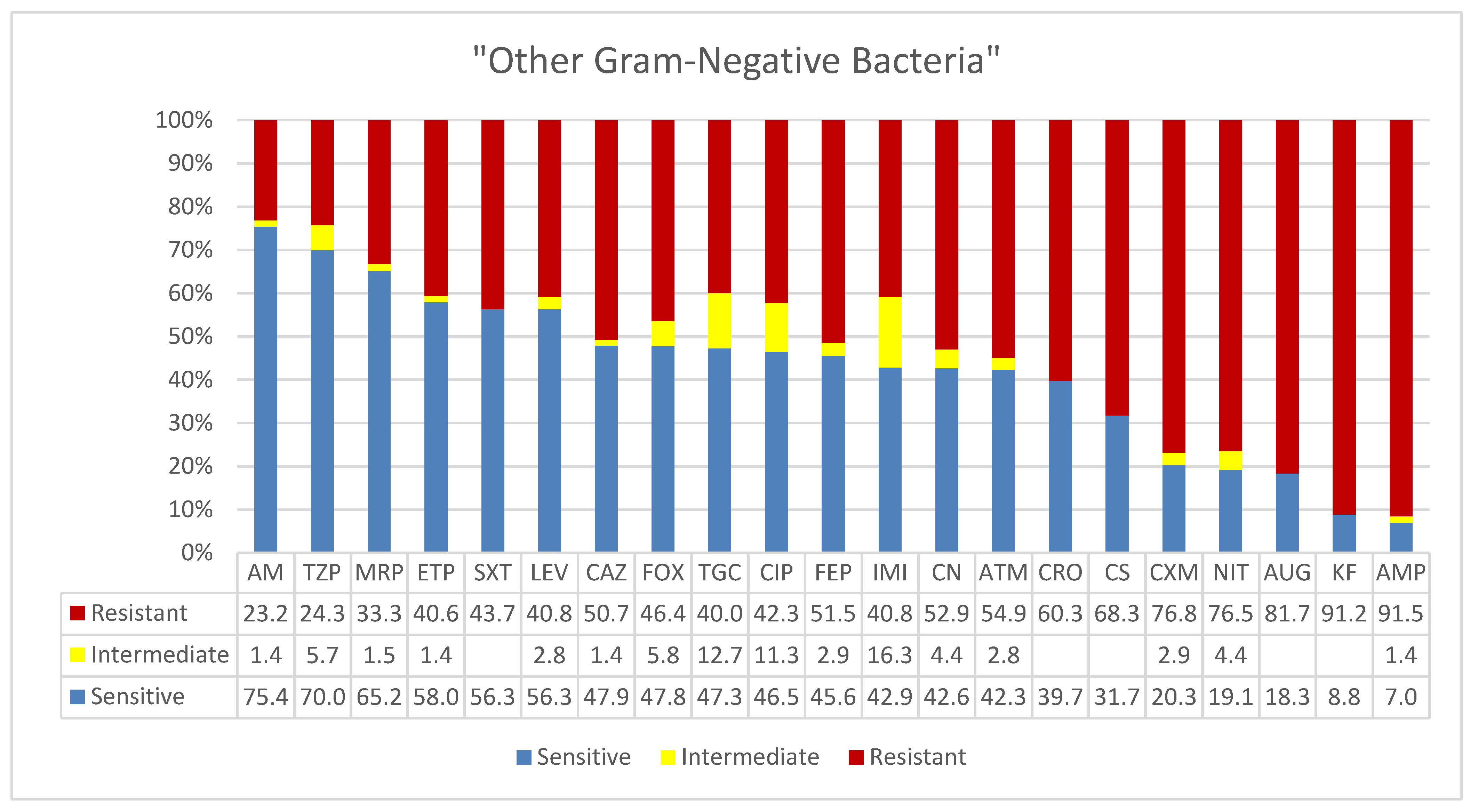
3.6. Other Gram-Negative Bacteria (MDR)
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mshana, S.E.; Gerwing, L.; Minde, M.; Hain, T.; Domann, E.; Lyamuya, E.; Chakraborty, T.; Imirzalioglu, C. Outbreak of a novel Enterobacter sp. carrying bla CTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int. J. Antimicrob. Agents 2011, 38, 265–269. [Google Scholar] [PubMed] [Green Version]
- Mshana, S.E.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T.; Imirzalioglu, C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect. Dis. 2013, 13, 466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willis, R.; Heslop, O.; Bodonaik, N.; Thame, M.; Smikle, M. Morbidity, mortality and antimicrobial resistance of pneumococcal infections in the Jamaican paediatric and adult populations. Hum. Antibodies 2019, 27, 155–160. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antimicrobial Resistance: Global Report on Surveillance. Available online: https://scholar.google.de/scholar?hl=en&q=Antimicrobial+resistance%3A+global+report+on+surveillance&btnG=&as_sdt=1%2C5&as_sdtp= (accessed on 17 October 2019).
