Cryptosporidium sciurinum n. sp. (Apicomplexa: Cryptosporidiidae) in Eurasian Red Squirrels (Sciurus vulgaris)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
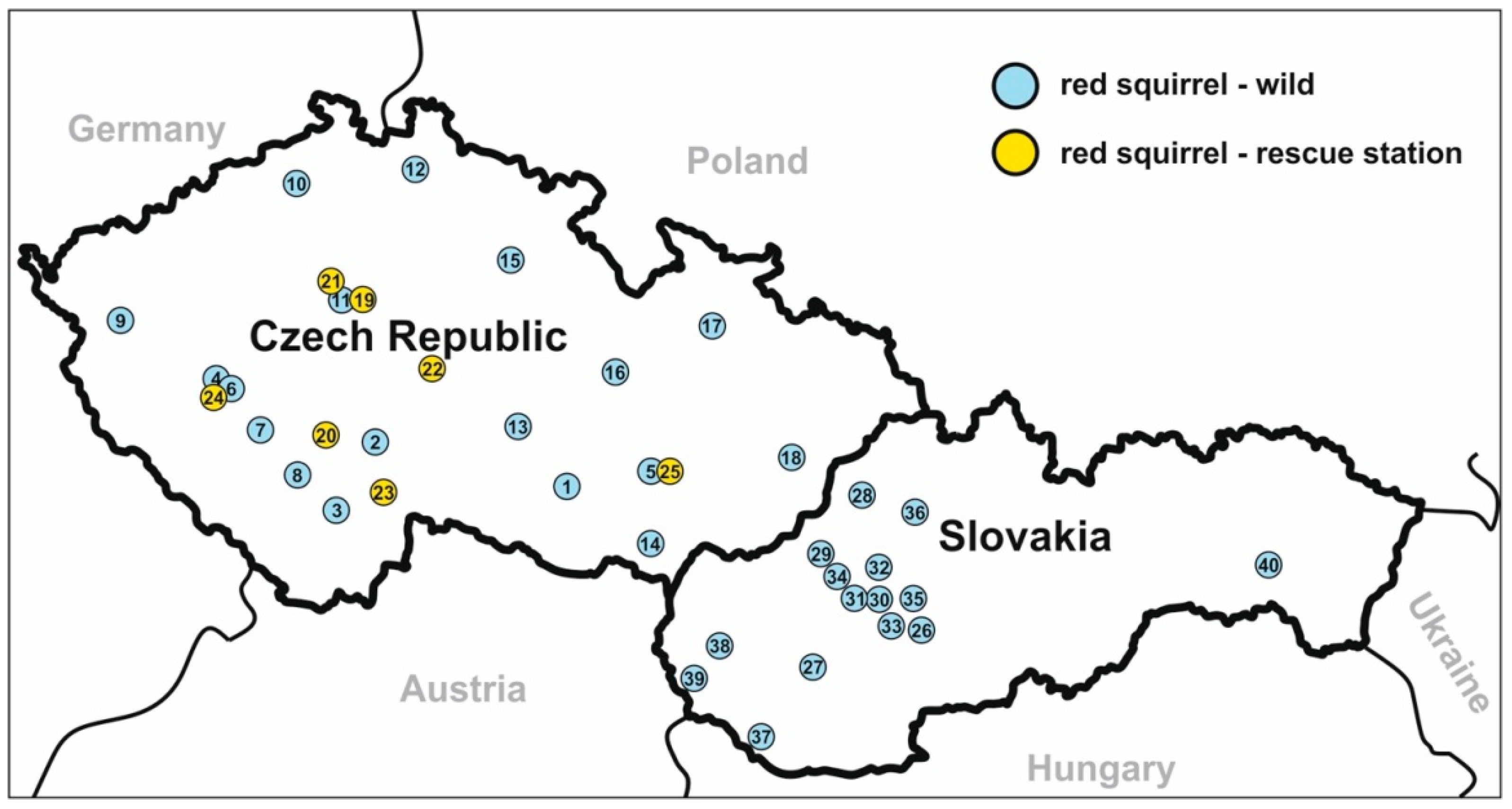
2.2. Trapping and Specimen Collection
2.3. Source of Oocysts of Cryptosporidium sp. Ferret Genotype
2.4. Molecular Characterization
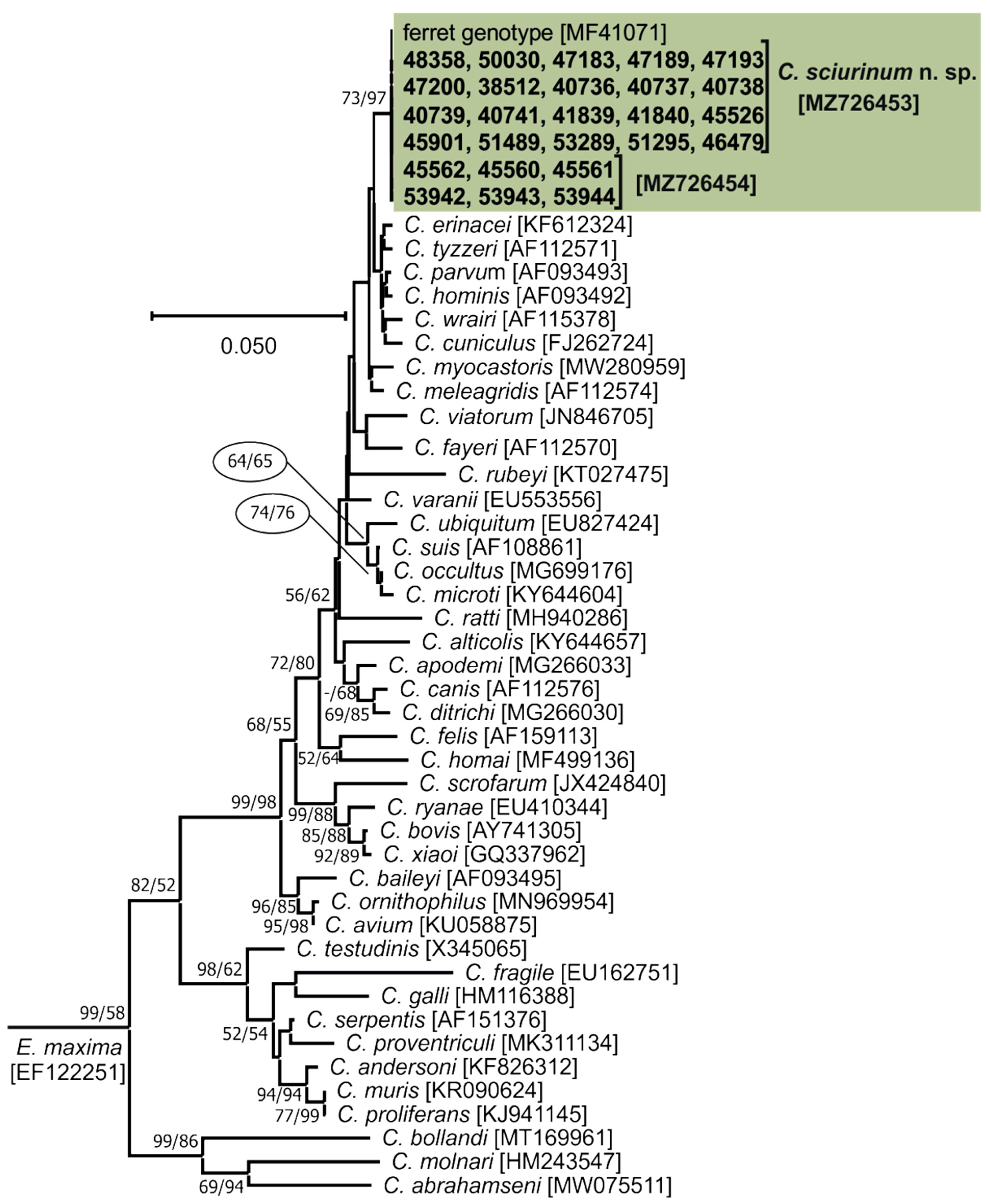
2.5. Sequence and Phylogenetic Analysis
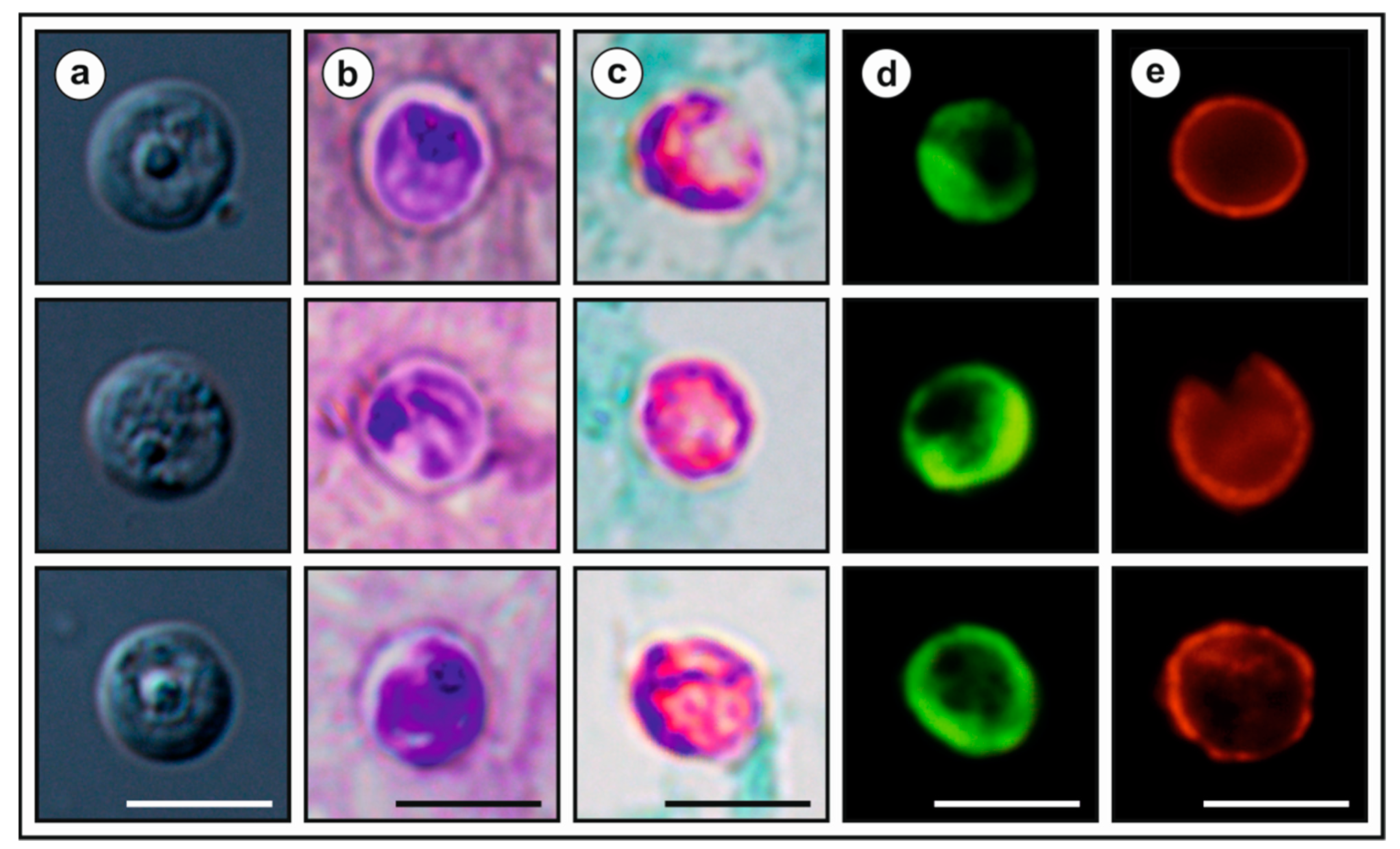
2.6. Oocyst Morphometry
2.7. Transmission Studies
2.8. Clinical and Pathomorphological Examinations
2.9. Statistical Analysis
3. Results
- Taxonomic summary
- Family Cryptosporidiidae Léger, 1911
- Genus Cryptosporidium Tyzzer, 1907
- Cryptosporidiumsciurinum n. sp.
- Syn: Cryptosporidium parvum ferret genotype ex black-footed ferret (Mustela nigripes) and Cryptosporidium sp. ferret genotype ex black-footed ferret (Mustela nigripes) of Xiao et al. [35].
- Type-host:Sciurus vulgaris Linnaeus, 1758 (Rodentia: Muridae) Eurasian red squirrel.
- Other natural hosts: black-footed ferret (Mustela nigripes), Siberian chipmunk (Tamias sibiricus), Eastern chipmunk (Tamias striatus), budgerigar (Melopsittacus undulatus).
- Type-locality: Třeboň, Czech Republic.
- Other localities: České Budějovice, Brno, Praha, Vlašim, whole Czech Republic; Povážská Bystrica, Handlová, Gabčíkovo, Bratislava, Košice, whole Slovakia; Guangdong, China; Northern Italy; Georgia, USA.
- Type-material: Faecal smear slides with oocysts stained by ACMV and ZN staining (nos. MV1-3/45901 and ZN1-2/45901) and gDNA isolated from faecal samples of a naturally infected Eurasian red squirrel (isolate 45901) are deposited at the Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Czech Republic.
- Site of infection: unknown.
- Distribution: As Cryptosporidium sp. ferret genotype ex Sciurus vulgaris: China, Italy [13,16,17]; as Cryptosporidium sp. ferret genotype or Cryptosporidium parvum ferret genotype ex Mustela nigripes: USA [35], as Cryptosporidium sp. ferret genotype ex Tamias striatus, T. sibiricus and Melopsittacus undulatus: China [13,36].
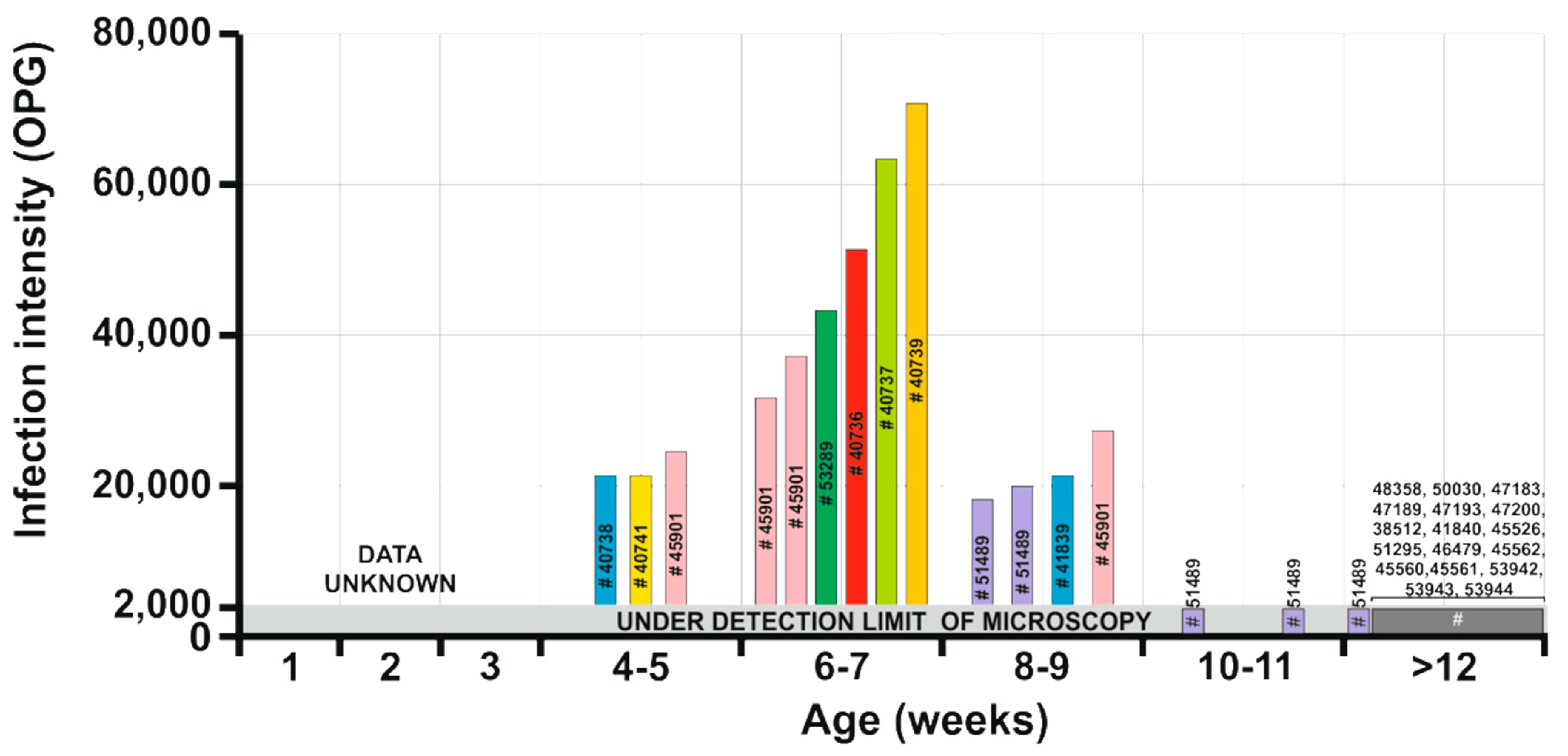
- Prepatent period: unknown.
- Patent period: At least 5 weeks in naturally infected Sciurus vulgaris (isolate 51489 in the present study).
- Representative DNA sequences: Representative nucleotide sequences of SSU [MZ726453], actin [MZ772035], HSP70 [MZ772047], TRAP-C1 [MZ772037], COWP [MZ772045], and gp60 [MZ772039–MZ772042] genes are deposited in the GenBank database.
- ZooBank registration: To comply with the regulations set out in Article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [37], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:83ABAD68-07C6-4E51-8234- A2AC4C0EE72C. The LSID for the new name Cryptosporidium sciurinum n. sp. is urn:lsid:zoobank.org:act:E13F2B9F-D9C6-4ED9-9EA8-B94AEB6189A2.