- Haque, M.; Sartelli, M.; McKimm, J.; Bakar, A.M. Health care-associated infections—An overview. Infect. Drug Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plachouras, D.; Kärki, T.; Hansen, S.; Hopkins, S.; Lyytikäinen, O.; Moro, M.L.; Reilly, J.; Zarb, P.; Zingg, W.; Kinross, P.; et al. Antimicrobial use in European acute care hospitals: Results from the second point prevalence survey (PPS) of healthcare-associated infections and antimicrobial use, 2016 to 2017. Eurosurveillance 2018, 23, 1800393. [Google Scholar] [CrossRef] [Green Version]
- Ricchizzi, E.; Latour, K.; Kärki, T.; Buttazzi, R.; Jans, B.; Moro, M.L.; Nakitanda, O.A.; Plachouras, D.; Monnet, D.L.; Suetens, C.; et al. Antimicrobial use in European long-term care facilities: Results from the third point prevalence survey of healthcare-associated infections and antimicrobial use, 2016 to 2017. Eurosurveillance 2018, 23, 1800394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suetens, C.; Latour, K.; Kärki, T.; Ricchizzi, E.; Kinross, P.; Moro, M.L.; Jans, B.; Hopkins, S.; Hansen, S.; Lyytikäinen, O.; et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: Results from two European point prevalence surveys, 2016 to 2017. Eurosurveillance 2018, 23, 1800516. [Google Scholar] [CrossRef] [Green Version]
- Hays, J.P.; Mitsakakis, K.; Luz, S.; van Belkum, A.; Becker, K.; van den Bruel, A.; Harbarth, S.; Rex, J.H.; Simonsen, G.S.; Werner, G.; et al. The successful uptake and sustainability of rapid infectious disease and antimicrobial resistance point-of-care testing requires a complex ‘mix-and-match’ implementation package. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1015–1022. [Google Scholar] [CrossRef] [Green Version]
- De Kraker, M.; Jarlier, V.; Monen, J.; Heuer, O.; Van De Sande, N.; Grundmann, H. The changing epidemiology of bacteraemias in Europe: Trends from the European antimicrobial resistance surveillance system. Clin. Microbiol. Infect. 2013, 19, 860–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenthal, V.D.; Bijie, H.; Maki, D.G.; Mehta, Y.; Apisarnthanarak, A.; Medeiros, E.A.; Leblebicioglu, H.; Fisher, D.; Álvarez-Moreno, C.; Khader, I.A. International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004–2009. Am. J. Infect. Control 2012, 40, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, L.E.; Herpichboehm, B.; Kost, G.J.; Kollef, M.H.; Stüber, F. Cost and mortality prediction using polymerase chain reaction pathogen detection in sepsis: Evidence from three observational trials. Crit. Care 2010, 14, R186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safdar, N.; Anderson, D.J.; Braun, B.I.; Carling, P.; Cohen, S.; Donskey, C.; Drees, M.; Harris, A.; Henderson, D.K.; Huang, S.S.; et al. The evolving landscape of healthcare-associated infections: Recent advances in prevention and a road map for research. Infect. Control Hosp. Epidemiol. 2014, 35, 480–493. [Google Scholar] [CrossRef] [Green Version]
- The Centers for Disease Control and Prevention (CDC); National Healthcare Safety Network (NHSN). Surveillance Definitions for Specific Types of Infections. Available online: https://www.cdc.gov/nhsn/pdfs/pscmanual/17pscnosinfdef_current.pdf) (accessed on 20 February 2021).
- Allegranzi, B.; Pittet, D. Role of hand hygiene in healthcare-associated infection prevention. J. Hosp. Infect. 2009, 73, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.J.; Oake, N.; Roth, V.; Suh, K.N.; Majewski, J.; Leeder, C.; Van Walraven, C. Patient-level factors associated with methicillin-resistant Staphylococcus aureus carriage at hospital admission: A systematic review. Am. J. Infect. Control. 2013, 41, 214–220. [Google Scholar] [CrossRef]
- Loveday, H.; Wilson, J.; Kerr, K.; Pitchers, R.; Walker, J.; Browne, J. Association between healthcare water systems and Pseudomonas aeruginosa infections: A rapid systematic review. J. Hosp. Infect. 2014, 86, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Allegranzi, B.; Pittet, D. Preventing infections acquired during health-care delivery. Lancet 2008, 372, 1719–1720. [Google Scholar] [CrossRef]
- Patel, J.; Cockerill, F., III; Eliopoulous, G.; Jenkins, S.; Lewis, J.S.; Brandi, L.; Mathers, A.J.; Mazzulli, T.; Patel, R.; Richter, S.S.; et al. Performance standards for antimicrobial susceptibility testing (CLSI document M100S-26). In Tweenty-Sixth Informational Supplement, Clinical and Laboratory Standards; Clinical Laboratory Standard Institute: Annapolis Junction, MA, USA, 2016. [Google Scholar]