- Etymology: The species name sciurinum is derived from the Latin noun sciurus, meaning squirrel.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nader, J.L.; Mathers, T.C.; Ward, B.J.; Pachebat, J.A.; Swain, M.T.; Robinson, G.; Chalmers, R.M.; Hunter, P.R.; van Oosterhout, C.; Tyler, K.M. Evolutionary genomics of anthroponosis in Cryptosporidium. Nat. Microbiol. 2019, 4, 826–836. [Google Scholar] [CrossRef]
- Caccio, S.M.; Putignani, L. Epidemiology of Human Cryptosporidiosis. In Cryptosporidium: Parasite and Disease, 1st ed.; Cacciò, S.M., Widmer, G., Eds.; Springer: Vienna, Austria, 2014; pp. 43–80. [Google Scholar]
- Kváč, M.; McEvoy, J.; Stenger, B.; Clark, M. Cryptosporidiosis in other vertebrates. In Cryptosporidium: Parasite and Disease, 1st ed.; Cacciò, S.M., Widmer, G., Eds.; Springer: Vienna, Austria, 2014; pp. 237–326. [Google Scholar]
- Zahedi, A.; Bolland, S.J.; Oskam, C.L.; Ryan, U. Cryptosporidium abrahamseni n. sp. (Apicomplexa: Cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae). Exp. Parasitol. 2021, 223, 108089. [Google Scholar] [CrossRef]
- Ryan, U.M.; Monis, P.; Enemark, H.L.; Sulaiman, I.; Samarasinghe, B.; Read, C.; Buddle, R.; Robertson, I.; Zhou, L.; Thompson, R.C.A.; et al. Cryptosporidium suis n. sp. (Apicomplexa: Cryptosporidiidae) in pigs (Sus scrofa). J. Parasitol. 2004, 90, 769–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bujila, I.; Troell, K.; Fischerstrom, K.; Nordahl, M.; Killander, G.; Hansen, A.; Soderlund, R.; Lebbad, M.; Beser, J. Cryptosporidium chipmunk genotype I—An emerging cause of human cryptosporidiosis in Sweden. Infect. Genet. Evol. 2021, 92, 104895. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, J.P.; Hill, D.; Ryan, M.J. Cryptosporidiosis in a gray squirrel. J. Am. Vet. Med. Assoc. 1982, 181, 1420–1422. [Google Scholar]
- Current, W.L. Cryptosporidium spp. In Parasitic Infections in the Compromised Host; Walzer, P.D., Genta, R.M., Eds.; Dekker: New York, NY, USA, 1990; pp. 281–341. [Google Scholar]
- Stenger, B.L.; Clark, M.E.; Kváč, M.; Khan, E.; Giddings, C.W.; Prediger, J.; McEvoy, J.M. North American tree squirrels and ground squirrels with overlapping ranges host different Cryptosporidium species and genotypes. Infect. Genet. Evol. 2015, 36, 287–293. [Google Scholar] [CrossRef]
- Ziegler, P.E.; Wade, S.E.; Schaaf, S.L.; Chang, Y.F.; Mohammed, H.O. Cryptosporidium spp. from small mammals in the New York City watershed. J. Wildl. Dis. 2007, 43, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, P.E.; Wade, S.E.; Schaaf, S.L.; Stern, D.A.; Nadareski, C.A.; Mohammed, H.O. Prevalence of Cryptosporidium species in wildlife populations within a watershed landscape in southeastern New York State. Vet. Parasitol. 2007, 147, 176–184. [Google Scholar] [CrossRef]