- Al-Tawfiq, J.A.; Memish, Z.A. The Hajj 2019 vaccine requirements and possible new challenges. J. Epidemiol. Glob. Health 2019, 9, 147–152. [Google Scholar] [CrossRef] [Green Version]
- Al-Abdely, H.M.; Alshehri, A.D.; Rosenthal, V.D.; Mohammed, Y.K.; Banjar, W.; Orellano, P.W.; Assiri, A.M.; Abedel Kader, N.M.; Al Enizy, H.A.; Mohammed, D.A.; et al. Prospective multicentre study in intensive care units in five cities from the Kingdom of Saudi Arabia: Impact of the International Nosocomial Infection Control Consortium (INICC) multidimensional approach on rates of central line-associated bloodstream infection. J. Infect. Prev. 2017, 18, 25–34. [Google Scholar]
- Al-Hajoj, S.; Varghese, B.; Shoukri, M.M.; Al-Omari, R.; Al-Herbwai, M.; Al Rabiah, F.; Alrajhi, A.A.; Abuljadayel, N.; Al-Thawadi, S.; Zumla, A.; et al. Epidemiology of antituberculosis drug resistance in Saudi Arabia: Findings of the first national survey. Antimicrob. Agents Chemother. 2013, 57, 2161–2166. [Google Scholar] [CrossRef] [Green Version]
- Al Watban, A.Z.; Al Salamah, A.; El Faki, M.G. Prevalence of suspected tuberculosis in the Kingdom of Saudi Arabia according to conventional and molecular methods. J. Fam. Commun. Med. 2014, 21, 182–185. [Google Scholar] [CrossRef] [Green Version]
- Abussaud, M.J. Incidence of wound infection in three different departments and the antibiotic sensitivity pattern of the isolates in a Saudi Arabian hospital. Acta Microbiol. Immunol. Hung. 1996, 43, 301–305. [Google Scholar]
- Tumala, R.B.; Almazan, J.; Alabdulaziz, H.; Felemban, E.M.; Alsolami, F.; Alquwez, N.; Alshammari, F.; Tork, H.M.M.; Cruz, J.P. Assessment of nursing students perceptions of their training hospital’s infection prevention climate: A multi-university study in Saudi Arabia. Nurse Educ. Today 2019, 81, 72–77. [Google Scholar] [CrossRef]
- Dandachi, I.; Chaddad, A.; Hanna, J.; Matta, J.; Daoud, Z. Understanding the epidemiology of multi-drug resistant gram-negative bacilli in the Middle East using a one health approach. Front. Microbiol. 2019, 10, 1941. [Google Scholar] [CrossRef] [Green Version]
- Memish, Z.A.; Assiri, A.; Almasri, M.; Roshdy, H.; Hathout, H.; Kaase, M.; Gatermann, S.G.; Yezli, S. Molecular characterization of carbapenemase production among gram-negative bacteria in Saudi Arabia. Microb. Drug Resist. 2015, 21, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Fayad, E.; El-Bazzal, B.; Daoud, Z.; Rolai, J.M. Prevalence of extended-spectrum beta-lactamase-producing gram-negative bacilli and emergence of mcr-1 colistin resistance gene in Lebanese swine farms. Microb. Drug Resist. 2019, 25, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Zaman, T.U.; Aldrees, M.; Al Johani, S.M.; Alrodayyan, M.; Faizah, A.; Aldughashem, F.A.; Balkhy, H.H. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2014, 28, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Zaman, T.U.; Alrodayyan, M.; Albladi, M.; Aldrees, M.; Siddique, M.I.; Aljohani, S.; Balkhy, H.H. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect. Dis. 2018, 18, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Balkhi, B.; Mansy, W.; Alghadeer, S.; Alnuaim, A.; AlShehri, A.; Somily, A. Antimicrobial susceptibility of microorganisms causing Urinary Tract Infections in Saudi Arabia. J. Infect. Dev. Ctries. 2018, 12, 220–227. [Google Scholar] [CrossRef] [Green Version]
- Duffa, Y.M.; Kitila, K.T.; Gebretsadik, D.M.; Bitew, A. Prevalence and Antimicrobial Susceptibility of Bacterial Uropathogens Isolated from Pediatric Patients at Yekatit 12 Hospital Medical College, Addis Ababa, Ethiopia. Int. J. Microbiol. 2018, 2018, 8492309. [Google Scholar] [CrossRef] [Green Version]
- Bandy, A.; Almaeen, A.H. Pathogenic spectrum of blood stream infections and resistance pattern in Gram-negative bacteria from Aljouf region of Saudi Arabia. PLoS ONE 2020, 15, e0233704. [Google Scholar] [CrossRef]
- Al-Zalabani, A.; AlThobyane, O.A.; AlShehri, A.H.; Al Rehaili, A.O.; Namankani, M.O.; Aljafri, O.H. Prevalence of Klebsiella pneumoniae antibiotic resistance in Medina, Saudi Arabia, 2014-2018. Cureus 2020, 12, e9714. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-F. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J. Clin. Cases 2014, 2, 787–814. [Google Scholar] [CrossRef]
- Sacco, F.; Visca, P.; Runci, F.; Antonelli, G.; Giammarco, G. Susceptibility testing of colistin for Acinetobacter baumannii: How Far Are We from the Truth? Antibiotics 2021, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Solgi, H.; Nematzadeh, S.; Giske, C.G.; Badmasti, F.; Westerlund, F.; Lin, Y.-L.; Goyal, G.; Nikbin, V.S.; Nemati, A.H.; Shahcheraghi, F. Molecular epidemiology of OXA-48 and NDM-1 producing enterobacterales species at a university hospital in Tehran, Iran, Between 2015 and 2016. Front. Microbiol. 2020, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Mehr, S.S.; Powell, C.V.E.; Curtis, N. Cephalosporin resistant urinary tract infections in young children. J. Paediatr. Child Health 2004, 40, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Cullen, I.M.; Manecksha, R.P.; Mccullagh, E.; Ahmad, S.; O’Kelly, F.; Flynn, R.; McDermott, T.E.D.; Murphy, P.; Grainger, R.; Fennell, J.P.; et al. An 11-year analysis of the prevalent uropathogens and the changing pattern of Escherichia coli antibiotic resistance in 38,530 community urinary tract infections, Dublin 1999–2009. Ir. J. Med Sci. 2012, 182, 81–89. [Google Scholar] [CrossRef]
- Gunduz, S.; Altun, H.U. Antibiotic resistance patterns of urinary tract pathogens in Turkish children. Glob. Health Res. Policy 2018, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryce, A.D.; Hay, A.D.; Lane, I.F.; Thornton, H.V.; Wootton, M.; Costelloe, C. Global prevalence of antibiotic resistance in paediatric urinary tract infections caused by Escherichia coli and association with routine use of antibiotics in primary care: Systematic review and meta-analysis. BMJ 2016, 352, i939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109e. [Google Scholar] [CrossRef] [PubMed]
- Azab, K.S.M.; Abdel-Rahman, M.A.; El-Sheikh, H.H.; Azab, E.; Gobouri, A.A.; Farag, M.M.S. Distribution of Extended-spectrum β-lactamase (ESBL)-encoding genes among multidrug-resistant gram-negative pathogens collected from three different countries. Antibiotics 2021, 10, 247. [Google Scholar] [CrossRef]
- Alonso, A.; Rojo, F.; Martõ, J.Â.L. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1999, 1, 421–430. [Google Scholar] [CrossRef]
- Grosso-Becerra, M.-V.; Santos-Medellín, C.; González-Valdez, A.; Méndez, J.-L.; Delgado, G.; Morales-Espinosa, R.; Servín-González, L.; Alcaraz, L.-D.; Soberón-Chávez, G. Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genom. 2014, 15, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.-C.; Wang, C.-Y.; Hsueh, P.-R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect. 2020, 53, 505–512. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Liu, H.G.; Liu, W.; Liu, J.; Liu, K.; Shang, J.; Deng, Y.; Wei, S. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jiehe He Huxi Zazhi 2020, 43, E005. [Google Scholar] [PubMed]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Ding, Q.; Lu, P.; Fan, Y.; Xia, Y.; Liu, M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol. 2020, 92, 1549–1555. [Google Scholar] [CrossRef]
- Xing, Q.; Li, G.; Xing, Y.; Chen, T.; Li, W.; Ni, W.; Deng, K.; Gao, R.-Q.; Chen, C.-Z.; Gao, Y.; et al. Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens. medRxiv 2020. [Google Scholar] [CrossRef]
- Li, H.; Chen, K.; Liu, M.; Xu, H.; Xu, Q. The profile of peripheral blood lymphocyte subsets and serum cytokines in children with 2019 novel coronavirus pneumonia. J. Infect. 2020, 81, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Arentz, M.; Yim, E.; Klaff, L.; Lokhandwala, S.; Riedo, F.X.; Chong, M.; Lee, M. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA 2020, 323, 1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Quinn, J.; Pinsky, B.; Shah, N.H.; Brown, I. Rates of Co-infection Between SARS-CoV-2 and other respiratory pathogens. JAMA 2020, 323, 2085–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, B.E.; Ong, S.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.-T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488. [Google Scholar] [CrossRef] [Green Version]