- Feng, Y.; Alderisio, K.A.; Yang, W.; Blancero, L.A.; Kuhne, W.G.; Nadareski, C.A.; Reid, M.; Xiao, L. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 2007, 73, 6475–6483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, C.; Zhang, L.; Wang, R.; Jian, F.; Zhang, S.; Ning, C.; Wang, H.; Feng, C.; Wang, X.; Ren, X.; et al. Cryptosporidium spp. in wild, laboratory, and pet rodents in china: Prevalence and molecular characterization. Appl. Environ. Microbiol. 2009, 75, 7692–7699. [Google Scholar] [CrossRef] [Green Version]
- Chai, Y.J.; Deng, L.; Liu, H.F.; Yao, J.X.; Zhong, Z.J.; Xiang, L.Q.; Fu, H.L.; Shen, L.H.; Zhou, Z.Y.; Deng, J.L.; et al. First detection of Cryptosporidium spp. in red-bellied tree squirrels (Callosciurus erythraeus) in China. Parasite 2019, 26, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Chai, Y.; Luo, R.; Yang, L.; Yao, J.; Zhong, Z.; Wang, W.; Xiang, L.; Fu, H.; Liu, H.; et al. Occurrence and genetic characteristics of Cryptosporidium spp. and Enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Sci. Rep. 2020, 10, 1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kváč, M.; Hofmannová, L.; Bertolino, S.; Wauters, L.; Tosi, G.; Modrý, D. Natural infection with two genotypes of Cryptosporidium in red squirrels (Sciurus vulgaris) in Italy. Folia Parasitol. 2008, 55, 95–99. [Google Scholar] [CrossRef] [Green Version]
- Prediger, J.; Horčičková, M.; Hofmannová, L.; Sak, B.; Ferrari, N.; Mazzamuto, M.V.; Romeo, C.; Wauters, L.A.; McEvoy, J.; Kváč, M. Native and introduced squirrels in Italy host different Cryptosporidium spp. Eur. J. Protistol. 2017, 61, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Wauters, L.; Dhondt, A.A. Body-Weight, Longevity and Reproductive Success in Red Squirrels (Sciurus vulgaris). J. Anim. Ecol. 1989, 58, 637–651. [Google Scholar] [CrossRef]
- Miláček, P.; Vítovec, J. Differential staining of cryptosporidia by aniline-carbol-methyl violet and tartrazine in smears from feces and scrapings of intestinal mucosa. Folia Parasitol. 1985, 32, 50. [Google Scholar]
- Kváč, M.; Ondráčková, Z.; Květoňová, D.; Sak, B.; Vítovec, J. Infectivity and pathogenicity of Cryptosporidium andersoni to a novel host, southern multimammate mouse (Mastomys coucha). Vet. Parasitol. 2007, 143, 229–233. [Google Scholar] [CrossRef]
- Arrowood, M.J.; Donaldson, K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J. Eukaryot. Microbiol. 1996, 43, 89S. [Google Scholar] [CrossRef]
- Sauch, J.F.; Flanigan, D.; Galvin, M.L.; Berman, D.; Jakubowski, W. Propidium iodide as an indicator of Giardia cyst viability. Appl. Environ. Microbiol. 1991, 57, 3243–3247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.; Escalante, L.; Yang, C.; Sulaiman, I.; Escalante, A.A.; Montali, R.J.; Fayer, R.; Lal, A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999, 65, 1578–1583. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, I.M.; Morgan, U.M.; Thompson, R.C.; Lal, A.A.; Xiao, L. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 2000, 66, 2385–2391. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, I.M.; Lal, A.A.; Xiao, L.H. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 2002, 88, 388–394. [Google Scholar] [CrossRef]