- Zangrillo, A.; Beretta, L.; Scandroglio, A.M.; Monti, G.; Fominskiy, E.; Colombo, S.; Morselli, F.; Belletti, A.; Silvanj, P.; Crivellari, M.; et al. Characteristics, treatment, outcomes and cause of death of invasively ventilated patients with COVID-19 ARDS in Milan, Italy. Crit. Care Resusc. 2020, 2, 200–211. [Google Scholar]
- Garcia-Vidal, C.; Sanjuan, G.; Moreno-García, E.; Puerta-Alcalde, P.; Garcia-Pouton, N.; Chumbita, M.; Fernandez-Pittol, M.; Pitart, C.; Inciarte, A.; Bodro, M.; et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: A retrospective cohort study. Clin. Microbiol. Infect. 2020, 27, 83–88. [Google Scholar] [CrossRef]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensiv. Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Vidal, C.; Barba, P.; Arnan, M.; Moreno, A.; Camps, I.R.; Gudiol, C.; Ayats, J.; Ortí, G.; Carratala, J. Invasive Aspergillosis Complicating Pandemic Influenza A (H1N1) Infection in Severely Immunocompromised Patients. Clin. Infect. Dis. 2011, 53, e16–e19. [Google Scholar] [CrossRef] [Green Version]
- Martin-Loeches, I.I.; Schultz, M.J.; Vincent, J.-L.; Alvarez-Lerma, F.F.; Bos, L.; Violán, J.S.J.; Torres, A.; Rodriguez, A.A. Increased incidence of co-infection in critically ill patients with influenza. Intensiv. Care Med. 2016, 43, 48–58. [Google Scholar] [CrossRef]
- Burrell, A.; Huckson, S.; Pilcher, D.V. ICU Admissions for sepsis or pneumonia in Australia and New Zealand in 2017. N. Engl. J. Med. 2018, 378, 2138–2139. [Google Scholar] [CrossRef] [PubMed]
- Schauwvlieghe, A.F.A.D.; Rijnders, A.B.J.; Philips, N.; Verwijs, R.; Vanderbeke, L.; Van Tienen, C.; Lagrou, K.; Verweij, P.; Van de Veerdonk, F.L.; Gommers, D.; et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: A retrospective cohort study. Lancet Respir. Med. 2018, 6, 782–792. [Google Scholar] [CrossRef]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Liu, D.; Zeng, X.; Peng, L.-S.; Yuan, Y.; Chen, Z.-F.; Zou, Q.-M.; Shi, Y. Aging exacerbates mortality of Acinetobacter baumannii pneumonia and reduces the efficacies of antibiotics and vaccine. Aging 2018, 10, 1597–1608. [Google Scholar] [CrossRef]
- Zeng, X.; Gu, H.; Cheng, Y.; Jia, K.-R.; Liu, D.; Yuan, Y.; Chen, Z.-F.; Peng, L.-S.; Zou, Q.-M.; Shi, Y. A lethal pneumonia model of Acinetobacter baumannii: An investigation in immunocompetent mice. Clin. Microbiol. Infect. 2018, 25, 516.e1–516.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, T.; Murray, C.K.; Vrahas, M.S.; Baer, D.G.; Tegos, G.P.; Hamblin, M.R. Ultraviolet C light for Acinetobacter baumannii wound infections in mice: Potential use for battlefield wound decontamination? J. Trauma Acute Care Surg. 2012, 73, 661–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armbruster, C.E.; Mobley, H.L.T.; Pearson, M.M. Pathogenesis of Proteus mirabilis infection. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Alamri, A.; Hamid, M.E.; Abid, M.; Alwahhabi, A.M.; Alqahtani, K.M.; Alqarni, M.S.; Abomughaid, M. Trend analysis of bacterial uropathogens and their susceptibility pattern: A 4-year (2013–2016) study from Aseer region, Saudi Arabia. Urol. Ann. 2018, 10, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.; Balshi, A. First literature review of carbapenem-resistant Providencia. New Microbes New Infect. 2018, 25, 16–23. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, C.M.; Brenner, F.W.; Miller, J.M. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev. 2000, 13, 534–546. [Google Scholar] [CrossRef]
- Abdallah, M.; Alhababi, R.; Alqudah, N.; Aldyyat, B.; Alharthy, A. First report of carbapenem-resistant Providencia stuartii in Saudi Arabia. New Microbes New Infect. 2018, 26, 107–109. [Google Scholar] [CrossRef] [PubMed]
- Zavascki, A.P.; Carvalhaes, C.G.; da Silva, G.L.; Tavares Soares, S.P.; de Alcântara, L.R.; Elias, L.S. Outbreak of carbapenem-resistant Providencia stuartii in an intensive care unit. Infect. Control Hosp. Epidemiol. 2012, 33, 627–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alghoribi, M.F.; Balkhy, H.H.; Woodford, N.; Ellington, M.J. The role of whole genome sequencing in monitoring antimicrobial resistance: A biosafety and public health priority in the Arabian Peninsula. J. Infect. Public Health 2018, 11, 784–787. [Google Scholar] [CrossRef] [PubMed]



| Ward a | Bacterial Clinical Isolates (n = 621) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae (n = 178) | E. coli (n = 151) | P. aeruginosa (n = 84) | A.baumannii (n = 82) | P. mirabilis (n = 46) | Other Gram- Negatives (n = 71) | |||||||
| n | % | n | % | n | % | n | % | n | % | n | % | |
|
Intensive Care Unit ICU (30%) |
42 Urine (16) Sputum (9) Blood (8) Wound/pus (7) AE (2) | 24 |
31 Urine (16), sputum (6) blood (6), wound/pus (3) | 21 |
34 Urine (5), Blood (5) Sputum (15) Wound swab/pus (2) Other (7) | 41 |
40 Sputum (20) Blood (8) Swab/wound/pus (8) Other (4) | 49 |
24 Blood (7) Sputum (6) Urine (5) Wound/pus/swab (6) | 52 |
15 Urine (3) Blood (2) Sputum (4) Wound/swab/pus (3) Other (3) | 21 |
|
COVID-19 Isolation Zones (ISO) or Ward (COW) 13.5% |
40 Blood (15) Sputum (12) Urine (6) Wound/pus/swab (5) Other (2) | 23 |
7 Blood (1) Sputum (1) Wound (1) Urine (4) | 5 | 0 | 0 |
29 Bloody sp. (12) Urine (2) Sputum (11) Wound (4) | 35 | 0 | 0 |
2 Blood Wound | 3 |
|
Surgical Ward (SW) 12% |
25 Wound/pus/swab (8) Urine (11) Blood (1) Other (5) | 14 |
19 Wound (7) Urine (8) Blood (1) Other (3) | 13 |
9 Wound (3) Sputum (4) Urine (2) | 11 |
3 Wound (3) | 4 |
3 Swab (2) Sputum (1) | 7 |
13 Sputum (6) Wound (4) Blood (1) Other (2) | 18 |
|
Male/Female Medical Ward (M/F MW) 17% |
21 Blood (3) Wound (3) Urine (11) Sputum (2) Other (2) | 12 |
44 Urine (26) Swabs (9) Blood (2) Sputum (1) Other (5) | 29 |
6 Wound swab (4) Urine (2) | 7 |
5 Wound (1) Urine (1) Other (3) | 6 |
13 Urine (10) Sputum (2) Blood (1) | 28 |
16 Urine (9) Swabs (3) Blood (2) Stool (2) | 23 |
|
AKU—AMR 4% |
8 Blood (4) Other (4) | 4.5 |
3 Urine (3) | 2 | 6 | 7 | 0 | 0 | 0 | 0 |
5 Blood (5) | 7 |
| Other (25%) | 42 | 24 | 47 | 30 | 29 | 35 | 5 | 6 | 6 | 13 | 24 | 34 |
|
Total n = 616 | 178 | 100% | 151 | 100% | 84 | 100% | 82 | 100% | 46 | 100% | 75 b | 100% |
| Age b | ||||||||||||
| Young 1–20 (16%) | 14 | 11 | 34 | 26 | 16 | 23 | 5 | 9 | 0 | 0 | 12 | 20 |
|
Adults (21–49 y) (30%) | 43 | 33 | 41 | 32 | 27 | 39 | 15 | 27 | 7 | 17 | 14 | 21 |
|
Seniors (>50 y) (54%) | 74 | 57 | 55 | 42 | 26 | 38 | 35 | 64 | 34 | 83 | 40 | 61 |
| Gender c | ||||||||||||
| Male (59%) | 72 | 53 | 57 | 43 | 59 | 75 | 43 | 74 | 29 | 66 | 45 | 63 |
| Female (41%) | 64 | 47 | 75 | 57 | 20 | 25 | 15 | 26 | 15 | 34 | 26 | 37 |
| MDR, XDR, PDR d | MDR | MDR | XDR | PDR | XDR | MDR | ||||||
| Organisms | Gender | Age a | Specimen | Ward b | AK | CN | ETP | IMI | MRP | KF | CXM | FOX | CAZ | CRO | FEP | ATM | AMP | AUG | TZP | CS | SXT | NIT | CIP | LEV | TGC | Hospital |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. stuartii | F | 71 | Swap | FMW | S | S | S | S | S | R | S | S | S | S | S | S | I | R | S | R | S | R | S | S | KKH | |
| P. stuartii | F | 74 | Sputum | ICU | R | R | S | R | R | R | R | S | R | R | R | R | R | R | R | R | R | R | R | R | KKH | |
| P. stuartii | M | 81 | Blood | AE | I | R | S | S | S | R | R | I | S | S | S | S | R | R | S | R | S | R | R | R | KKH | |
| P. stuartii | M | 31 | Sputum2-B | MSW | S | R | S | S | R | R | I | S | S | S | S | S | R | R | S | R | S | R | I | R | KKH | |
| P. stuartii | M | 38 | Wound | ICU | S | R | S | S | S | R | S | R | R | R | R | R | R | R | R | R | R | R | R | KKH | ||
| P. stuartii | M | 68 | Sputum | MSW | S | R | S | S | R | R | S | R | R | R | R | R | R | S | R | R | R | R | R | KKH | ||
| P. stuartii | M | 33 | Urine | EAB | S | R | R | R | S | R | R | R | R | R | R | R | R | R | S | R | R | R | R | R | X | KKH |
| P. stuartii | M | 61 | Sputum | ICU | S | R | R | R | R | R | R | S | R | R | R | R | R | R | S | R | R | R | R | R | X | KKH |
| P. stuartii | F | 61 | Dialysis | ICU | S | R | S | X | X | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | X | KKH |
| P. stuartii | F | 52 | Urine | HND | S | R | S | S | R | S | S | S | S | S | S | R | R | S | R | R | R | R | R | R | X | KKH |
| P. stuartii | M | 31 | Sputum | MSW | S | R | S | R | R | I | S | S | S | S | S | R | R | S | R | S | R | I | R | KKH | ||
| P. stuartii | F | 71 | Sputum | ICU | S | R | R | S | R | R | R | R | R | S | R | R | R | S | R | R | R | R | R | KKH | ||
| P. stuartii | F | 52 | Urine | LAB | R | R | S | S | R | R | S | R | R | R | R | R | R | S | R | R | R | R | R | KKH | ||
| P. stuartii | F | 42 | Wound | FMW | S | S | S | X | S | R | S | S | S | S | S | S | S | R | S | R | S | R | I | I | KKH | |
| P. stuartii | M | 40 | Wound | S | R | R | X | R | R | R | S | R | S | S | R | R | R | R | R | R | R | R | R | R | KSSH | |