- Spano, F.; Putignani, L.; Naitza, S.; Puri, C.; Wright, S.; Crisanti, A. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol. Biochem. Parasitol. 1998, 92, 147–162. [Google Scholar] [CrossRef]
- Jiang, J.; Alderisio, K.A.; Xiao, L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 2005, 71, 4446–4454. [Google Scholar] [CrossRef] [Green Version]
- Spano, F.; Puri, C.; Ranucci, L.; Putignani, L.; Crisanti, A. Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual parasite development. Parasitology 1997, 114 Pt 5, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.; Xiao, L.H.; Sulaiman, I.; Lal, A.A.; Matos, O.; Antunes, F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 2003, 41, 2744–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Henriksen, S.A.; Pohlenz, J.F. Staining of cryptosporidia by a modified Ziehl-Neelsen technique. Acta Vet. Scand. 1981, 22, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Kváč, M.; Vítovec, J. Prevalence and pathogenicity of Cryptosporidium andersoni in one herd of beef cattle. J. Vet. Med. B 2003, 50, 451–457. [Google Scholar] [CrossRef]
- Holubová, N.; Zikmundová, V.; Limpouchová, Z.; Sak, B.; Konečný, R.; Hlásková, L.; Rajský, D.; Kopacz, Z.; McEvoy, J.; Kváč, M. Cryptosporidium proventriculi sp. n. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur. J. Protistol. 2019, 69, 70–87. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Xiao, L.; Morgan, U.M.; Limor, J.; Escalante, A.; Arrowood, M.; Shulaw, W.; Thompson, R.C.; Fayer, R.; Lal, A.A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 1999, 65, 3386–3391. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, L.; Tao, W.; Jiang, Y.; Wan, Q.; Lin, Y.; Li, W. Molecular investigation of Cryptosporidium in small caged pets in northeast China: Host specificity and zoonotic implications. Parasitol. Res. 2016, 115, 2905–2911. [Google Scholar] [CrossRef]
- ICZN. International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the international code of zoologicalnomenclature to expand and refine methods of publication. Bull. Zool. Nomencl. 2012, 69, 161–169. [Google Scholar] [CrossRef]
- Stenger, B.L.S.; Horčičková, M.; Clark, M.E.; Kváč, M.; Čondlová, S.; Khan, E.; Widmer, G.; Xiao, L.; Giddings, C.W.; Pennil, C.; et al. Cryptosporidium infecting wild cricetid rodents from the subfamilies Arvicolinae and Neotominae. Parasitology 2017, 145, 326–334. [Google Scholar] [CrossRef]
- Li, F.L.; Zhang, Z.J.; Hu, S.H.; Zhao, W.T.; Zhao, J.G.; Kváč, M.; Guo, Y.Q.; Li, N.; Feng, Y.Y.; Xiao, L.H. Common occurrence of divergent Cryptosporidium species and Cryptosporidium parvum subtypes in farmed bamboo rats (Rhizomys sinensis). Parasites Vectors 2020, 13, 149. [Google Scholar] [CrossRef]
- Ježková, J.; Prediger, J.; Holubová, N.; Sak, B.; Konečný, R.; Feng, Y.; Xiao, L.; Rost, M.; McEvoy, J.; Kváč, M. Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology 2021, 148, 84–97. [Google Scholar] [CrossRef]
- Murakoshi, F.; Fukuda, Y.; Matsubara, R.; Kato, Y.; Sato, R.; Sasaki, T.; Tada, C.; Nakai, Y. Detection and genotyping of Cryptosporidium spp. in large Japanese field mice, Apodemus speciosus. Vet. Parasitol. 2013, 196, 184–188. [Google Scholar] [CrossRef]
- Čondlová, S.; Horčičková, M.; Havrdová, N.; Sak, B.; Hlásková, L.; Perec-Matysiak, A.; Kicia, M.; McEvoy, J.; Kváč, M. Diversity of Cryptosporidium spp. in Apodemus spp. in Europe. Eur. J. Protistol. 2019, 69, 1–13. [Google Scholar] [CrossRef]
- Čondlová, S.; Horčičková, M.; Sak, B.; Květoňová, D.; Hlásková, L.; Konečný, R.; Stanko, M.; McEvoy, J.; Kváč, M. Cryptosporidiumapodemi sp. n. and Cryptosporidium ditrichi sp. n. (Apicomplexa: Cryptosporidiidae) in Apodemus spp. Eur. J. Protistol. 2018, 63, 1–12. [Google Scholar] [CrossRef]
- Horčičková, M.; Čondlová, S.; Holubová, N.; Sak, B.; Květoňová, D.; Hlásková, L.; Konečný, R.; Sedláček, F.; Clark, M.; Giddings, C.; et al. Diversity of Cryptosporidium in common voles and description of Cryptosporidium alticolis sp. n. and Cryptosporidium microti sp. n. (Apicomplexa: Cryptosporidiidae). Parasitology 2019, 146, 220–233. [Google Scholar] [CrossRef]
- Holubová, N.; Sak, B.; Hlásková, L.; Květoňová, D.; Hanzal, V.; Rajský, D.; Rost, M.; McEvoy, J.; Kváč, M. Host specificity and age-dependent resistance to Cryptosporidium avium infection in chickens, ducks and pheasants. Exp. Parasitol. 2018, 191, 62–65. [Google Scholar] [CrossRef]
- Pavlásek, I. Findings of cryptosporidia in the stomach of hens and of exotic and wild birds. Veterinářství (Czech) 2001, 51, 103–108. [Google Scholar]
- Sreter, T.; Varga, I.; Bekesi, L. Age-dependent resistance to Cryptosporidium baileyi infection in chickens. J. Parasitol. 1995, 81, 827–829. [Google Scholar] [CrossRef]
- Rhee, J.K.; So, W.S.; Kim, H.C. Age-dependent resistance to Cryptosporidium muris (strain MCR) infection in golden hamsters and mice. Korean J. Parasitol. 1999, 37, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Kváč, M.; Sak, B.; Květoňová, D.; Secor, W.E. Infectivity of gastric and intestinal Cryptosporidium species in immunocompetent Mongolian gerbils (Meriones unguiculatus). Vet. Parasitol. 2009, 163, 33–38. [Google Scholar] [CrossRef]