| P. stuartii | M | 6 | Sputum | MSW | S | S | S | S | R | S | S | R | R | R | S | R | S | S | R | R | R | R | R | KKH | ||
| P. stuartii | M | 40 | Sputum | MSW | S | S | R | S | S | R | S | R | R | S | R | R | R | S | S | R | S | R | S | R | X | KKH |
| P. stuartii | M | 51 | Other | LAB | S | S | R | X | S | R | R | S | S | R | R | R | S | R | S | R | S | R | R | S | X | KKH |
| P. stuartii | F | 65 | Urine | ICU | S | R | S | R | R | R | R | S | R | S | R | R | S | R | R | R | R | R | S | R | X | KKH |
| P. stuartii | F | 52 | Urine | HND | S | R | S | S | R | S | S | S | S | S | S | R | R | S | R | R | R | R | R | R | X | KKH |
| P. stuartii | F | 30 | Sputum | MSW | S | R | S | R | S | I | S | R | S | R | S | R | R | S | R | S | S | R | R | KKH | ||
| P. stuartii | M | 72 | Wound | S | R | R | X | S | R | R | S | S | S | S | S | R | R | S | R | S | R | S | S | R | KSSH | |
| M. morganii | M | 54 | Pus | MSW | S | S | S | S | R | R | S | R | R | S | S | R | R | S | R | R | I | I | I | |||
| M. morganii | M | AE–AN | ICU | S | S | S | S | S | R | R | S | S | S | S | S | R | R | S | R | R | R | S | S | R | ||
| M. morganii | M | 61 | Blood | ICU | S | R | S | S | S | R | R | S | R | R | R | I | R | R | S | R | R | R | R | R | R | |
| M. morganii | S | S | S | S | S | S | S | S | R | S | S | S | S | S | s | MCH | ||||||||||
| M. morganii | F | 33 | S | S | S | X | S | R | R | S | S | S | S | S | R | R | S | R | S | R | S | S | S | KKH | ||
| M. morganii | F | 54 | Wound | FSW | S | S | S | X | S | R | R | S | S | S | S | S | R | R | S | R | S | R | S | S | R | KKH |
| M. morganii | M | 51 | Blood | MSW | S | S | S | X | S | R | R | S | R | R | R | S | R | R | S | R | S | R | S | S | R | KKH |
| M. morganii | F | 79 | Urine | S | R | R | R | S | R | R | I | S | R | R | R | R | R | S | R | R | R | R | R | R | KSH | |
| S. fonticola | M | 85 | Urine | MMW | S | S | R | I | R | R | R | S | S | R | R | R | R | S | S | S | S | R | S | S | R | |
| S. fonticola | M | 88 | Urine | MMW | S | S | R | R | R | R | R | R | S | S | S | R | R | S | R | R | R | S | S | R | ||
| Haemophilus sp. | M | 3m | Eyes swap | PICU | R | S | S | S | R | S | S | S | S | S | S | S | R | |||||||||
| C. koseri | F | 62 | Blood | AKU | S | S | S | S | S | S | R | S | S | S | S | S | R | S | S | S | S | R | S | S | S | |
| C. koseri | F | 28 | FSW | S | S | S | S | S | S | S | S | S | S | S | S | R | S | S | S | S | S | S | S | S | KKH | |
| C. koseri | M | 3m | Urine | PW | S | S | R | S | S | R | R | S | R | R | R | R | R | S | S | S | S | R | S | S | R | |
| C. koseri | M | 8M | Urine | PICU | R | I | X | I | S | R | R | S | R | R | R | R | R | S | S | R | R | I | S | S | I | KKH |
| K. oxytoca | M | 3 | Urine | PW | S | S | R | S | S | R | R | R | R | R | R | S | R | S | S | S | R | S | S | S | S | |
| K. oxytoca | M | 6M | Urine | UR | S | S | S | S | S | S | S | S | S | S | S | S | R | S | S | S | S | R | S | S | S | |
| K. oxytoca | M | 7M | Urine | UPO | S | S | S | S | S | S | S | S | S | S | S | S | R | S | S | S | S | S | S | S | S | KKH |
| K. oxytoca | M | 96 | Urine | LAB | S | S | R | R | R | R | R | S | R | R | R | R | R | R | R | S | S | S | S | S | S | KKH |
| K. ozae | M | 64 | Urine | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | KSSH | |
| S. enterica | Urine | AE/ER | S | S | S | S | S | S | S | S | s | s | S | S | s | MCH | ||||||||||
| S. typhi | S | S | S | S | S | S | S | S | S | S | S | S | S | |||||||||||||
| Salmonella sp. | M | 4M | Stool | PW | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | KKH |
| P. fluorescens | M | 24 | Blood | MMW | S | I | S | S | S | R | R | R | S | R | S | R | R | R | S | S | S | R | I | S | S | KKH |
| P. putida | F | 65 | Urine | FMW | S | S | R | S | S | R | R | R | S | S | I | I | R | R | S | S | R | R | I | R | R | KKH |
| S. maltophilia | F | 18 | Wound | FSW | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | R | S | I | KKH | |
| S. maltophilia | M | 70 | Blood | AMR | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | I | S | I | KKH | ||
| S. maltophilia | M | 64 | Blood | AMR | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | X | S | R | I | S | I | KKH |
| S. maltophilia | M | 64 | Blood | MMW | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | S | S | S | KKH | |