- Kváč, M.; Kestřánová, M.; Květoňová, D.; Kotková, M.; Ortega, Y.; McEvoy, J.; Sak, B. Cryptosporidium tyzzeri and Cryptosporidium muris originated from wild West-European house mice (Mus musculus domesticus) and East-European house mice (Mus musculus musculus) are non-infectious for pigs. Exp. Parasitol. 2012, 131, 107–110. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Cranfield, M.R. Experimental transmission of Cryptosporidium oocyst isolates from mammals, birds and reptiles to captive snakes. Vet. Res. 1998, 29, 187–195. [Google Scholar]
- Graczyk, T.K.; Cranfield, M.R.; Fayer, R. Oocysts of Cryptosporidium from snakes are not infectious to ducklings but retain viability after intestinal passage through a refractory host. Vet. Parasitol. 1998, 77, 33–40. [Google Scholar] [CrossRef]
- Crawshaw, G.J.; Mehren, K.G. Cryptosporidiosis in Zoo and Wild Animals. In Erkrankungen der Zootiere, Verhandlungsbericht des 29, Proceedings of the Internationalen Symposiums Uber die Erkrankungen der Zootiere, Cardiff, UK, 20–24 May 1987; Internationales Symposium uber die Erkrankungen der Zoo- und Wildtiere: Berlin, Germany, 1987; pp. 353–362. [Google Scholar]
- Stenger, B.L.; Clark, M.E.; Kváč, M.; Khan, E.; Giddings, C.W.; Dyer, N.W.; Schultz, J.L.; McEvoy, J.M. Highly divergent 18S rRNA gene paralogs in a Cryptosporidium genotype from eastern chipmunks (Tamias striatus). Infect. Genet. Evol. 2015, 32, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Le Blancq, S.M.; Khramtsov, N.V.; Zamani, F.; Upton, S.J.; Wu, T.W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 1997, 90, 463–478. [Google Scholar] [CrossRef]
- Ng-Hublin, J.S.; Singleton, G.R.; Ryan, U. Molecular characterization of Cryptosporidium spp. from wild rats and mice from rural communities in the Philippines. Infect. Genet. Evol. 2013, 16, 5–12. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Xiao, L.; Alderisio, K.; Elwin, K.; Cebelinski, E.; Chalmers, R.; Santin, M.; Fayer, R.; Kváč, M.; Ryan, U.; et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014, 20, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Ježková, J.; Limpouchová, Z.; Prediger, J.; Holubová, N.; Sak, B.; Konečný, R.; Květoňová, D.; Hlásková, L.; Rost, M.; McEvoy, J.; et al. Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus). Microorganisms 2021, 9, 813. [Google Scholar] [CrossRef] [PubMed]
- Kváč, M.; McEvoy, J.; Loudová, M.; Stenger, B.; Sak, B.; Květoňová, D.; Ditrich, O.; Rašková, V.; Moriarty, E.; Rost, M.; et al. Coevolution of Cryptosporidium tyzzeri and the house mouse (Mus musculus). Int. J. Parasitol. 2013, 43, 805–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010, 124, 80–89. [Google Scholar] [CrossRef]

| Country | Locality | Type | No. of Screened/No. of Positive | ID of Positive Animal | Microscopical Positivity | Sex/Age | Genotyping at the Gene Loci | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSU | Actin | HSP70 | COWP | TRAP-C1 | gp60 | |||||||
| Czech Republic | (1) | W | 2/0 | - | - | - | - | - | - | - | - | - |
| (2) | W | 1/0 | - | - | - | - | - | - | - | - | - | |
| (3) | W | 5/2 | 48358 | No | F/A | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | VIIIbA11G1R1 | |
| 50030 | No | M/A | C. sciurinum | C. sciurinum | - | C. sciurinum | - | VIIIbA11G1R1 | ||||
| (4) | W | 2/0 | - | - | - | - | - | - | - | - | - | |
| (5) | W | 15/4 | 47183 | No | M/J | C. sciurinum | C. sciurinum | C. sciurinum | - | C. sciurinum | VIIIbA11G1R1 | |
| 47189 | No | F/A | C. sciurinum | C. sciurinum | - | ferret genotype | - | VIIIbA11G1R1 | ||||
| 47193 | No | M/A | C. sciurinum | C. sciurinum | C. sciurinum | - | C. sciurinum | VIIIbA11G1R1 | ||||
| 47200 | No | F/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | - | VIIIbA11G1R1 | ||||
| (6) | W | 1/0 | - | - | - | - | - | - | - | - | - | |
| (7) | W | 2/0 | - | - | - | - | - | - | - | - | - | |
| (8) | W | 4/0 | - | - | - | - | - | - | - | - | - | |
| (9) | W | 3/0 | - | - | - | - | - | - | - | - | - | |
| (10) | W | 3/0 | - | - | - | - | - | - | - | - | - | |
| (11) | W | 7/1 | 38512 | No | F/A | C. sciurinum | C. sciurinum | C. sciurinum | - | - | VIIIcA10G2R1 | |
| (12) | W | 3/0 | - | - | - | - | - | - | - | - | - | |
| (13) | W | 6/0 | - | - | - | - | - | - | - | - | - | |
| (14) | W | 5/0 | - | - | - | - | - | - | - | - | - | |
| (15) | W | 5/0 | - | - | - | - | - | - | - | - | - | |
| (16) | W | 3/0 | - | - | - | - | - | - | - | - | - | |
| (17) | W | 4/0 | - | - | - | - | - | - | - | - | - | |
| (18) | W | 5/0 | - | - | - | - | - | - | - | - | - | |
| (19) J | RC | 14/0 | - | - | - | - | - | - | - | - | - | |
| (20) | RC | 4/0 | - | - | - | - | - | - | - | - | - | |
| (21) | RC | 37/5 | 40736 | Yes | F/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | - | VIIIcA10G2R1 | |
| 40737 | Yes | M/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | VIIIcA10G2R1 | ||||
| 40738 | Yes | F/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | - | VIIIcA10G2R1 | ||||
| 40739 | Yes | M/J | C. sciurinum | C. sciurinum | - | C. sciurinum | C. sciurinum | VIIIcA10G2R1 | ||||
| 40741 | Yes | M/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | VIIIcA10G2R1 | ||||
| (22 ) | RC | 23/2 | 41839 | Yes | F/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | VIIIcA10G2R1 | |
| 41840 | No | M/A | C. sciurinum | C. sciurinum | - | - | C. sciurinum | VIIIcA10G2R1 | ||||
| (23) | RC | 21/5 | 45526 | No | M/A | C. sciurinum | C. sciurinum | C. sciurinum | - | C. sciurinum | VIIIbA11G1R1 | |
| 45901 | Yes | F/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | VIIIbA11G1R1 | ||||
| 51489 | Yes | M/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | - | VIIIbA11G1R1 | ||||
| 53289 | Yes | F/J | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | VIIIbA11G1R1 | ||||