| S. maltophilia | M | 70 | Blood | AMR | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | X | S | R | R | S | S | KKH |
| S. maltophilia | M | 45 | Bloody Sputum | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | KSSH | |
| P. rettgeri | F | 61 | Blood | ICU | S | I | I | S | R | S | S | S | S | S | S | R | R | S | R | S | R | S | S | KKH | ||
| P. rettgeri | Wound | S | S | S | I | S | R | S | S | S | S | S | S | R | R | S | R | S | S | S | S | R | KSSH | |||
| P. rettgeri | M | 70 | Wound | MSW | S | R | S | R | S | R | R | S | R | R | R | R | R | R | S | R | R | R | R | R | R | KKH |
| *C. freundii | M | 56 | Blood | COW | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | S | R | S | S | KKH |
| C. freundii | M | 65 | Wound | COW | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | R | S | KKH |
| C. freundii | M | 2M | Urine | PW | S | R | S | S | S | R | R | R | R | R | R | R | R | R | S | S | S | S | S | S | S | KKH |
| C. freundii | F | 81 | Wound | ICU | S | S | S | S | S | R | R | R | R | R | R | R | R | R | R | S | R | S | R | R | S | KKH |
| C. freundii | F | 62 | wound | FMW | S | R | S | S | S | R | R | R | R | R | R | R | R | R | S | S | S | S | S | S | S | KKH |
| C. freundii | M | 75 | ICU | S | R | S | R | S | R | R | R | R | R | R | R | R | R | I | S | R | S | R | R | S | KKH | |
| C. koseri | F | 62 | Blood | AKU | S | S | S | S | S | S | R | S | S | S | S | S | R | S | S | S | S | R | S | S | S | |
| C. koseri | F | 28 | FSW | S | S | S | S | S | S | S | S | S | S | S | S | R | S | S | S | S | S | S | S | S | KKH | |
| C. koseri | M | 3m | Urine | PW | S | S | R | S | S | R | R | S | R | R | R | R | R | S | S | S | S | R | S | S | R | |
| C. koseri | M | 8M | Urine | PICU | R | I | X | I | S | R | R | S | R | R | R | R | R | S | S | R | R | I | S | S | I | KKH |
| B. cepacia | F | 49 | Urine | FMW | R | R | R | R | R | R | R | S | R | R | R | R | S | R | S | R | R | R | S | |||
| B. cepacia | M | 63 | Sputum | ICU | S | S | R | I | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | |||
| P. vulgaris | M | 66 | Urine | R | R | I | R | I | R | R | I | R | R | R | R | R | R | I | R | S | R | R | R | R | KHH | |
| Shigella sp. | M | Stool | LAB | R | S | S | S | S | S | S | R | S | S | S | S | S | S | MCH | ||||||||
| Shigella sp. | M | Stool | LAB | S | S | S | S | S | S | S | I | S | S | S | S | S | S | MCH | ||||||||
| Shigella sp. | F | Stool | FMW | R | S | S | S | S | S | S | R | S | S | S | S | S | S | KKH | ||||||||
| Shigella sp. | F | Stool | LAB | S | R | S | S | R | S | R | S | I | S | R | S | S | I | S | KKH | |||||||
| C. lapagei | M | 58 | Urine | MMW | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | R | R | R | R | S | KKH |
| C. werkmanii | F | 61 | Urine | LAB | S | S | S | S | S | R | S | R | S | S | S | S | R | R | S | S | S | S | S | S | S | KKH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Said, K.B.; Alsolami, A.; Khalifa, A.M.; Khalil, N.A.; Moursi, S.; Osman, A.; Fahad, D.; Rakha, E.; Rashidi, M.; Moussa, S.; et al. A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha’il Hospitals, Saudi Arabia. Microorganisms 2021, 9, 2024. https://doi.org/10.3390/microorganisms9102024
Said KB, Alsolami A, Khalifa AM, Khalil NA, Moursi S, Osman A, Fahad D, Rakha E, Rashidi M, Moussa S, et al. A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha’il Hospitals, Saudi Arabia. Microorganisms. 2021; 9(10):2024. https://doi.org/10.3390/microorganisms9102024
Chicago/Turabian StyleSaid, Kamaleldin B., Ahmed Alsolami, Amany M. Khalifa, Nuha A. Khalil, Soha Moursi, Abuzar Osman, Dakheel Fahad, Ehab Rakha, Musleh Rashidi, Safia Moussa, and et al. 2021. "A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha’il Hospitals, Saudi Arabia" Microorganisms 9, no. 10: 2024. https://doi.org/10.3390/microorganisms9102024
APA StyleSaid, K. B., Alsolami, A., Khalifa, A. M., Khalil, N. A., Moursi, S., Osman, A., Fahad, D., Rakha, E., Rashidi, M., Moussa, S., Bashir, A. I., Alfouzan, F., Hammam, S., Taha, T. E., Al-hazimi, A., Al Jadani, A., & On behalf of the Ha’il COM Research Unit Group. (2021). A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha’il Hospitals, Saudi Arabia. Microorganisms, 9(10), 2024. https://doi.org/10.3390/microorganisms9102024