| 51295 | No | M/A | C. sciurinum | C. sciurinum | - | - | C. sciurinum | VIIIbA11G1R1 | ||||
| (24) | RC | 15/1 | 46479 | No | F/A | C. sciurinum | C. sciurinum | C. sciurinum | - | - | VIIIbA11G1R1 | |
| (25) | RC | 15/0 | - | - | - | - | - | - | - | - | - | |
| Slovakia | (26) | W | 8/0 | - | - | - | - | - | - | - | - | - |
| (27) | W | 2/0 | - | - | - | - | - | - | - | - | - | |
| (28) | W | 5/1 | 45562 | No | M/A | C. sciurinum | C. sciurinum | - | C. sciurinum | C. sciurinum | VIIIcA10G2R1 | |
| (29) | W | 2/0 | - | - | - | - | - | - | - | - | - | |
| (30) | W | 2/0 | - | - | - | - | - | - | - | - | - | |
| (31) | W | 5/1 | 45560 | No | F/J | C. sciurinum | C. sciurinum | C. sciurinum | - | C. sciurinum | VIIIcA10G2R1 | |
| (32) | W | 4/0 | - | - | - | - | - | - | - | - | - | |
| (33) | W | 2/0 | - | - | - | - | - | - | - | - | - | |
| (34) | W | 6/0 | - | - | - | - | - | - | - | - | - | |
| (35) | W | 1/0 | - | - | - | - | - | - | - | - | - | |
| (36) | W | 4/0 | - | - | - | - | - | - | - | - | - | |
| (37) | W | 1/1 | 45561 | No | F/A | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | - | VIIIcA10G2R1 | |
| (38) | W | 1/0 | - | - | - | - | - | - | - | - | - | |
| (39) | W | 7/2 | 53942 | No | F/A | C. sciurinum | C. sciurinum | - | - | C. sciurinum | VIIIcA10G2R1 | |
| 53943 | No | M/A | C. sciurinum | C. sciurinum | C. sciurinum | C. sciurinum | - | VIIIcA10G2R1 | ||||
| (40) | W | 8/1 | 53944 | No | M/A | C. sciurinum | C. sciurinum | C. sciurinum | - | - | VIIIcA10G2R1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prediger, J.; Ježková, J.; Holubová, N.; Sak, B.; Konečný, R.; Rost, M.; McEvoy, J.; Rajský, D.; Kváč, M. Cryptosporidium sciurinum n. sp. (Apicomplexa: Cryptosporidiidae) in Eurasian Red Squirrels (Sciurus vulgaris). Microorganisms 2021, 9, 2050. https://doi.org/10.3390/microorganisms9102050
Prediger J, Ježková J, Holubová N, Sak B, Konečný R, Rost M, McEvoy J, Rajský D, Kváč M. Cryptosporidium sciurinum n. sp. (Apicomplexa: Cryptosporidiidae) in Eurasian Red Squirrels (Sciurus vulgaris). Microorganisms. 2021; 9(10):2050. https://doi.org/10.3390/microorganisms9102050
Chicago/Turabian StylePrediger, Jitka, Jana Ježková, Nikola Holubová, Bohumil Sak, Roman Konečný, Michael Rost, John McEvoy, Dušan Rajský, and Martin Kváč. 2021. "Cryptosporidium sciurinum n. sp. (Apicomplexa: Cryptosporidiidae) in Eurasian Red Squirrels (Sciurus vulgaris)" Microorganisms 9, no. 10: 2050. https://doi.org/10.3390/microorganisms9102050
APA StylePrediger, J., Ježková, J., Holubová, N., Sak, B., Konečný, R., Rost, M., McEvoy, J., Rajský, D., & Kváč, M. (2021). Cryptosporidium sciurinum n. sp. (Apicomplexa: Cryptosporidiidae) in Eurasian Red Squirrels (Sciurus vulgaris). Microorganisms, 9(10), 2050. https://doi.org/10.3390/microorganisms9102050





